Abstract
A bacterial strain S-51T was isolated from rhizospheric forest soil at Kyonggi University during the study of previously uncultured bacterium. The phylogenetic analysis was based on 16S rRNA gene sequences that indicated that strain S-51T belonged to the genus Rhizobium within the family Rhizobiaceae. The closest members of strain S-51T were Rhizobium naphthalenivorans TSY03bT (98.2% sequence similarity) and Rhizobium selenitireducens ATCC BAA-1503T (98.1%). The sequence similarities of other members were <97.7%. The sole respiratory quinone was Q-10 and the major polar lipids were phosphatidylethanolamine, diphosphatidylglycerol, phosphatidylcholine, and unidentified glycolipid. The principal fatty acids were summed feature 8 (C18:1ω7c and/or C18:1ω6c), cyclo-C19:0ω8c, and C18:0. The DNA G+C content was 63.1 mol%. The genome was 4930044 bp long and contained N2-fixing genes, such as fixF, ntrC, and ptsN, in addition to respiratory nitrate reductase genes, such as narC, narG, narH, narI, and narJ. The average nucleotide identity (ANIu), average amino acid identity (AAI), and digital DNA-DNA hybridization (dDDH) relatedness between strain S-51T and phylogenetically related species were ≤82.6, ≤83.6 and ≤25.3%, respectively, much lower than the species delineation thresholds. Based on the polyphasic taxonomic study, strain S-51T represents a new species in the genus Rhizobium, for which the name Rhizobium terricola is proposed. The type strain is S-51T (=KACC 19117T = KEMB 9005-539T = NBRC 112711T).
1. Introduction
Advances in technology and metagenomic studies have revealed that most of the environmental microorganisms are ‘unculturable’, ‘uncultured’, or ‘not-yet cultured’ on synthetic media in the laboratory [1]. Uncultured microbes encompass most of the microbial diversity on this planet, as around 99% of microorganisms are still uncultivated [2]. There are various normal to complex reasons for failing to cultivate these microbes in the laboratory, such as lack of exact or correct nutrients, unsuitable pH, excessive use of rich-nutrient media, insufficient or improper incubation time, inappropriate temperature, specific growth signal, dependence on other microbes, and specially the lack of natural growth environment in the laboratory setup [1,2,3].
The genus Rhizobium has been described as root and/or stem-nodule bacteria by Frank [4]. To date, there are 117 species of the genus Rhizobium with validly published names (https://lpsn.dsmz.de/genus/rhizobium (accessed on 25 June 2022)). Representatives of the genus Rhizobium are characterized as motile, non-spore-forming, Gram-stain-negative, rod-shaped, and containing DNA G+C between 57 and 66 mol% and C18:1ω7c as a principal fatty acid [5,6]. The members of the genus Rhizobium are well distributed in soil and are significant for their use in agriculture, as most of the species are able to fix nitrogen in legume plants [5]. Most of the Rhizobium species have been isolated from the root nodule of leguminous plants [5,7]. However, in recent years, novel members of the genus Rhizobium have also been isolated from non-legume niches, such as activated sludge, bioreactor, sand dunes, pesticide-contaminated sites, effluent treatment plant, sediment, sea water, fresh water, and groundwater [6,8,9,10,11,12,13,14]. This study describes the polyphasic and taxonomic characteristics of strain S-51T isolated from forest rhizospheric soil during the study of uncultured soil bacteria.
Due to their nitrogen fixing capability, the members of the genus Rhizobium are agriculturally and environmentally valuable [15,16]. The species of Rhizobium primarily inhabit the root or stem nodules of plants and provide a nitrogen source to their host plants [16]. However, the free-living nitrogen fixing species such as S-51T isolated in this study are equally important, as these species could contribute adequate amounts of elemental nitrogen to the soil.
2. Material and Methods
2.1. Isolation and Ecology
Strain S-51T was isolated from the rhizospheric loamy sandy soil in Kyonggi University’s forest, located at Suwon, Gyeonggi-do, Republic of Korea (37°18′5″ N and 127°1′56″ E). A modified culture method using a six-well transwell plate (Corning Inc., Corning, NY, US) was used for isolation. Debris-free sieved soil (~3 g) was kept on the bottom of the transwell plate and 3 mL of diluted (1/10) R2A (Reasoner’s 2A; KisanBio, Seoul, Republic of Korea) broth was added to the insert. After that, 100 µL of soil suspension (1 g of soil in 9 mL of distilled water) was added to the insert. Then, the transwell plate was incubated in a shaker at 130 rpm at 28 °C for 6 weeks. After 6 weeks, the enriched culture was serially diluted, and then 100 µL of each dilution was spread on 1/10 R2A agar and incubated at 28 °C for 4 weeks [17]. The short-term maintenance and long-term preservation were performed as described previously [18].
