Composition Diversity and Expression Specificity of the TPS Gene Family among 24 Ficus Species
Abstract
1. Introduction
2. Results
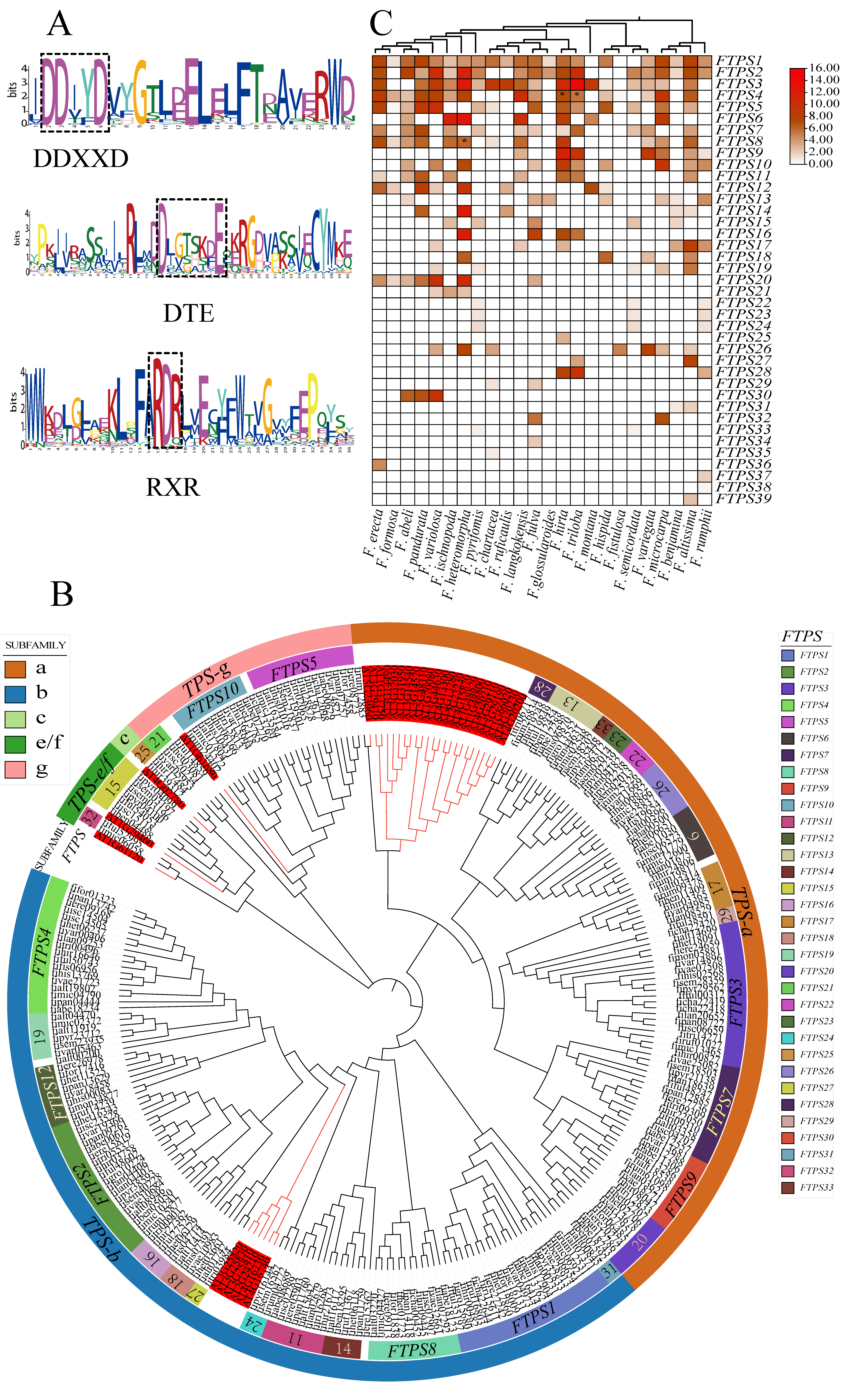
2.1. Characteristics of the TPS Genes in the Receptive Phase of Male Figs of 24 Ficus Species
2.2. Subfamily Analysis of the FTPS Genes
2.3. Expression Analysis of the FTPS Genes
2.4. Positive Selection Analysis
3. Discussion
3.1. Numbers of TPS Genes in 24 Ostiole Bract Transcriptomes
3.2. Sequence Characteristics and Subfamily Categories of the FTPS Genes
3.3. Expression Profiles of the FTPS Genes
3.4. Positive Selection of FTPSs
4. Materials and Methods
4.1. Sample Collection and RNA Sequencing
4.2. RNA-Seq Assembly and Identification of TPS Genes
4.3. Phylogenetic Relationship, Subfamily Classification, and Motif Analysis of the Ficus TPS Genes
4.4. Clustering, Annotation, and Expression Analysis of the Ficus TPS Genes
4.5. Positive Selection Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ehrlich, P.R.; Raven, P.H. Butterflies and Plants: A Study in Coevolution. Evolution 1964, 18, 586–608. [Google Scholar] [CrossRef]
- Oku, H.; Inafuku, M.; Ishikawa, T.; Takamine, T.; Ishmael, M.; Fukuta, M. Molecular cloning and biochemical characterization of isoprene synthases from the tropical trees Ficus virgata, Ficus septica, and Casuarina equisetifolia. J. Plant Res. 2015, 128, 849–861. [Google Scholar] [CrossRef] [PubMed]
- Berg, C.C. Classification and distribution of Ficus. Experientia 1989, 45, 605–611. [Google Scholar] [CrossRef]
- Herre, E.A.; Jander, K.C.; Machado, C.A. Evolutionary Ecology of Figs and Their Associates: Recent Progress and Outstanding Puzzles. Annu. Rev. Ecol. Evol. Syst. 2008, 39, 439–458. [Google Scholar] [CrossRef]
- Souza, C.D.; Pereira, R.A.S.; Marinho, C.R.; Kjellberg, F.; Teixeira, S.P. Diversity of fig glands is associated with nursery mutualism in fig trees. Am. J. Bot. 2015, 102, 1564–1577. [Google Scholar] [CrossRef]
- Hu, R.; Sun, P.; Yu, H.; Cheng, Y.F.; Wang, R.; Chen, X.Y.; Kjellberg, F. Similitudes and differences between two closely related Ficus species in the synthesis by the ostiole of odors attracting their host-specific pollinators: A transcriptomic based investigation. Acta Oecologica-Int. J. Ecol. 2020, 105, 103554. [Google Scholar] [CrossRef]
- Galil, J. Fig biology. Endeavour 1977, 1, 52–56. [Google Scholar] [CrossRef]
- Weiblen, G.D. How to be a fig wasp. Annu. Rev. Entomol. 2002, 47, 299–330. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, G.; Zhang, S.; Chen, S.; Wang, Y.; Wen, P.; Ma, X.; Shi, Y.; Qi, R.; Yang, Y.; et al. Genomes of the Banyan Tree and Pollinator Wasp Provide Insights into Fig-Wasp Coevolution. Cell 2020, 183, 875–889. [Google Scholar] [CrossRef]
- Hossaert-McKey, M.; Soler, C.; Schatz, B.; Proffit, M. Floral scents: Their roles in nursery pollination mutualisms. Chemoecology 2010, 20, 75–88. [Google Scholar] [CrossRef]
