Abstract
Heterobasidion root rot is one of the most economically important conifer diseases in the Northern Hemisphere, and stump removal is considered to be one of most effective control methods. However, the impact of stump removal on the diversity of mycorrhizal and soil fungi should be evaluated. From 2011 to 2012, a stump removal trial was established in six different sample plots in two regions of Latvia. The stump removal sites and control sites were replanted with spruce seedlings in 2012. Seven years later, soil samples were collected in the stump removal and control sample plots to compare the diversity of mycorrhizal and soil fungi. Fungal communities were analyzed using next-generation sequencing methods. Our results showed that there are no significant differences in mycorrhizal and soil fungal communities between the stump removal area and the clear-felled control area seven years later. The mycorrhizal fungi were the most commonly sequenced fungal ecological group, and their diversity was similar to clear-felled control sites. However, there were some differences in the fungal species composition.
1. Introduction
Fungi from the Heterobasidion annosum species complex can cause root rot and stem the decay of many coniferous and some deciduous trees in the Northern Hemisphere, mainly in Europe, causing economic losses of more than 500 million EU [1,2]. The species complex comprises five fungal species: Heterobasidion annosum s.s., H. parviporum, H. abietinum, H. occidentale, and H. irregulare, which all have different geographical ranges and host preferences [2]. Heterobasidion infection can spread in two ways: by air-borne basidiospores, which infect freshly cut conifer stumps, as well as by infecting bark-peeling wounds of living trees, usually close to the stem base; and by mycelium, infecting closely-standing trees or newly-planted seedlings through root contact [3]. The viable Heterobasidion mycelium can survive in conifer stumps for decades, which means that infection tends to accumulate in forest areas over time [4,5].
In Latvia, the Norway spruce (Picea abies (L.) Karst) is one of the most economically significant and widespread tree species [6]. A country-scale inventory made in 2005–2006 revealed that approximately 23% of all Latvian spruces contained butt and stem rot, mainly caused by H. parviporum [7]. Stump removal is considered to be the most efficient method to reduce Heterobasidion infection incidences in already-infected forest stands [8,9,10,11]. However, this method could have a negative impact on stand biodiversity, as it causes site disturbances due to stump extraction, which may lead to changes in soil properties [12]. Soil and mycorrhizal fungi play a crucial role in degrading organic materials, as they positively impact tree growth and resistance to soil-borne diseases in tree seedlings [13,14]. The high diversity of soil microorganisms contributes to the resilience of ecosystems against disturbances, which is becoming more relevant in the context of global change [15]. Many of these soil microorganisms are affected by changes in land use, as well as management practices [16,17,18]. Therefore, it is important to evaluate the impact of stump removal on the diversity of soil fungi, as well as on the mycorrhization of spruce seedlings planted after stump removal.
Data about the impact of stump removal on the diversity of mycorrhizal [19,20,21,22,23] and soil fungi [24] are scarce, and most of the research was conducted 2–5 years after seedling planting. However, new technologies for the analysis of environmental DNA samples have expanded the understanding of the diversity and abundance of soil-dwelling organisms, including fungi [25]. The Pacific Bioscience SMRT Sequencing Platform allows long sequence reads to be obtained, allowing for more accurate identification of soil fungi and thus, a clearer description of their diversity. Recent studies have shown the usefulness of this method for the study of microscopic fungi in forest soils [26].
The aim of the current research was to evaluate the impact of stump removal on the diversity of mycorrhizal and soil fungi seven years after the replanting of the stump removal site.
2. Materials and Methods
2.1. Field Work
In 2011–2012, a long-term field trial using stump removal as a Heterobasidion root rot control method was established [27]. For this study, we selected sites where stump removal was carried out under similar conditions and in the same period, but without previous evaluation of the Heterobasidion infection rate. The territory of Latvia is characterized by a temperate climate. Sampling areas were located in Central Latvia, where the average annual temperature ranges from 6.8 to 7.9 °C and annual precipitation rate ranges from 574 to 636 mm. In total, six sample plots in two Latvian regions (Figure 1) were analyzed. All sample plots were clear-felled in 2010–2011, and the area of each stand was more than 1 ha. Three forest stands were of a Hylocomiosa forest site type, and three were of a Oxalidosa forest site type [28].