2.2. 16S rRNA Phylogeny
Genomic DNA of strain S-51T was extracted using the InstaGene Matrix kit (Bio-Rad; Hercules, CA, US) according to the manufacturer’s instructions. Amplification of the 16S rRNA gene was performed with PCR using the primers 27F and 1492R [19]. Sequencing was carried out using an Applied Biosystems 3770XL DNA analyzer with a BigDye Terminator cycle sequencing Kit v.3.1 (Applied Biosystems, Foster City, CA, US). Nearly complete sequences of 16S rRNA genes were assembled with the SeqMan software (DNASTAR Inc., Madison, WI, US). For type material sequences, the nearest phylogenetic neighbours were identified using the EzBioCloud database [20]. In addition, non-type material sequences were compared with the top hits of Megablast (GenBank). All the 16S rRNA gene sequences of the closest phylogenetic members were retrieved from the ncbi GenBank and aligned using silva alignment (https://www.arb-silva.de (accessed on 14 March 2018)). Phylogenetic trees were reconstructed using the three tree making methods: neighbor-joining [21], maximum-likelihood [22], and maximum-parsimony [23] algorithms with mega7 [24]. The evolutionary distances were calculated according to the Kimura 2-parameter model [25] and bootstrap analysis was based on 1000 replications [26].
2.3. Genome Features
Whole-genome-based approaches were used for the further analysis of taxonomic status of novel strain. The genomic DNA was extracted using DNeasy Blood and Tissue kits (Qiagen, Hilden, Germany). Whole genome shotgun sequencing of strains was performed using Macrogen (Seoul, Korea) using the Illumina HiSeq platform and assembled by SPAdes version 3.13.0 [27]. The Whole Genome Shotgun project of strain S-51T was deposited in the NCBI repository (or DDBJ/ENA/GenBank) and the sequences are available at https://www.ncbi.nlm.nih.gov/bioproject (accessed on 28 April 2020) under accession numbers PRJNA626510 and SAMN14642770. The authenticity of the genome assembly was checked by comparing 16S rRNA gene sequence using ncbi Align Sequences Nucleotide BLAST tool [28], and the potential contamination was checked using the ContEst16S algorithm [29]. The completeness of genome was checked using searches of conserved single-copy orthologous BUSCO V3.0.2 [30]. After analysis, the genome sequence was annotated using the ncbi Prokaryotic Genome Annotation Pipeline version 4.11 [31] and Rapid Annotations using Subsystems Technology (RAST) server version 2.0 [32]. Genome-based relatedness between strain S-51T and closely related strains was determined using whole genome sequences based on average nucleotide identity (ANI) using the OrthoANIu algorithm [33]. DNA-DNA hybridization (DDH) was calculated using whole genome sequences in silico with the Genome-to-Genome Distance Calculator (GGDC version 2.1) using the blast method [34]. The average amino acid identity (AAI) was determined using a web server (http://enve-omics.ce.gatech.edu/aai/index (accessed on 18 July 2022)). In addition, a genomic feature map was constructed using CGView server [35]. Moreover, the annotation and analysis of secondary metabolite biosynthesis genes were carried out using anti-SMASH server version 5.0 [36]. The COG (Clusters of Orthologous Group) functional categories were assigned by searching against the KEGG (Kyoto Encyclopedia of Genes and Genomes) database [37]. Furthermore, CRISPR gene and Cas cluster were analyzed using the CRISPRCasFinder online server (https://crisprcas.i2bc.paris-saclay.fr/ (accessed on 18 July 2022)). The Venn diagram of whole genome orthologous genes was constructed using the Orthovenn2 web server [38] (https://orthovenn2.bioinfotoolkits.net/home (accessed on 18 July 2022)).
2.4. Physiology and Chemotaxonomy
The cell morphology of strain S-51T, grown on R2A agar for 1 week at 28 °C, was examined by transmission electron microscopy (Talos L120C; FEI). Colony morphology was observed with the Zoom Stereo Microscope (SZ61; Olympus, Tokyo, Japan). Gram staining was performed as described previously [39]. Motility of strain S-51T was determined in R2A medium containing 0.4% (w/v) agar. Oxidase activity was observed using 1% (w/v) tetra-methyl-p-phenylenediamine dihydrochloride. Catalase activity was assessed using 3% (v/v) hydrogen peroxide (H2O2). Growth at various temperatures from 4 to 40 °C on R2A agar plates was monitored for 10 days. Growth was determined on various media, including R2A agar, nutrient agar (NA; Oxoid, Hampshire, UK), sorbitol MacConkey agar (MA; Oxoid), tryptone soya agar (TSA; Oxoid), marine agar 2216 (Becton), potato dextrose agar (PDA; Becton), brain heart infusion agar (BHI; Oxoid), veal infusion agar (Becton), and Luria-Bertani agar (LBA; Oxoid). Salt tolerance was examined in R2A broth supplemented with NaCl (0–5%, at 0.5% intervals). The pH range was determined at 28 °C in R2A broth adjusted to pH 4–12 (in increments of 0.5 pH units) [40]. Hydrolysis of Tween 40, Tween 60, and Tween 80 was assessed using the method of Smibert and Krieg [41]. Anaerobic growth was monitored on the R2A agar plate at 28 °C for 10 days using the BD GasPak EZ Gas Generating Pouch System with nitrate as a final electron acceptor. Hydrolysis of xanthine, hypoxanthine, starch, chitin, CM-cellulose, tyrosine, and casein was tested as previously described [42]. A DNase activity assay was performed with DNase agar (Oxoid). Presence of spore was examined by staining with malachite green. For the nitrogen-fixation test, strain S-51T was grown on N2-free media with bromothymol blue (NfB) indicator [43]. Other biochemical and physiological tests were performed using the API ID 32GN and API 20NE test kits (bioMérieux; Marcy-l’Étoile, France). Enzyme activities were determined using an API ZYM kit (bioMérieux), as per the manufacturer’s instructions.