- Proffit, M.; Johnson, S.D. Specificity of the signal emitted by figs to attract their pollinating wasps: Comparison of volatile organic compounds emitted by receptive syconia of Ficus sur and F. sycomorus in Southern Africa. S. Afr. J. Bot. 2009, 75, 771–777. [Google Scholar] [CrossRef]
- Gu, D.; Compton, S.G.; Peng, Y.; Yang, D. ‘Push’ and ‘pull’ responses by fig wasps to volatiles released by their host figs. Chemoecology 2012, 22, 217–227. [Google Scholar] [CrossRef]
- Borges, R.M.; Bessiere, J.M.; Ranganathan, Y. Diel variation in fig volatiles across syconium development: Making sense of scents. J. Chem. Ecol. 2013, 39, 630–642. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Zhao, C.; Zhang, G.; Teixeira da Silva, J.A.; Duan, J. Genome-Wide Identification and Expression Profile of TPS Gene Family in Dendrobium officinale and the Role of DoTPS10 in Linalool Biosynthesis. Int. J. Mol. Sci. 2020, 21, 5419. [Google Scholar] [CrossRef] [PubMed]
- Tholl, D. Biosynthesis and Biological Functions of Terpenoids in Plants. Adv. Biochem. Eng. Biotechnol. 2015, 148, 63–106. [Google Scholar] [CrossRef]
- Nawade, B.; Shaltiel-Harpaz, L.; Yahyaa, M.; Kabaha, A.; Kedoshim, R.; Bosamia, T.C.; Ibdah, M. Characterization of terpene synthase genes potentially involved in black fig fly (Silba adipata) interactions with Ficus carica. Plant Sci. 2020, 298, 110549. [Google Scholar] [CrossRef]
- Dhifi, W.; Bellili, S.; Jazi, S.; Bahloul, N.; Mnif, W. Essential Oils’ Chemical Characterization and Investigation of Some Biological Activities: A Critical Review. Medicines 2016, 3, 25. [Google Scholar] [CrossRef]
- Zhou, H.C.; Shamala, L.F.; Yi, X.K.; Yan, Z.; Wei, S. Analysis of Terpene Synthase Family Genes in Camellia sinensis with an Emphasis on Abiotic Stress Conditions. Sci. Rep. 2020, 10, 933. [Google Scholar] [CrossRef]
- Gao, F.; Liu, B.; Li, M.; Gao, X.; Fang, Q.; Liu, C.; Ding, H.; Wang, L.; Gao, X. Identification and characterization of terpene synthase genes accounting for volatile terpene emissions in flowers of Freesia x hybrida. J. Exp. Bot. 2018, 69, 4249–4265. [Google Scholar] [CrossRef]
- Zhou, S.S.; Xing, Z.; Liu, H.; Hu, X.G.; Gao, Q.; Xu, J.; Jiao, S.Q.; Jia, K.H.; Jin, Y.Q.; Zhao, W.; et al. In-depth transcriptome characterization uncovers distinct gene family expansions for Cupressus gigantea important to this long-lived species’ adaptability to environmental cues. BMC Genom. 2019, 20, 213. [Google Scholar] [CrossRef]
- Chen, F.; Tholl, D.; Bohlmann, J.; Pichersky, E. The family of terpene synthases in plants: A mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 2011, 66, 212–229. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.Y.; Jin, J.; Sarojam, R.; Ramachandran, S. A Comprehensive Survey on the Terpene Synthase Gene Family Provides New Insight into Its Evolutionary Patterns. Genome Biol. Evol. 2019, 11, 2078–2098. [Google Scholar] [CrossRef] [PubMed]