Figure 1.
Locations of stump removal sample plots (1—Dursupe, 2—Stende, 3—Jaunpils, 4—Tinuzi, 5—Rembate, 6—Nitaure).
Each sample plot was divided in two parts, 0.5 ha each: one left intact, and one where section stumps were removed using New Holland E21B caterpillar excavators, fitted with a MCR500 stump head and a Komatsu PC210LC with a CBI stump head [29]. The soil of the clear-felled control area was prepared prior to planting by using a disc-trenching method. All sample plots were replanted in 2012 with Picea abies seedlings (both containerized and bare-rooted).
During September and October of 2019, soil samples were collected for the analysis of fungal community and diversity using molecular methods in all six stump removal sites. Soil sampling was done as follows: 10 circular sub-sample plots with a radius of 5 m were established within each long-term sample plot, with six plots in the stump removal areas and six in the control areas. In each sample plot, the average growth rates of saplings (height and diameter), as well as tree species composition were assessed, and soil samples were collected from five spruce saplings growing in each sub-sample plot. Soil samples were collected at a depth of 20 cm using a soil probe [30]. A total of 60 soil samples were collected: 30 samples from the stump removal plots and 30 samples from the control plots.
2.2. Sample Preparation and Sequencing
In the laboratory, the soil samples were sorted by removing litter, and the roots of woody plants and forbs, all of which were discarded. Root samples were washed and mechanically cleaned to remove soil particles before freezing. Samples of both soil and roots were lyophilized and stored at −20 °C for fungal diversity analysis. A portion of the collected soil was analyzed in the LSFRI Silava Environmental Laboratory where soil chemical composition was determined.
Sample preparation and total DNA extraction was performed at the LSFRI Silava Forest Phytopathology and Mycology Laboratory, as well as at the LSFRI Silava Genetic Resources Center. Further laboratory processes were done at the end of May 2021, at the University of Tartu, Estonia. Amplification of DNA samples was performed with sample-specific, double-labeled primers. The primer pair used in this study was the primer F-ITS1CATTA and the universal primer R-ITS4ngsUni [31,32]. This pair of primers enables the generation of complete fungal species-specific ITS region sequences that allow detailed identification of fungal species. The polymerase chain reaction mixture was a total of 25 µL for each reaction; 5 µL of 5× HOT FIREPol Blend Master Mix (Solis Biodyne, Tartu, Estonia) and 18 µL of ddH2O was used per sample. 23 µL of the mixture was added to the PCR plate, and then 1 μL of the DNA sample and 0.5 µL of each primer (20 mM) were added. The PCR thermal cycling conditions were as follows: initial denaturation for 15 min at 95 °C, for 30–35 cycles; DNA denaturation at 95 °C for 30 s; primer hybridization at 57 °C for 30 s; DNA synthesis at 72 °C for 1 min; and the final synthesis at 72 °C for 10 min. The polymerase chain reaction products (5 µL) were tested on a 1% agarose gel to assess the amount of amplified DNA. The volume of PCR products corresponding to the concentration of each sample was combined in one sample library. The library was purified using the FavorPrep PCR Purification Kit (Favorgen Biotech. Corp., Vienna, Austria).
The prepared samples were sent to the University of Oslo in Norway for sequencing using the Pacific Bioscience SequelII sequencing platform (Pacific Biosciences, Inc. Menlo Park, CA, USA). Libraries were prepared according to the manufacturer’s protocol. Polymerase chain reaction products were normalized during library preparation. Sequencing was performed using anSMRT cell (SMRT cell 1M, v2, Sequel polymerase v2.1, sequencing chemical reagents v2.1, using a diffuse sampling method with an exposure time of 600 min, resulting in a final synthesis time 45 min).