For fatty acid analysis, cells of strain S-51T and reference strains were harvested from identical culture condition (on R2A agar plates at 28 °C for 4 days). The cellular fatty acids were extracted using the MIDI protocol (Sherlock Microbial Identification System, version 6.0B) and analyzed with a gas chromatograph (HP 6890 Series GC System; Hewlett Packard). The fatty acids (%of totals) were identified using the TSBA6 database of the Microbial Identification System [44]. Polar lipids and isoprenoid quinones were extracted from freeze-dried cells according to the procedures described by Minnikin et al. [45]. The biomass was harvested from R2A agar plates incubated at 28 °C for 6 days. The polar lipids were analyzed with two-dimensional TLC (thin layer chromatography) using chloroform:methanol:water (65:25:4; v/v/v) in the first dimension and chloroform:methanol:aceteic acid:water (40:7.5:6:2; v/v/v/v) in the second direction.
3. Results and Discussion
3.1. Isolation of Uncultured Strain
In general, the isolation of bacterial strains involves enrichment culture, seral dilution, plating, and pure culture in this order using known media. However, in this study, a modified method using a transwell plate and diluted R2A medium was applied to isolate uncultured soil bacteria, including strain S-51T, in the step of enrichment culture and in the step of plating, respectively.
3.2. Phylogenetic Analysis
The nucleotide sequence of 16S rRNA gene of strain S-51T was deposited in the GenBank/EMBL/DDBJ database under the accession number KY117474. Preliminary comparisons with 16S rRNA gene sequences indicated that strain S-51T showed top hits with previously uncultured bacterial clones in the ncbi GenBank database (99.65–97.72%) for non-type material sequences. The phylogenetic tree based on 16S rRNA gene sequences also revealed that strain S-51T clustered with uncultured bacterium clones with strong bootstrap (99%) values (Figure 1). In addition, when the 16S rRNA gene sequence of strain S-51T was analyzed with the EzBioCloud server for type-material sequences, it showed that strain S-51T belonged to the family Rhizobiaceae and was most closely related to Rhizobium naphthalenivorans TSY03bT (98.2% sequence similarity) and Rhizobium selenitireducens ATCC BAA-1503T (98.1%). The sequence similarities of other members were <97.7%. As a result, the strain S-51T showed 98.2–97.7% 16S rRNA gene sequence similarities to its closest members of the genus Rhizobium. These values lie within the range needed to specify a strain as a novel species of the genus [46].
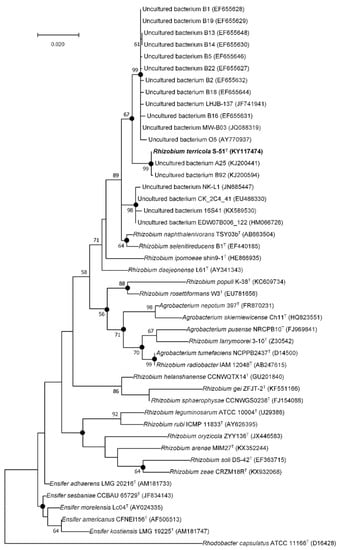
Figure 1.
Maximum-likelihood phylogenetic tree based on 16S rRNA gene sequences of strain S-51T among closely related representatives of the family Rhizobiaceae with previously uncultured bacterium clones. Filled circles indicate nodes recovered by different tree-making algorithms (neighbor–joining and maximum–likelihood). The numbers at the nodes indicate the percentage of 1000 bootstrap replicates yielding this topology; only values > 50% are shown. Rhodobacter capsulatus ATCC 11166T was used as an out-group. GenBank accession numbers are given in parentheses. Bar, 0.02 substitutions/nucleotide position.
Separate lineage and the strong bootstrap values with closest members (Rhizobium naphthalenivorans TSY03bT and Rhizobium selenitireducens ATCC BAA-1503T) formed in the neighbor-joining (NJ), maximum-parsimony (MP), and maximum-likelihood (ML) trees displayed that strain S-51T belonged to a novel species within the genus Rhizobium (Figure 2, Figures S1 and S2).
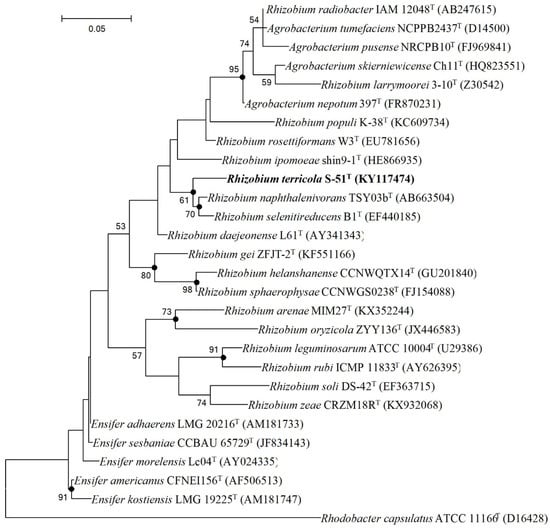
Figure 2.