- Alicandri, E.; Paolacci, A.R.; Osadolor, S.; Sorgona, A.; Badiani, M.; Ciaffi, M. On the Evolution and Functional Diversity of Terpene Synthases in the Pinus Species: A Review. J. Mol. Evol. 2020, 88, 253–283. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.M.; Aubourg, S.; Schouwey, M.B.; Daviet, L.; Schalk, M.; Toub, O.; Lund, S.T.; Bohlmann, J. Functional annotation, genome organization and phylogeny of the grapevine (Vitis vinifera) terpene synthase gene family based on genome assembly, FLcDNA cloning, and enzyme assays. BMC Plant Biol. 2010, 10, 226. [Google Scholar] [CrossRef]
- Hattan, J.; Shindo, K.; Sasaki, T.; Misawa, N. Isolation and Functional Characterization of New Terpene Synthase Genes from Traditional Edible Plants. J. Oleo Sci. 2018, 67, 1235–1246. [Google Scholar] [CrossRef]
- Booth, J.K.; Yuen, M.M.S.; Jancsik, S.; Madilao, L.L.; Page, J.E.; Bohlmann, J. Terpene Synthases and Terpene Variation in Cannabis sativa. Plant Physiol. 2020, 184, 130–147. [Google Scholar] [CrossRef]
- Huang, L.M.; Huang, H.; Chuang, Y.C.; Chen, W.H.; Wang, C.N.; Chen, H.H. Evolution of Terpene Synthases in Orchidaceae. Int. J. Mol. Sci. 2021, 22, 6947. [Google Scholar] [CrossRef]
- He, J.; Verstappen, F.; Jiao, A.; Dicke, M.; Bouwmeester, H.J.; Kappers, I.F. Terpene synthases in cucumber (Cucumis sativus) and their contribution to herbivore-induced volatile terpenoid emission. New Phytol. 2022, 233, 862–877. [Google Scholar] [CrossRef]
- Huang, X.Z.; Xiao, Y.T.; Kollner, T.G.; Jing, W.X.; Kou, J.F.; Chen, J.Y.; Liu, D.F.; Gu, S.H.; Wu, J.X.; Zhang, Y.J.; et al. The terpene synthase gene family in Gossypium hirsutum harbors a linalool synthase GhTPS12 implicated in direct defence responses against herbivores. Plant Cell Environ. 2018, 41, 261–274. [Google Scholar] [CrossRef]
- Zhou, F.; Pichersky, E. The complete functional characterisation of the terpene synthase family in tomato. New Phytol. 2020, 226, 1341–1360. [Google Scholar] [CrossRef]
- Berg, C.C.; Wiebes, J.T. African Fig Trees and Fig Wasps; KNAW: Amsterdam, The Netherlands; New York, NY, USA, 1992; p. 298. [Google Scholar]
- Chen, F.; Ro, D.K.; Petri, J.; Gershenzon, J.; Bohlmann, J.; Pichersky, E.; Tholl, D. Characterization of a root-specific Arabidopsis terpene synthase responsible for the formation of the volatile monoterpene 1,8-cineole. Plant Physiol. 2004, 135, 1956–1966. [Google Scholar] [CrossRef] [PubMed]
- Allen, K.D.; McKernan, K.; Pauli, C.; Roe, J.; Torres, A.; Gaudino, R. Genomic characterization of the complete terpene synthase gene family from Cannabis sativa. PLoS ONE 2019, 14, e0222363. [Google Scholar] [CrossRef] [PubMed]