2.3. Data Analyses
Bioinformatics data analyses were performed using a variety of applications connected to the Pipecraft (v1.0) analysis platform [33]. Sequence quality was tested with the Mothur (v1.36.1) program [34], excluding sequences shorter than 100 bp. Sequence chimeras were excluded from the analysis using UCHIME [35]. The full-length ITS (Internal Transcribed Spacer) region was isolated from the genes encoding rRNA by ITSx (v1.0.11) [36]. The resulting sequences were grouped into taxonomic units (“Operational Taxonomic Units”) based on 98% similarity with already published taxa (BLASTn: e-value = 0.001, word size = 7, reward = 1, penalty = −1, gap open = 1, gap extend = 2) [37], using the CD-HIT (v4.6) program [38]. Groups of taxonomic units that included only one sequence were excluded from further analysis. The taxonomic affiliation of the sequences was determined in the UNITE platform [39]. The functional groups of the fungi were determined in the FungalTraits database [40].
Statistical calculations were performed in R (v.4.0.3) [41]. The Shannon diversity index and the occurrence of taxonomic units in the samples [42], calculated with the estimate_richness function in the phyloseq package [43], were used as parameters for characterizing fungal diversity. In addition, the diversity of taxonomic units of ectomycorrhizal, saprotrophic, and pathogenic fungi in the samples was calculated.
The number of taxonomic units and the Shannon diversity index were compared between stump removal and control samples using ANOVAs if the data were normally distributed, or Wilcoxon’s tests if the distribution of the data was not normally distributed.
In addition, whether certain species can be distinguished from the most common taxa as indicator species for stump removal or control plots was assessed. The significance of these species as indicator species was tested with the indicspecies package in R [44].
The Bray–Curtis distinction has also been tested in mushroom societies [45]. These models are visualized using the NMDS (Non-metric multidimensional scaling (NMDS)) ordination method, which is based on grouping companies according to the Bray–Curtis difference. This analysis was performed using the phyloseq package and, in addition to the graphs, the ellipse of the 95% confidence interval was calculated using the stat_ellipse function for visualization [43]. The effects of stump removal, forest type, and stand on fungal communities were tested by multivariate analysis of variance with permutations (PERMANOVA) using the adonis function (999 permutations) in the vegan package [46].
3. Results
3.1. Impact of Stump Removal on Diversity of Soil and Mycorrhizal Fungi
The initial data included 132,098 sequences. After the quality control phase, the number of sequences decreased to 120,981. After control of chimeras, it further decreased to 115,326. Finally, after extraction of the ITS region, the number of sequences was 111,511, ranging from 277 to 3348 bp. Seven samples were excluded from the analysis due to the small number of sequences obtained compared to the mean in the data set. The final data table consisted of 105,208 sequences divided into taxonomic units, and represented 53 out of 60 samples (27 from stump removal sample plots, and 26 from control sample plots).
The fungal community as a whole was characterized by a pronounced dominance of individual taxa. The hundred most frequently sequenced species accounted for 75% of all sequences obtained. The twenty most frequently sequenced species accounted for almost half or 49% of the sequences.
The functional group of the fungi could be determined for 71.8% of the sequences. The samples were mainly ectomycorrhizal fungi (40.3% of all sequences), saprotrophs of various substrates (litter, soil, and wood degrading fungi) (22.4% of all sequences), and plant pathogens (4.0% of all sequences).
The most commonly sequenced fungal taxa are shown in Table 1. The most frequently sequenced species were two mycorrhizal fungi; Thelephora terrestris (11.2% of all sequences) and Inocybe lacera (10.4% of all sequences) (Table 1). Other common mycorrhizal species were Trichophaea sp. (2.2%), Lactarius sp. (1.5%), and Amphinema sp. (1.4%). The most frequently sequenced species from the saprotrophic genera were Hyaloscypha sp. (1.5%), Mycena sp. (1.3%), and Solicoccozyma sp. (1.0%).

Table 1.
Most common fungal taxa in the analyzed samples.
The data from our study showed that the total number of fungal taxonomic units and the Shannon diversity indices did not differ between stump removal and control samples (Figure 2 and Figure 3, respectively). The number of taxa of the functional groups of fungi (ectomycorrhizal, saprophytic, and plant pathogenic fungi) and the Shannon diversity did not differ between the groups in the stump removal samples. The analysis of the fungal community also did not show significant differences between the fungal diversity in the stump removal and control samples (Figure 4).