Maximum-likelihood tree based on 16S rRNA gene sequences showing the phylogenetic position of strain S-51T among closely related members of the family Rhizobiaceae. Filled circles indicate nodes recovered by all three treeing methods (neighbor-joining, maximum-likelihood, and maximum-parsimony). The numbers at the nodes indicate the percentage of 1000 bootstrap replicates yielding this topology; only values > 50% are shown. Rhodobacter capsulatus ATCC 11166T was used as an out-group. GenBank accession numbers are given in parentheses. Bar, 0.05 substitutions per nucleotide position.
Based on sequence similarities and phylogenetic analyses, Rhizobium naphthalenivorans KCTC 23252T and Rhizobium selenitireducens KCTC 23231T were selected for comparative analysis and were used as reference strains for physiology, biochemical, quinone, and fatty acid analyses.
3.3. Genome Analysis
The Whole-Genome Shotgun project of strain S-51T was deposited at DDBJ/ENA/GenBank under the accession JABBGK000000000. The version described in this manuscript is version JABBGK010000000. The genome of strain S-51T was 4,930,044 bp long. DNA G+C content calculated from whole genome sequence was 63.1 mol%. The genome contained 19 scaffolds, 327 subsystems, 254.0× genome coverage, and 1,092,255 N50 value (Table 1). The ANI, AAI, and dDDH values of strain S-51T with the closely related type species of the genus Rhizobium were ≤82.6%, ≤83.6, and ≤25.3%, respectively (Table 2). These values were below the threshold values for ANI and AAI (95–96%) [47,48] and dDDH (70%) [34], suggesting that strain S-51T is a species of a different type than those that exist within the genus Rhizobium.

Table 1.
Genome features of strain S-51T.

Table 2.
ANI, AAI, and dDDH values (%) of strain S-51T and other members of the genus Rhizobium.
Anti-SMASH analysis revealed that the genome of strain S-51T consists of six biosynthetic gene clusters (BGCs), which are supposedly responsible for the biosynthesis of secondary metabolites such as hserlactones, terpene, bactetiocin, N-acyl amino acid, NRPS, and T1PKS (Table S1). Moreover, NRPS (non-ribosomal synthesized peptide) BGC showed a 17% similarity to entolysin (Table S1). The genomic map of strain S-51T represents 3rRNAs, 50 tRNAs, and a tmRNA (Figure 3). In addition, 88 genes for the biosynthesis, transport, and catabolism of secondary metabolites and 1347 genes used for unknown functions were detected in COG functional categories (Figure 4). The Venn diagram was constructed based on whole-genome orthologous genes of strain S-51T and the reference genome showed that the genome of S-51T consisted of 75 unique genes and 2847 common genes for all four strains (Figure 5). The functions of these common genes may include carbohydrate metabolism for host plant recognition, lectin binding, and root attachment, membrane transport, secretion machinery for metabolites and signals important in plant-microbe interactions, nodulation (nod), and symbiotic N2 fixation (nif and fix). Interestingly, the number of unique genes in the reference strains is reversely proportional to the values of ANI, AAI, and dDDH in order. Furthermore, various N2-fixation regulatory proteins have been detected in the genome of strain S-51T (Table S2).
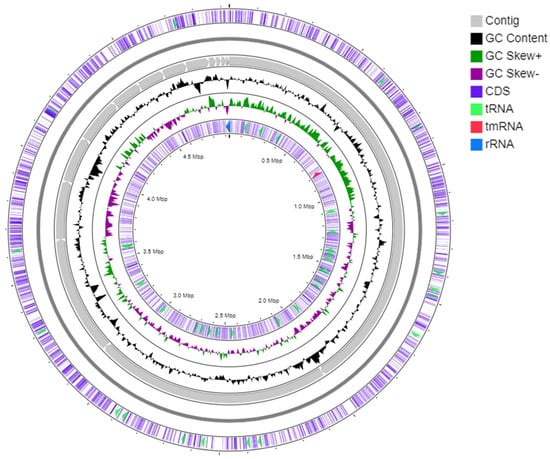
Figure 3.
Graphical genome map of strain S-51T. RNA genes (tRNAs light green, rRNAs blue, tmRNA red), GC content, and GC skew.
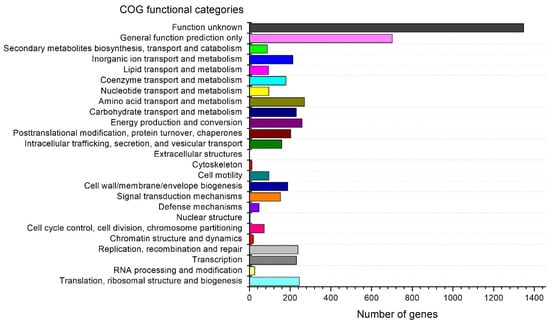
Figure 4.