- Souto-Vilaros, D.; Machac, A.; Michalek, J.; Darwell, C.T.; Sisol, M.; Kuyaiva, T.; Isua, B.; Weiblen, G.D.; Novotny, V.; Segar, S.T. Faster speciation of fig-wasps than their host figs leads to decoupled speciation dynamics: Snapshots across the speciation continuum. Mol. Ecol. 2019, 28, 3958–3976. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Feng, X.; Zheng, Y.; Huang, C.H.; Nakano, C.; Hoshino, T.; Bogue, S.; Ko, T.P.; Chen, C.C.; Cui, Y.; et al. Structure, function and inhibition of ent-kaurene synthase from Bradyrhizobium japonicum. Sci. Rep. 2014, 4, 6214. [Google Scholar] [CrossRef]
- Kruppa, J.; Jo, W.K.; van der Vries, E.; Ludlow, M.; Osterhaus, A.; Baumgaertner, W.; Jung, K. Virus detection in high-throughput sequencing data without a reference genome of the host. Infect. Genet. Evol. 2018, 66, 180–187. [Google Scholar] [CrossRef]
- Jia, Q.; Brown, R.; Kollner, T.G.; Fu, J.; Chen, X.; Wong, G.K.; Gershenzon, J.; Peters, R.J.; Chen, F. Origin and early evolution of the plant terpene synthase family. Proc. Natl. Acad. Sci. USA 2022, 119, e2100361119. [Google Scholar] [CrossRef]
- Aubourg, S.; Lecharny, A.; Bohlmann, J. Genomic analysis of the terpenoid synthase ( AtTPS) gene family of Arabidopsis thaliana. Mol. Genet. Genom. 2002, 267, 730–745. [Google Scholar] [CrossRef]
- Luck, K.; Chen, X.L.; Norris, A.M.; Chen, F.; Gershenzon, J.; Kollner, T.G. The reconstruction and biochemical characterization of ancestral genes furnish insights into the evolution of terpene synthase function in the Poaceae. Plant Mol. Biol. 2020, 104, 203–215. [Google Scholar] [CrossRef]
- Dudareva, N.; Klempien, A.; Muhlemann, J.K.; Kaplan, I. Biosynthesis, function and metabolic engineering of plant volatile organic compounds. New Phytol. 2013, 198, 16–32. [Google Scholar] [CrossRef]
- Green, S.; Friel, E.N.; Matich, A.; Beuning, L.L.; Cooney, J.M.; Rowan, D.D.; MacRae, E. Unusual features of a recombinant apple alpha-farnesene synthase. Phytochemistry 2007, 68, 176–188. [Google Scholar] [CrossRef]
- Danner, H.; Boeckler, G.A.; Irmisch, S.; Yuan, J.S.; Chen, F.; Gershenzon, J.; Unsicker, S.B.; Kollner, T.G. Four terpene synthases produce major compounds of the gypsy moth feeding-induced volatile blend of Populus trichocarpa. Phytochemistry 2011, 72, 897–908. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Wang, D.; Chen, X.; Kollner, T.G.; Mazarei, M.; Guo, H.; Pantalone, V.R.; Arelli, P.; Stewart, C.N., Jr.; Wang, N.; et al. An (E,E)-alpha-farnesene synthase gene of soybean has a role in defence against nematodes and is involved in synthesizing insect-induced volatiles. Plant Biotechnol. J. 2017, 15, 510–519. [Google Scholar] [CrossRef] [PubMed]
- Williams, D.C.; McGarvey, D.J.; Katahira, E.J.; Croteau, R. Truncation of limonene synthase preprotein provides a fully active ‘pseudomature’ form of this monoterpene cyclase and reveals the function of the amino-terminal arginine pair. Biochemistry 1998, 37, 12213–12220. [Google Scholar] [CrossRef] [PubMed]