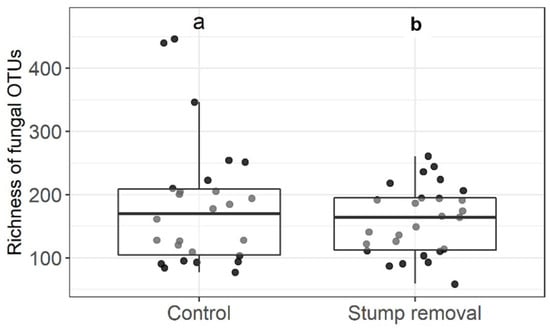
Figure 2.
Effect of stump removal on richness of fungal OTUs. Middle lines, boxes, whiskers, and circles represent medians, quartiles, 90% quantiles, and all sampled trees, respectively. Different letters above whiskers indicate statistically different groups (p < 0.05). Number of stands: stump removal—27; control—26.
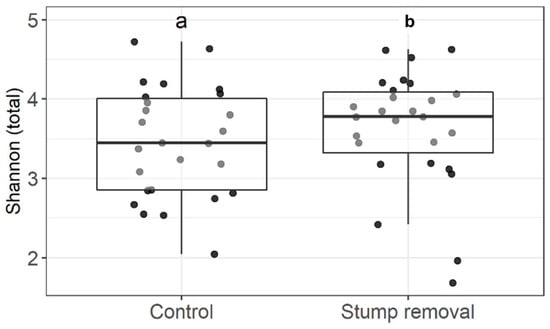
Figure 3.
Effect of stump removal on the Shannon diversity of fungal OTUs. Middle lines, boxes, whiskers, and circles represent medians, quartiles, 90% quantiles, and all sampled trees, respectively. Different letters above whiskers indicate statistically different groups (p < 0.05). Number of stands: stump removal—27; control—26.
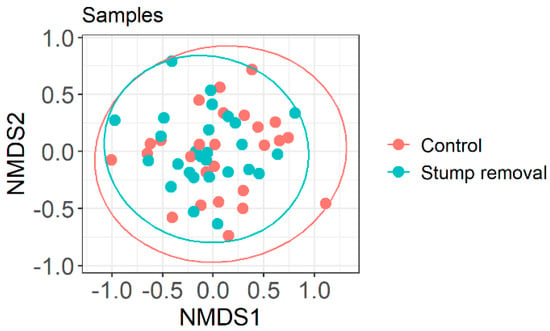
Figure 4.
Non-metric multidimensional scaling (NMDS) ordination for fungal communities (presence/absence data of fungal OTUs) as a function of sampling site management history (stump removal (red) vs. control (blue) samples). Ellipses indicate 95% confidence intervals for each sample group.
3.2. Impact of Forest Site Type on Diversity of Soil and Mycorrhizal Fungi
In addition to stump removal, the physio-chemical and biological processes in the soil, which are characteristic of the stand and forest type, also affect fungal communities. Our data show that the forest type and stand significantly influenced the number of taxonomic units of fungi. More fungal taxa in total, as well as separate functional groups such as saprophytic fungal taxa were found in Oxalidosa stands compared to samples from Hylocomiosa type stands (p < 0.05) (Figure 5). Moreover, a lower number of fungal taxa in general were observed in the two long-term sample plots, including the functional groups of fungi, namely saprophytic and ectomycorrhizal fungi. Shannon’s diversity index did not differ significantly between stands and forest types.
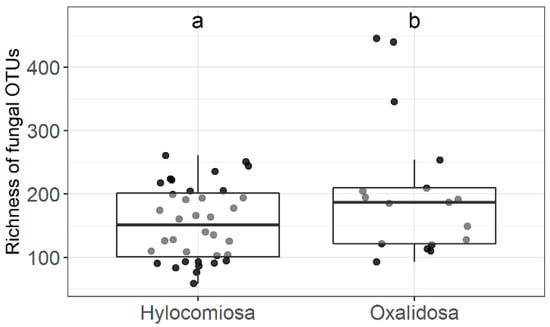
Figure 5.