COG functional classification of proteins in strain S-51T genome.
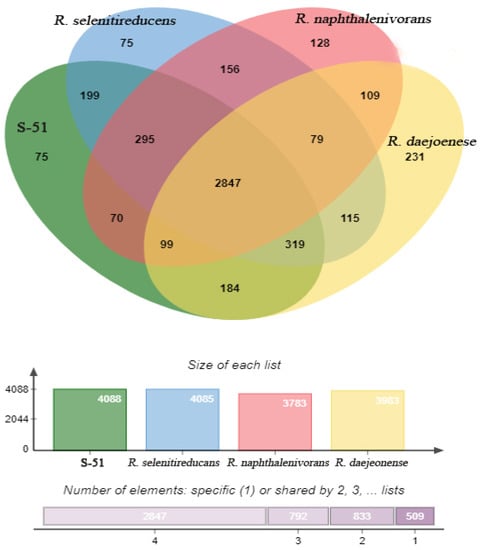
Figure 5.
Venn diagram of whole-genome orthologous genes in S-51T and three type species of reference strains (Rhizobium selenitireducens, Rhizobium naphthalenivorans, and Rhizobium daejeonense). The numbers in the diagram indicate overlapped conserved genes or non-overlapped unique genes in each species.
3.4. Physiology and Chemotaxonomic Characteristics
Cells of strain S-51T (Figure S3) were rod-shaped, Gram-stain-negative, aerobic, and motile. Cells were 1.2–1.9 µm long and 0.6–0.8 µm wide. Colonies on R2A agar were straw-color, circular, entire, and convex. It could grow well on R2A agar, but would be difficult to grow on other media. The differential phenotypic characteristic features of strain S-51T are presented in Table 3 with the closest reference of the genus Rhizobium; highest salt tolerance (5.0%), weakly assimilation of l-alanine, adipic acid and propionic acid, positive activity of β-glucosidase, and negative activity of β-galactosidase clearly distinguished strain S-51T from closely related type strains. The free-living strain S-51T was able to grow in the nitrogen-free media, revealing the ability to fix atmospheric nitrogen by discoloration of the NfB indicator [43].

Table 3.
Differential phenotypic characteristics of strain S-51T with closely related species of the genus Rhizobium. Strains: 1, S-51T; 2, Rhizobium naphthalenivorans KCTC 23252T [8]; 3, Rhizobium selenitireducens KCTC 23231T [14]. DNA G+C content was calculated using whole genome sequences. All the data were from this study, except the data in parentheses. +, positive; w, weakly positive; −, negative.
The major cellular fatty acids of strain S-51T were summed feature 8 (C18:1ω7c and/or C18:1ω6c), cyclo-C19:0 ω8c and C18:0 (Table 4). The major fatty acids of closely related type strains were similar. However, the compositions of primary and secondary fatty acids showed characteristic differences for strain S-51T compared to other references. The only respiratory quinone was ubiquinone-10 (Q-10), the same as the reported major respiratory quinone of the genus Rhizobium. The principal polar lipids for strain S-51T were phosphatidylethanolamine (PE), diphosphatidylglycerol (DPG), phosphatidylcholine (PC), and unidentified glycolipid (GL). The presence of unidentified glycolipid (GL) as a major polar lipid, along with the presence of unidentified aminophospholipid (APL) and other minor polar lipids (L1-L3), distinguished strain S-51T from other closely related type strains (Figure 6) [6,14].

Table 4.
Cellular fatty acid profiles (% of total) of strain S-51T and closely related reference strains. Strains: 1, S-51T; 2, Rhizobium naphthalenivorans KCTC 23252T; 3, Rhizobium selenitireducens KCTC 23231T. All data were obtained from this study. tr, trace amount (<0.5%); –, not detected.
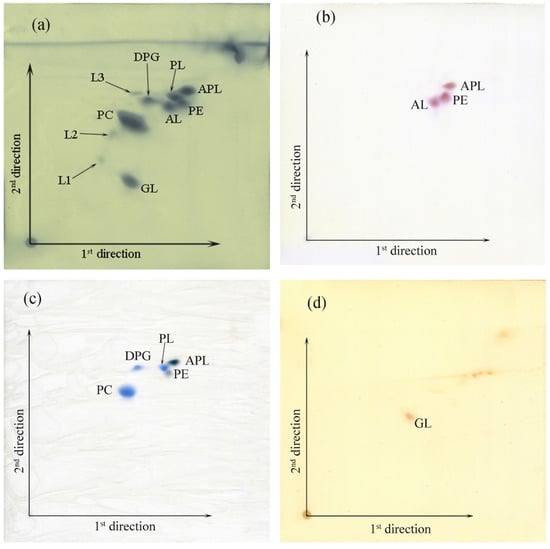
Figure 6.