- Degenhardt, J.; Kollner, T.G.; Gershenzon, J. Monoterpene and sesquiterpene synthases and the origin of terpene skeletal diversity in plants. Phytochemistry 2009, 70, 1621–1637. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- García, P.A.; de Oliveira, A.B.; Batista, R. Occurrence, Biological Activities and Synthesis of Kaurane Diterpenes and their Glycosides. Molecules 2007, 12, 455–483. [Google Scholar] [CrossRef]
- Morris, B.D.; Foster, S.P.; Grugel, S.R.; Charlet, L.D. Isolation of the diterpenoids, ent-kauran-16alpha-ol and ent-atisan-16alpha-ol, from sunflowers, as oviposition stimulants for the banded sunflower moth, Cochylis hospes. J. Chem. Ecol. 2005, 31, 89–102. [Google Scholar] [CrossRef]
- Pichersky, E.; Lewinsohn, E.; Croteau, R. Purification and characterization of S-linalool synthase, an enzyme involved in the production of floral scent in Clarkia breweri. Arch. Biochem. Biophys. 1995, 316, 803–807. [Google Scholar] [CrossRef]
- Grison-Pige, L.; Hossaert-McKey, M.; Greeff, J.M.; Bessiere, J.M. Fig volatile compounds--a first comparative study. Phytochemistry 2002, 61, 61–71. [Google Scholar] [CrossRef]
- Prosser, I.; Altug, I.G.; Phillips, A.L.; Konig, W.A.; Bouwmeester, H.J.; Beale, M.H. Enantiospecific (+)- and (-)-germacrene D synthases, cloned from goldenrod, reveal a functionally active variant of the universal isoprenoid-biosynthesis aspartate-rich motif. Arch. Biochem. Biophys. 2004, 432, 136–144. [Google Scholar] [CrossRef]
- Stranden, M.; Borg-Karlson, A.K.; Mustaparta, H. Receptor neuron discrimination of the germacrene D enantiomers in the moth Helicoverpa armigera. Chem. Senses 2002, 27, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Rostelien, T.; Borg-Karlson, A.K.; Faldt, J.; Jacobsson, U.; Mustaparta, H. The plant sesquiterpene germacrene D specifically activates a major type of antennal receptor neuron of the tobacco budworm moth Heliothis virescens. Chem. Senses 2000, 25, 141–148. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yu, H.; Zhang, Z.; Liu, L.; Cheng, Y.; Deng, X.; Segar, S.T.; Compton, S.G. Asymmetric sharing of pollinator fig wasps between two sympatric dioecious fig trees: A reflection of supply and demand or differences in the size of their figs? Bot. Stud. 2022, 63, 7. [Google Scholar] [CrossRef] [PubMed]
- Cornille, A.; Underhill, J.G.; Cruaud, A.; Hossaert-McKey, M.; Johnson, S.D.; Tolley, K.A.; Kjellberg, F.; van Noort, S.; Proffit, M. Floral volatiles, pollinator sharing and diversification in the fig-wasp mutualism: Insights from Ficus natalensis, and its two wasp pollinators (South Africa). Proc. Biol. Sci. 2012, 279, 1731–1739. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.H.; Sasaki, A.; Kusumi, J.; Chou, P.A.; Tzeng, H.Y.; Li, H.Q.; Yu, H. Pollinator sharing, copollination, and speciation by host shifting among six closely related dioecious fig species. Commun. Biol. 2022, 5, 284. [Google Scholar] [CrossRef]