Effect of forest type on richness of fungal OTUs. Middle lines, boxes, whiskers, and circles represent medians, quartiles, 90% quantiles, and all sampled trees, respectively. Different letters above whiskers indicate statistically different groups (p < 0.05). Number of stands: Hylocomiosa—36; Oxalidosa—17.
In our study, we also evaluated the association of the most common fungal taxa with stump removal or control specimens, thus determining stump removal indicator species. Lactarius tabidus was more frequently found in the stump removal sites, while saproxylic, and sometimes pathogenic fungi of the genus Armillaria sp. were more typical in the control sample plots (p < 0.05).
4. Discussion
Mycorrhizal fungi are an important group of soil microorganisms that form mycorrhizae with tree roots, thus promoting tree growth and vitality [47,48]. Our results showed that there are no significant differences in mycorrhizal and soil fungal communities between stump removal plots and clear-felled control plots seven years after stump removal. Mycorrhizal fungi were the most commonly sequenced fungal ecological group, and their diversity was similar to the clear-felled control site. However, there were some differences in species composition. One of the dominant species in this assessment was the mycorrhizal fungus Thelephora terrestris, which is consistent with the results of our previous study performed in similar sites after the first growing season following seedling planting [23]. The prevalence of this fungal species in the samples did not differ significantly between the stump removal and control sample plots. This basidiomycete is the most common species in tree nurseries worldwide [49]. However, despite its ability to adapt to nursery conditions [50], this species does not promote seedling growth and vitality after planting in forest sites [51,52]. The mycorrhizal fungus Lactarius tabidus was more frequently found in the stump removal sites, which could be related to the formation of more pronounced microrelief depressions as a result of stump extraction and, consequently, a higher humidity level, which has a positive impact on the development of this ectomycorrhizal fungus [53]. Fungi of the genus Armillaria were more common in control sample plots, which could be related to higher availability of wood, as stumps are left intact. Armillaria spp. spread through the soil using rhizomorphs, and stump removal was suggested as a control method to reduce the damage made by pathogenic Armillaria species [10]. The research results showed that stump removal has little impact on fungal diversity in soil seven years after seedling planting, however, more research is needed to evaluate the possible long-term impact of stump removal on mycorrhization of planted trees.
Author Contributions
Conceptualization, D.K., N.B. and T.G.; methodology, D.K. and K.P.; software, D.K.; formal analysis, K.P. and D.K.; writing, N.B.; supervision, T.G. and J.J. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by JSC “Latvia’s State Forests” project “Investigation of the factors limiting the spread of root rot” and project No. 5-5.9.1_007q_101_21_79.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article.
Acknowledgments
We are grateful to Rein Drenkhan, Leho Tedersoo and Dainis Ruņģis for the lab facilities used to prepare samples for sequencing, and Brigita Javoiša for assistance in field work.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Woodward, S.; Stenlid, J.; Karjalainen, R.A. Heterobasidion Annosum: Biology, Ecology, Impact and Control; CAB International: Wallingford, UK, 1998; 616p. [Google Scholar]