Thin-layer chromatograms of the polar lipids from strain S-51T. The components were seen by staining with 5% molybdophosphoric acid in ethanol and heating them at 180 °C for 15 min (a); ninhydrin at 110 °C for 15 min (b); molybdenum at room temperature (c); and α-naphthol-sulphuric acid at 110 °C for 15 min (d). Abbreviations: PE, phosphatidylethanolamine; PL, unidentified phospholipid; DPG, diphosphatidylglycerol; PC, phosphatidylcholine; GL, unidentified glycolipid; AL, unidentified aminolipid; APL, aminophospholipid; L1–L3, unidentified polar lipids.
4. Conclusions
Based on the above discussed genomic, phylogenetic, phenotypic, and chemotaxonomic characteristic differences, strain S-51T represents a novel member in the genus Rhizobium, for which the name Rhizobium terricola sp. nov. is proposed. In particular, it has 75 unique genes different from the closest reference strains, although two strains share many more genes. The presence of unidentified glycolipid (GL) and unidentified aminophospholipid (APL) as major polar lipids is also distinguished from other known closely related type strains. In addition, strain S-51T is differentiated by isolation source, wider pH range, and higher salt tolerance than the closest two type strains, which may derivate uncultivable property.
So far, many microbiologists have isolated a great number of cultivable bacteria and characterized them for identification, mechanism, interaction, and function, but these bacteria are only tiny portion of all bacteria known through metagenomic data. Therefore, the isolation of uncultured strains including strain S-51T may increase the possibility of new functions or improve functions in various areas.
Description of Rhizobium terricola sp. nov.
Rhizobium terricola [ter.ri’co.la. L. fem. n. terra soil; l- suff. -cola (from L. masc. or fem. n. incola) inhabitant, dweller; N.L. n. terricola a dweller of soil, referring to the isolation of the type strain from soil].
Cells (0.6–0.8 µm wide and 1.2–1.9 µm long) are non-spore-forming, aerobic, rod-shaped, motile, and Gram-stain-negative. No growth occurs on NA, LBA, BHI, PDA, LBA, MA, and veal infusion agar, but poorly grows on TSA and marine 2216 agar. Cells are able to fix nitrogen. Colonies on R2A agar are straw colored, entire, convex, and circular. Colony size of the cells are 0.5–1 mm on R2A agar for seven days at 28 °C. Cells grow optimally in the absence of NaCl, but tolerate up to 5.0% (w/v). Cells grow at 15–37 °C (optimum, 25–32 °C) and pH 5.5–11.0 (optimum, 7.0–9.5). Catalase and oxidase tests are positive. Urea and aesculin are hydrolyzed, but casein, tyrosine, xanthine, hypoxanthine, CM-cellulose, starch, DNA, chitin, Tween 80, Tween 60, and Tween 40 are not. The flexirubin test is negative. Nitrate is reduced to nitrite, but nitrite is not reduced to nitrogen. The glucose is not fermented. The PNPG (4-nitrophenyl-β-d-galactopyranoside) is positive. The type strain shows the following enzyme activities: positive for esterase lipase (C8), leucine arylamidase, valine arylamidase, cystine arylamidase, trypsin, α-glucosidase, and β-glucosidase; weakly positive for alkaline phosphatase, esterase (C4), and acid phosphatase; negative for lipase (C14), α-chymotrypsin, naphthol-AS-BI-phosphohydrolase, α-galactosidase, β-galactosidase, β-glucuronidase, N-acetyl-β-glucosaminidase, α-mannosidase, and α-fucosidase. In API 20NE and ID 32GN tests, 3-hydroxybutyric acid, d-glucose, d-maltose, d-mannitol, d-mannose, d-ribose, d-saccharose, d-sorbitol, inositol, lactic acid, l-arabinose, l-fucose, l-proline, l-rhamnose, malic acid, sodium acetate, and valeric acid are assimilated; adipic acid, l-alanine, and propionic acid are weakly assimilated. The sole respiratory quinone is Q-10. The principal cellular fatty acids are summed feature 8 (C18:1ω7c and/or C18:1ω6c), cyclo-C19:0 ω8c and C18:0. The major polar lipids are phosphatidylethanolamine, diphosphatidylglycerol, phosphatidylcholine, and unidentified glycolipid. The DNA G+C content of the type strain is 63.1 mol%.
The type strain, S-51T (=KACC 19117T = KEMB 9005-539T = NBRC 112711T), was isolated from forest soil, geographically located at Suwon, Gyeonggi-do, Republic of Korea (37°18′5″ N and 127°1′56″ E). The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA gene sequence and the whole genome sequence of strain S-51T are KY117474 and JABBGK000000000, respectively.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d14090733/s1: Figure S1: Neighbor–joining tree based on 16S rRNA gene sequences showing the phylogenetic position of strain S-51T among closely related members of the family Rhizobiaceae; Figure S2: Maximum–parsimony tree based on 16S rRNA gene sequences showing the phylogenetic position of strain S-51T among closely related members of the family Rhizobiaceae; Figure S3: Transmission electron microscopic image of strain S-51T grown on R2A agar. Table S1: The distribution of biosynthetic gene clusters in strain S-51T; Table S2. N2-fixation regulatory proteins in S-51T genome.