- Wang, G.; Compton, S.G.; Chen, J. The mechanism of pollinator specificity between two sympatric fig varieties: A combination of olfactory signals and contact cues. Ann. Bot. 2013, 111, 173–181. [Google Scholar] [CrossRef]
- Wang, G.; Cannon, C.H.; Chen, J. Pollinator sharing and gene flow among closely related sympatric dioecious fig taxa. Proc. Biol. Sci. 2016, 283, 20152963. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644-U130. [Google Scholar] [CrossRef]
- Wheeler, T.J.; Eddy, S.R. nhmmer: DNA homology search with profile HMMs. Bioinformatics 2013, 29, 2487–2489. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; Hernandez-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef]
- Katoh, K.; Kuma, K.; Toh, H.; Miyata, T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994, 2, 28–36. [Google Scholar] [PubMed]
- Fischer, S.; Brunk, B.P.; Chen, F.; Gao, X.; Harb, O.S.; Iodice, J.B.; Shanmugam, D.; Roos, D.S.; Stoeckert, C.J., Jr. Using OrthoMCL to assign proteins to OrthoMCL-DB groups or to cluster proteomes into new ortholog groups. Curr. Protoc. Bioinform. 2011, 12, 11–19. [Google Scholar] [CrossRef]
- Mahmoudinia, S.; Niapour, A.; Ghasemi Hamidabadi, H.; Mazani, M. 2,4-D causes oxidative stress induction and apoptosis in human dental pulp stem cells (hDPSCs). Environ. Sci. Pollut. Res. 2019, 26, 26170–26183. [Google Scholar] [CrossRef]
- Borodina, T.; Adjaye, J.; Sultan, M. Chapter five—A Strand-Specific Library Preparation Protocol for RNA Sequencing. In Methods in Enzymology; Jameson, D., Verma, M., Westerhoff, H.V., Eds.; Academic Press: San Diego, CA, USA, 2011; Volume 500, pp. 79–98. [Google Scholar]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Yang, Z. Statistical properties of a DNA sample under the finite-sites model. Genetics 1996, 144, 1941–1950. [Google Scholar] [CrossRef]
- Jovanovic, V.M.; Sarfert, M.; Reyna-Blanco, C.S.; Indrischek, H.; Valdivia, D.I.; Shelest, E.; Nowick, K. Positive Selection in Gene Regulatory Factors Suggests Adaptive Pleiotropic Changes During Human Evolution. Front. Genet. 2021, 12, 662239. [Google Scholar] [CrossRef]

| Subgenus | Section | Subsection | Species | Name Abbreviations | TPS No. | Site | Latitude (N) | Longitude (E) |
|---|---|---|---|---|---|---|---|---|
| Ficus | Eriosycea | Eriosycea | F. chartacea | ficha | 8 | China—Guangdong | 8.776 | 99.724 |
| F. fulva | fiful | 14 | China—Guangdong | 8.776 | 99.724 | |||
| F. grossularoides | figro | 3 | Thailand—Narathiwat | 5.799 | 101.762 | |||
| F. hirta | fihir | 16 | China—SCBG | 23.171 | 113.349 | |||
| F. langkokensis | filan | 11 | China—Guangdong | 24.227 | 112.006 | |||
| F. ruficaulis var. antaoensis | firuf | 4 | Taiwan | 21.962 | 120.811 | |||
| F. triloba | fitri | 13 | China—SCBG | 23.180 | 112.537 | |||
| Ficus | Frutescentiae | F. abeli | fiabe | 12 | China—Guangdong | 23.636 | 113.780 | |
| F. erecta var. beecheyana | fiere | 11 | China—Guangdong | 23.765 | 113.915 | |||
| F. formosa | fifor | 6 | China—Guangdong | 23.623 | 113.811 | |||