- Gonthier, P.; Thor, M. Annosus root and butt rots. In Infectious Forest Diseases; Gonthier, P., Nicolotti, G., Eds.; CAB International: Wallingford, UK, 2013; pp. 128–158. [Google Scholar]
- Stenlid, J.; Redfern, D.B. Spread within the tree and stand. In Heterobasidion Annosum: Biology, Ecology, Impact and Control; Woodward, S., Stenlid, J., Karjalainen, R., Hüttermann, A., Eds.; CAB International: Wallingford, UK, 1998; pp. 125–141. [Google Scholar]
- Piri, T. The spreading of the S type of Heterobasidion annosum from Norway spruce stumps to subsequent tree stands. Eur. J. For. Pathol. 1998, 26, 193–204. [Google Scholar] [CrossRef]
- Gaitnieks, T.; Bruna, L.; Zaluma, A.; Burnevica, N.; Klavina, D.; Legzdina, L.; Jansons, J.; Piri, T. Development of Heterobasidion spp. fruit bodies on decayed Picea abies. For. Ecol. Manag. 2021, 482, 118835. [Google Scholar] [CrossRef]
- Laivins, M. Geography of Norway spruce (Picea abies) stands in Latvia. Proc. Latv. Univ. Agric. 2005, 14, 1–9, (In Latvian with English Abstract). [Google Scholar]
- Arhipova, N.; Gaitnieks, T.; Donis, J.; Stenlid, J.; Vasaitis, R. Butt rot incidence, causal fungi, and related yield loss in Picea abies stands of Latvia. Can. J. For. Res. 2011, 41, 2337–2345. [Google Scholar] [CrossRef]
- Korhonen, K.; Stenlid, J. Biology of Heterobasidion annosum. In Heterobasidion Annosum: Biology, Ecology, Impact and Control; Woodward, S., Stenlid, J., Karjalainen, R., Hüttermann, A., Eds.; CAB International: Wallingford, UK, 1998; pp. 43–70. [Google Scholar]
- Vasaitis, R.; Stenlid, J.; Thomsen, I.M.; Berklund, P.; Dahlberg, A. Stump removal to control root rot in forest stands. A literature study. Silva Fenn. 2008, 42, 457–483. [Google Scholar] [CrossRef]
- Cleary, M.R.; Arhipova, N.; Morrison, D.J.; Thomsen, I.M.; Sturrock, R.N.; Vasaitis, R.; Gaitnieks, T.; Stenlid, J. Stump removal to control root disease in Canada and Scandinavia: A synthesis of results from long-term trials. For. Ecol. Manag. 2013, 290, 5–14. [Google Scholar] [CrossRef]
- Aosaar, J.; Drenkhan, T.; Adamson, K.; Aun, K.; Becker, H.; Buht, M.; Drenkhan, R.; Fjodorov, M.; Jürimaa, K.; Morozov, G.; et al. The effect of stump harvesting on tree growth and the infection of root rot in young Norway spruce stands in hemiboreal Estonia. For. Ecol. Manag. 2020, 475, 118425. [Google Scholar] [CrossRef]
- Hope, G.D. Changes in soil properties, tree growth, and nutrition over a period of a 10 years after stump removal and scarification on moderately coarse soils in interior British Columbia. For. Ecol. Manag. 2007, 242, 625–635. [Google Scholar] [CrossRef]
- Frąc, M.; Hannula, S.E.; Bełka, M.; Jędryczka, M. Fungal biodiversity and their role in soil health. Front. Microbiol. 2018, 9, 707. [Google Scholar] [CrossRef]
- Powell, J.R.; Rillig, M.C. Biodiversity of arbuscular mycorrhizal fungi and ecosystem function. New Phytol. 2018, 220, 1059–1075. [Google Scholar] [CrossRef]
- Bardgett, R.D.; Van der Putten, W.H. Belowground biodiversity and ecosystem functioning. Nature 2014, 515, 505–511. [Google Scholar] [CrossRef] [PubMed]
- Van der Putten, W.H.; Bardgett, R.D.; Bever, J.D.; Bezemer, T.M.; Casper, B.B.; Fukami, T.; Kardol, P.; Klironomus, J.N.; Kulmatiski, A.; Schweitzer, J.A.; et al. Plant-soil feedbacks: The past, the present and future challenges. J. Ecol. 2013, 101, 265–276. [Google Scholar] [CrossRef]
- Makiola, A.; Dickie, I.A.; Holdaway, R.J.; Wood, J.R.; Orwin, K.H.; Glare, T.R. Land use is a determinant of plant pathogen alpha-but not beta-diversity. Mol. Ecol. 2019, 28, 3786–3798. [Google Scholar] [CrossRef] [PubMed]