Author Contributions
R.H.D., D.K.C. conceived, designed and conducted all the experiments. D.-U.K. interpreted the data. J.K. (Jungmin Kim) and J.K. (Jaisoo Kim) coordinated and supervised the study. R.H.D., D.K.C. and D.-U.K. analyzed all the data and prepared the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (No. 2022R1A2C1010877).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Chaudhary, D.K.; Khulan, A.; Kim, J. Development of a novel cultivation technique for uncultured soil bacteria. Sci. Rep. 2019, 9, 6666. [Google Scholar] [CrossRef] [PubMed]
- Vartoukian, S.R.; Palmer, R.M.; Wade, W.G. Strategies for culture of “unculturable” bacteria. FEMS Microbiol. Lett. 2010, 309, 1–7. [Google Scholar] [CrossRef]
- Pulschen, A.A.; Bendia, A.G.; Fricker, A.D.; Pellizari, V.H.; Galante, D.; Rodrigues, F. Isolation of uncultured bacteria from Antarctica using long incubation periods and low nutritional media. Front. Microbiol. 2017, 8, 1346. [Google Scholar] [CrossRef] [PubMed]
- Frank, B. Über die Pilzsymbiose der Leguminosen. Ber. Dtsch. Bot. Ges. 1889, 7, 332–346. [Google Scholar]
- Young, J.M.; Kuykendall, L.D.; Martínez-Romero, E.; Kerr, A.; Sawada, H. A revision of Rhizobium Frank 1889, with an emended description of the genus, and the inclusion of all species of Agrobacterium Conn 1942 and Allorhizobium undicola de Lajudie et al. 1998 as new combinations: Rhizobium radiobacter, R. rhizogenes, R. rubi, R. undicola and R. vitis. Int. J. Syst. Evol. Microbiol. 2001, 51, 89–103. [Google Scholar] [PubMed]
- Mohapatra, B.; Sarkar, A.; Joshi, S.; Chatterjee, A.; Kazy, S.K.; Maiti, M.K.; Satyanarayana, T.; Sar, P. An arsenate-reducing and alkane-metabolizing novel bacterium, Rhizobium arsenicireducens sp. nov., isolated from arsenic-rich groundwater. Arch. Microbiol. 2017, 199, 191–201. [Google Scholar] [CrossRef]
- Zhang, X.; Harper, R.; Karsisto, M.; Lindstrom, K. Diversity of Rhizobium bacteria isolated from the root nodules of leguminous trees. Int. J. Syst. Evol. Microbiol. 1991, 41, 104–113. [Google Scholar] [CrossRef]
- Kaiya, S.; Rubaba, O.; Yoshida, N.; Yamada, T.; Hiraishi, A. Characterization of Rhizobium naphthalenivorans sp. nov. with special emphasis on aromatic compound degradation and multilocus sequence analysis of housekeeping genes. J. Gen. Appl. Microbiol. 2012, 58, 211–224. [Google Scholar] [CrossRef]
- Quan, Z.X.; Bae, H.S.; Baek, J.H.; Chen, W.F.; Im, W.T.; Lee, S.T. Rhizobium daejeonense sp. nov. isolated from a cyanide treatment bioreactor. Int. J. Syst. Evol. Microbiol. 2005, 55, 2543–2549. [Google Scholar] [CrossRef][Green Version]
- Kaur, J.; Verma, M.; Lal, R. Rhizobium rosettiformans sp. nov., isolated from a hexachlorocyclohexane dump site, and reclassification of Blastobacter aggregatus Hirsch and Müller 1986 as Rhizobium aggregatum comb. nov. Int. J. Syst. Evol. Microbiol. 2011, 61, 1218–1225. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, R.P.; Ren, C.; Lai, Q.L.; Zeng, R.Y. Rhizobium marinum sp. nov., a malachitegreen-tolerant bacterium isolated from seawater. Int. J. Syst. Evol. Microbiol. 2015, 65, 4449–4454. [Google Scholar] [CrossRef] [PubMed]
- Sheu, S.Y.; Huang, H.W.; Young, C.C.; Chen, W.M. Rhizobium alvei sp. nov., isolated from a freshwater river. Int. J. Syst. Evol. Microbiol. 2015, 65, 472–478. [Google Scholar] [CrossRef] [PubMed]
- Ramana, C.V.; Parag, B.; Girija, K.R.; Raghu Ram, B.; Venkata Ramana, V.; Sasikala, C. Rhizobium subbaraonis sp. nov., an endolithic bacterium isolated from beach sand. Int. J. Syst. Evol. Microbiol. 2013, 63, 581–585. [Google Scholar] [CrossRef] [PubMed]
- Hunter, W.J.; Kuykendall, L.D.; Manter, D.K. Rhizobium selenireducens sp. nov.: A selenite-reducing α- Proteobacteria isolated from a bioreactor. Curr. Microbiol. 2007, 55, 455–460. [Google Scholar] [CrossRef]
- Mus, F.; Crook, M.B.; Garcia, K.; Costas, A.G.; Geddes, B.A.; Kouri, E.D.; Paramasivan, P.; Ryu, M.H.; Oldroyd, G.E.D.; Poole, P.S.; et al. Symbiotic nitrogen fixation and the challenges to its extension to nonlegumes. Appl. Environ. Microbiol. 2016, 82, 3698–3710. [Google Scholar] [CrossRef] [PubMed]
- Zahran, H.H. Rhizobium-legume symbiosis and nitrogen fixation under severe conditions and in an arid climate. Microbiol. Mol. Biol. Rev. 1999, 63, 968–989. [Google Scholar] [CrossRef]