| F. heteromorpha | fihet | 15 | China—Guangdong | 24.918 | 113.033 | |||
| F. ischnopoda | fiisc | 11 | Thailand—Chiang Mai | 18.504 | 98.665 | |||
| F. pandurata | fipan | 13 | China—Guangdong | 24.252 | 112.036 | |||
| F. pyrifomis | fipyr | 12 | Thailand—Chiang Mai | 18.504 | 98.665 | |||
| F. variolosa | fivar | 13 | China—Guangdong | 23.624 | 113.797 | |||
| Sycidium | Sycidium | Sycidium | F. montana | fimon | 3 | Thailand | 7.557 | 99.776 |
| Sycomorus | Sycomorus | Sycocarpus | F. fistulosa | fifis | 3 | China—Guangdong | 23.156 | 112.511 |
| F. hispida | fihis | 8 | China—SCBG | 23.180 | 113.350 | |||
| Neomorphe | F. variegata | fivae | 9 | China—Guangdong | 23.176 | 112.538 | ||
| Hemicardia | F. semicordata | fisem | 11 | Thailand—Chiang Mai | 19.362 | 98.922 | ||
| Urostigma | Urostigma | Conosycea | F. altissima | fialt | 19 | China—SCBG | 23.188 | 113.363 |
| F. benjamina | fiben | 7 | China—SCBG | 23.186 | 113.358 | |||
| F. microcarpa | fimic | 13 | China—SCBG | 23.178 | 113.352 | |||
| Urostigma | F. rumphii | firum | 13 | Myanmar | 21.966 | 96.069 |
| Gene | Model | np | Mates of Parameter | lnL | df | 2ΔlnL | p-Value a |
|---|---|---|---|---|---|---|---|
| FTPS1 | Free-ratio | 79 | Variable ω | −4424.469 | 38 | 35.058 | 0.606 |
| One-ratio | 41 | 0.522 | −4441.998 | ||||
| FTPS2 | Free-ratio | 67 | Variable ω | −5413.296 | 32 | 33.835 | 0.379 |
| One-ratio | 35 | 0.449 | −5430.214 | ||||
| FTPS3 | Free-ratio | 67 | Variable ω | −5872.822 | 32 | 37.559 | 0.229 |
| One-ratio | 35 | 0.679 | −5891.602 | ||||
| FTPS4 | Free-ratio | 59 | Variable ω | −5024.747 | 28 | 21.298 | 0.813 |
| One-ratio | 31 | 0.496 | −5035.396 | ||||
| FTPS5 | Free-ratio | 51 | Variable ω | −3504.033 | 24 | 19.138 | 0.745 |
| One-ratio | 27 | 0.345 | −3513.602 | ||||
| FTPS6 | Free-ratio | 23 | Variable ω | −3252.649 | 20 | 3.613 | 0.999 |
| One-ratio | 13 | 0.465 | −3254.456 | ||||
| FTPS7 | Free-ratio | 43 | Variable ω | −3883.714 | 20 | 19.883 | 0.465 |
| One-ratio | 23 | 0.503 | −3893.656 | ||||
| FTPS8 | Free-ratio | 43 | Variable ω | −6311.560 | 20 | 14.685 | 0.794 |
| One-ratio | 23 | 0.504 | −6318.902 | ||||
| FTPS9 | Free-ratio | 23 | Variable ω | −3433.342 | 10 | 8.137 | 0.615 |
| One-ratio | 13 | 0.479 | −3437.411 | ||||
| FTPS10 | Free-ratio | 35 | Variable ω | −4081.220 | 16 | 26.337 | 0.049 * |
| One-ratio | 19 | 0.426 | −4094.389 | ||||
| FTPS11 | Free-ratio | 27 | Variable ω | −3358.780 | 12 | 18.882 | 0.091 |
| One-ratio | 15 | 0.499 | −3368.221 | ||||
| FTPS12 | Free-ratio | 27 | Variable ω | −3715.070 | 12 | 23.205 | 0.026 * |
| One-ratio | 15 | 0.517 | −3726.673 | ||||
| FTPS13 | Free-ratio | 23 | Variable ω | −4886.043 | 10 | 6.841 | 0.740 |
| One-ratio | 13 | 0.382 | −4889.464 | ||||
| FTPS14 | Free-ratio | 15 | Variable ω | −3614.771 | 6 | 10.329 | 0.111 |
| One-ratio | 9 | 0.637 | −3619.935 | ||||