- Sterkenburg, E.; Clemmensen, K.E.; Lindahl, B.D.; Dahlberg, A. The significance of retention trees for survival of ectomycorrhizal fungi in clear-cut Scots pine forests. J. Appl. Ecol. 2019, 56, 1367–1378. [Google Scholar] [CrossRef]
- Page-Dumroese, D.S.; Harvey, A.E.; Jurgensen, M.F.; Amaranthus, M.P. Impacts of soil compaction and tree stump removal on soil properties and outplanted seedlings in northern Idaho, USA. Can. J. Soil Sci. 1998, 78, 29–34. [Google Scholar] [CrossRef]
- Menkis, A.; Uotila, A.; Arhipova, N.; Vasaitis, R. Effects of stump and slash removal on growth and mycorrhization of Picea abies seedlings outplanted on a forest clear-cut. Mycorrhiza 2010, 20, 505–509. [Google Scholar] [CrossRef]
- Kataja-aho, S.; Pennanen, T.; Lensu, A.; Haimi, J. Does stump removal affect early growth and mycorrhizal infection of spruce (Picea abies) seedlings in clear-cuts? Scand. J. For. Res. 2012, 27, 746–753. [Google Scholar] [CrossRef]
- Huusko, K.; Tarvainen, O.; Saravesi, K.; Pennanen, T.; Fritze, H.; Kubin, E.; Markkola, A. Short-term impacts of energy woods harvesting on ectomycorrhizal fungal communities of Norway spruce saplings. ISME J. 2015, 9, 581–591. [Google Scholar] [CrossRef]
- Kļaviņa, D.; Menkis, A.; Gaitnieks, T.; Pennanen, T.; Lazdiņš, A.; Velmala, S.; Vasaitis, R. Low impact of stump removal on mycorrhization and growth of replanted Picea abies: Data from three types of Hemiboreal forest. Balt. For. 2016, 22, 16. [Google Scholar]
- Modi, D.; Simard, S.; Bérubé, J.; Lavkulich, L.; Hamelin, R.; Grayston, S.J. Long-term effect of stump removal and tree species composition on the diversity and structure of soil fungal communities. FEMS Microbiol. Ecol. 2020, 96, fiaa061. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Anslan, S.; Bahram, M.; Wurzbacher, C.; Baldrian, P.; Tedersoo, L. Mycobiome diversity: High-throughput sequencing and identification of fungi. Nat. Rev. Microbiol. 2019, 17, 95–109. [Google Scholar] [CrossRef]
- Tedersoo, L.; Anslan, S.; Bahram, M.; Drenkhan, R.; Pritsch, K.; Buegger, F.; Padari, A.; Hagh-Doust, N.; Mikryukov, V.; Gohar, D.; et al. Regional-Scale In-Depth Analysis of Soil Fungal Diversity Reveals Strong pH and Plant Species Effects in Northern Europe. Front. Microbiol. 2020, 11, 1953. [Google Scholar] [CrossRef]
- Burņeviča, N.; Zaļuma, A.; Kļaviņa, D.; Brūna, L.; Legzdiņa, L.; Gaitnieks, T. Initial and long-term fungal diversity and occurrence of Heterobasidion spp. in Norway spruce root fragments remaining in soil after stump extraction. Scand. J. For. Res. 2021, 36, 117–125. [Google Scholar] [CrossRef]
- Bušs, K. Forest ecosystem classification in Latvia. Proc. Latv. Acad. Sci. 1997, 51, 204–218. [Google Scholar]
- Zimelis, A.; Lazdiņš, A.; Sarmulis, Z. Comparison of productivity of CBI and MCR—500 stump lifting buckets in Latvia. In Proceedings of the 19th International Scientific Conference “Research for Rural Development 2013”, Jelgava, Latvia, 15–17 May 2013; pp. 59–66. [Google Scholar]
- Klavina, D.; Tadersoo, L.; Agan, A.; Adamson, K.; Bitinieks, K.; Gaitnieks, T.; Drenkhan, R. Soil fungal communities in young Norway-spruce dominant stands: Footprints of former agricultural land use and selective thinning. Eur. J. For. Res. 2022, 141, 503–516. [Google Scholar] [CrossRef]
- Tedersoo, L.; Lindahl, B. Fungal identification biases in microbiome projects. Environ. Microbiol. Rep. 2016, 8, 774–779. [Google Scholar] [CrossRef] [PubMed]