- Dahal, R.H.; Kim, J. Fluviicola kyonggii sp. nov., a bacterium isolated from forest soil and emended description of the genus Fluviicola. Int. J. Syst. Evol. Microbiol. 2018, 68, 1885–1889. [Google Scholar] [CrossRef]
- Dahal, R.H.; Chaudhary, D.K.; Kim, J. Pinisolibacter ravus gen. nov., sp. nov., isolated from pine forest soil and allocation of the genera Ancalomicrobium and Pinisolibacter to the family Ancalomicrobiaceae fam. nov., and emendation of the genus Ancalomicrobium Staley 1968. Int. J. Syst. Evol. Microbiol. 2018, 68, 1955–1962. [Google Scholar] [CrossRef]
- Frank, J.A.; Reich, C.I.; Sharma, S.; Weisbaum, J.S.; Wilson, B.A.; Olsen, G.J. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl. Environ. Microbiol. 2008, 74, 2461–2470. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Fitch, W.M. Toward defining the course of evolution: Minimum change for a specific tree topology. Syst. Zool. 1971, 20, 406. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Zhang, Z.; Schwartz, S.; Wagner, L.; Miller, W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000, 7, 203–214. [Google Scholar] [CrossRef]
- Lee, I.; Chalita, M.; Ha, S.-M.; Na, S.-I.; Yoon, S.-H.; Chun, J. ContEst16S: An algorithm that identifies contaminated prokaryotic genomes using 16S RNA gene sequences. Int. J. Syst. Evol. Microbiol. 2017, 67, 2053–2057. [Google Scholar] [CrossRef]
- Seppey, M.; Manni, M.; Zdobnov, E.M. BUSCO: Assessing Genome Assembly and Annotation Completeness. Methods Mol. Biol. 2019, 1962, 227–245. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.-H.; Ha, S.-M.; Lim, J.; Kwon, S.; Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 2017, 110, 1281–1286. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef]
- Grant, J.R.; Stothard, P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, W181–W184. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Xu, L.; Dong, Z.; Fang, L.; Luo, Y.; Wei, Z.; Guo, H.; Zhang, G.; Gu, Y.Q.; Coleman-Derr, D.; Xia, Q.; et al. OrthoVenn2: A web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2019, 47, W52–W58. [Google Scholar] [CrossRef]
- Doetsch, R.N. Determinative Methods of Light Microscopy. In Manual of Methods for General Bacteriology; Gerdhardt, P., Murray, R.G.E., Costilow, R.N., Nester, E.W., Wood, W.A., Krieg, N.R., Phillips, G.B., Eds.; American Society for Microbiology: Washington, DC, USA, 1981; pp. 21–33. [Google Scholar]
- Breznak, J.A.; Costilow, R.N. Physicochemical factors in growth. In Methods for General and Molecular Bacteriology; Reddy, C.A., Beveridge, T.J., Breznak, J.A., Marzluf, G.A., Schmidt, T.M., Snyder, L.R., Eds.; American Society of Microbiology: Washington, DC, USA, 2007; pp. 309–329. [Google Scholar]
- Smibert, R.M.; Krieg, N.R. Phenotypic characterization. In Methods for General and Molecular Bacteriology; Gerhardt, P., Murray, R.G.E., Wood, W.A., Krieg, N.R., Eds.; American Society for Microbiology: Washington, DC, USA, 1994; pp. 607–654. [Google Scholar]
- Dahal, R.H.; Kim, J. Dyadobacter flavus sp. nov. and Dyadobacter terricola sp. nov., two novel members of the family Cytophagaceae isolated from forest soil. Arch. Microbiol. 2018, 200, 1067–1074. [Google Scholar] [CrossRef]
- Miladiarsi; Mubarik, N.R.; Widyastuti, R. Selection, characterization and application of rhizobacteria and its effect on chili (Capsicum annuum L.) Plant Growth. Res. J. Microbiol. 2017, 12, 161–169. [Google Scholar] [CrossRef]
- Sasser, M. Bacterial Identification by Gas Chromatographic Analysis of Fatty Acid Methyl Esters (GC-FAME); MIDI Technical Note 101; MIDI Inc.: Newark, DE, USA, 1990. [Google Scholar]
- Minnikin, D.E.; O’Donnell, A.G.; Goodfellow, M.; Alderson, G.; Athalye, M.; Schaal, A.; Parlett, J.H. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 1984, 2, 233–241. [Google Scholar] [CrossRef]
- Stackebrandt, E.; Goebel, B.M. Taxonomic note: A place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 1994, 44, 846–849. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef] [PubMed]
- Konstantinidis, K.T.; Tiedje, J.M. Towards a genome-based taxonomy for prokaryotes. J. Bacteriol. 2005, 187, 6258–6264. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).