| FTPS15 | Free-ratio | 15 | Variable ω | −3086.012 | 6 | 4.530 | 0.605 |
| One-ratio | 9 | 0.250 | −3088.278 | ||||
| FTPS16 | Free-ratio | 15 | Variable ω | −3125.859 | 6 | 6.115 | 0.410 |
| One-ratio | 9 | 0.758 | −3128.916 | ||||
| FTPS17 | Free-ratio | 19 | Variable ω | −3127.501 | 8 | 3.009 | 0.934 |
| One-ratio | 11 | 0.447 | −3129.006 | ||||
| FTPS19 | Free-ratio | 15 | Variable ω | −2834.753 | 6 | 4.801 | 0.570 |
| One-ratio | 9 | 0.407 | −2837.153 | ||||
| FTPS20 | Free-ratio | 23 | Variable ω | −3453.672 | 7 | 3.639 | 0.820 |
| one-ratio | 13 | 0.548 | −3455.491 | ||||
| FTPS26 | Free-ratio | 23 | Variable ω | −5711.069 | 8 | 14.155 | 0.078 |
| one-ratio | 13 | 0.489 | −5718.147 |
| Gene | Clade | Models Compared | np | p-Value a | lnL | ω Values | Positive Sites (BEB) b |
|---|---|---|---|---|---|---|---|
| FTPS3 | fimon | Model A | 38 | 1.490 × 10−2 * | −5595.802 | p0 = 0.023 ω1 = 1.000 ω2 = 44.817 | |
| Model A null | 37 | −5598.162 | p0 = 0.023 ω1 = 1.000 ω2 = 1.000 | ||||
| fipyr | Model A | 38 | 6.105 × 10−4 ** | −5490.893 | p0 = 0.015 ω1 = 1.000 ω2 = 315.194 | ||
| Model A null | 37 | −5496.038 | p0 = 0.018 ω1 = 1.000 ω2 = 1.000 | ||||
| FTPS4 | fihir-fitri | Model A | 34 | 0.051 | −4860.385 | p0 = 0.000 ω1 = 1.000 ω2 = 92.201 | 258S * 345Y * 424M * |
| Model A null | 33 | −4861.730 | p0 = 0.000 ω1 = 1.000 ω2 = 1.000 | ||||
| FTPS8 | fihet | Model A | 26 | 0.059 | −5773.442 | p0 = 0.047 ω1 = 1.000 ω2 = 28.837 | 217H ** |
| Model A null | 25 | −5776.607 | p0 = 0.047 ω1 = 1.000 ω2 = 1.000 | ||||
| FTPS16 | fifis | Model A | 13 | 1.129 × 10−3 ** | −3118.162 | p0 = 0.417 ω1 = 1.000 ω2 = 314.584 | |
| Model A null | 12 | −3122.826 | p0 = 0.408 ω1 = 1.000 ω2 = 1.000 | ||||
| FTPS20 | fifor | Model A | 16 | 6.005 × 10−7 ** | −3399.251 | p0 = 0.588 ω1 = 1.000 ω2 = 246.640 | |
| Model A null | 15 | −3427.039 | p0 = 0.626 ω1 = 1.000 ω2 = 1.000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, P.; Chen, X.; Chantarasuwan, B.; Zhu, X.; Deng, X.; Bao, Y.; Yu, H. Composition Diversity and Expression Specificity of the TPS Gene Family among 24 Ficus Species. Diversity 2022, 14, 721. https://doi.org/10.3390/d14090721
Sun P, Chen X, Chantarasuwan B, Zhu X, Deng X, Bao Y, Yu H. Composition Diversity and Expression Specificity of the TPS Gene Family among 24 Ficus Species. Diversity. 2022; 14(9):721. https://doi.org/10.3390/d14090721
Chicago/Turabian StyleSun, Peng, Xiaoyong Chen, Bhanumas Chantarasuwan, Xueying Zhu, Xiaoxia Deng, Ying Bao, and Hui Yu. 2022. "Composition Diversity and Expression Specificity of the TPS Gene Family among 24 Ficus Species" Diversity 14, no. 9: 721. https://doi.org/10.3390/d14090721
APA StyleSun, P., Chen, X., Chantarasuwan, B., Zhu, X., Deng, X., Bao, Y., & Yu, H. (2022). Composition Diversity and Expression Specificity of the TPS Gene Family among 24 Ficus Species. Diversity, 14(9), 721. https://doi.org/10.3390/d14090721