- Tedersoo, L.; Anslan, S. Towards PacBio-based pan-eukaryote metabarcoding using full-length ITS sequences. Environ. Microbiol. Rep. 2019, 11, 659–668. [Google Scholar] [CrossRef]
- Anslan, S.; Bahram, M.; Hiiesalu, I.; Tedersoo, L. PipeCraft: Flexible open-source toolkit for bioinformatics analysis of custom high-throughput amplicon sequencing data. Mol. Ecol. Resour. 2017, 17, 234–240. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, J.B.; Clemente, C.J.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Bengtsson-Palme, J.; Ryberg, M.; Hartmann, M.; Branco, S.; Wang, Z.; Godhe, A.; Wit, P.D.; Sánchez-García, M.; Ebersberger, I.; de Sousa, F.; et al. Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol. Evol. 2013, 4, 914–919. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Larsson, K.-H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2018, 47, D259–D264. [Google Scholar] [CrossRef]
- Põlme, S.; Abarenkov, K.; Nilsson, R.H.; Lindahl, B.D.; Engelbrecht Clemmensen, K.; Kauserud, H.; Nguyen, N.; Kjøller, R.; Bates, S.T.; Baldrian, P.; et al. FungalTraits: A user-friendly traits database of fungi and fungus-like stramenopiles. Fungal Divers. 2020, 105, 1–16. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.R-project.org/ (accessed on 14 August 2022).
- Shannon, C.E. A mathematical theory of communication. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, 11. [Google Scholar] [CrossRef]
- De Caceres, M.; Legendre, P. Associations between species and groups of sites: Indices and statistical inference. Ecology 2009, 90, 3566–3574. [Google Scholar] [CrossRef]
- Bray, J.R.; Curtis, J.T. An ordination of the upland forest communities of Southern Wisconsin. Ecol. Monogr. 1957, 27, 325–349. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. R Package Version 2.5-7. 2020. Available online: https://CRAN.R-project.org/package=vegan (accessed on 14 August 2022).
- Ferlian, O.; Cesarz, S.; Craven, D.; Hines, J.; Barry, K.E.; Bruelheide, H.; Buscot, F.; Hayder, S.; Heklau, H.; Herrmann, S.; et al. Mycorrhiza in tree diversity–ecosystem function relationships: Conceptual framework and experimental implementation. Ecosphere 2018, 9, e02226. [Google Scholar] [CrossRef]
- Wagner, K.; Krause, K.; Gallegos-Monterrosa, R.; Sammer, D.; Kovacs, A.T.; Kothe, E. The Ectomycorrhizospheric Habitat of Norway Spruce and Tricholoma vaccinum: Promotion of Plant Growth and Fitness by a Rich Microorganismic Community. Front. Microbiol. 2019, 10, 307. [Google Scholar] [CrossRef] [PubMed]
- Marx, D.H.; Cordell, C.E.; Kenney, D.S.; Mexal, J.G.; Artman, J.D.; Riffle, J.W.; Molina, R. Commercial vegetative inoculum of Pisolithus tinctorius and inoculation techniques for development of ectomycorrhizae on bare root tree seedlings. For. Sci. Monogr. 1984, 25, 101. [Google Scholar]
- Perry, A.D.; Molina, R.; Amaranthus, P.M. Mycorrhizae, mycorrhizospheres, and reforestation: Current knowledge and research needs. Can. J. For. Res. 1987, 17, 929–940. [Google Scholar] [CrossRef]
- Ivory, M.; Munga, F. Growth and survival of container-grown Pinus caribaea infected with various ectomycorrhizal fungi. Plant Soil 1983, 71, 339–344. [Google Scholar] [CrossRef]
- Lee, K.J. A ten-year result of artificial inoculation of pines with ectomycorrhizal fungi, Pisolithus tinctorius and Thelephora terrestris. J. Korean For. Soc. 1992, 81, 156–163. [Google Scholar]
- Breitenbach, J.; Kränzlin, F. Fungi of Switzerland 6: Russulacea (Lactarius and Russula); Verlag Mycologia: Lucern, Switzerland, 2005; 320p. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).