Land Use, Microorganisms, and Soil Organic Carbon: Putting the Pieces Together
Abstract
:1. Introduction
- It is not easy to circumscribe the concept of biodiversity. It is closely linked to those of life and evolution: (1) to that of life, because we think that to live means “to complexify”, (a) in the sense of Lynn Margulis [1], to become more and more complex by symbiosis with other living beings; or (b) even to increase at ever larger scales, from cell to individual, from ecosystem to the entire planet or universe, in the spirit of Gaia [2]; (2) to that of evolution, on the scale of the individual in the population of a species, to conform to a changing niche [3]. In this context, the definition of biodiversity should be considered in parallel with those of complex organism or biotic community [4], climax [5], and ecosystem [6]. In such a confrontation of concepts, biodiversity acquires a finality, as it was a living and growing complex entity. Tansley accepted the concepts of climax and biome, but decisively rejected the similar ones of complex organism and biotic community. His judgment is accepted as a definitive choice by the scientific community [7,8] and is generally considered to be a fundamental law for modern ecology. However, it still divides at least the authors of this article, who are of different nationalities and representatives of a wide range of human activities, such as research, economic and ecological management, manufacturing, commerce, art and teaching, and some students in forest sciences. A fundamental fact that depends on this comparison of concepts has remained unresolved: on planet Earth, since its abiotic origin and up to the present day, despite well-known periods of crisis, biodiversity continued to increase [9,10]. Is the fact of growing, on average (and not linearly), a fundamental law, or a consequence linked to abiotic factors? Let us not forget that life on the planet started from a very inhospitable environment [11], adapting it to its needs [2]. The answer to this question of intrinsic growth determines how to sustain biodiversity in the coming years and in a changing climate. The way out seems to inevitably involve soil living storage power and management [12,13,14,15,16,17,18,19].
- Ample theoretical and empirical work has shown that interactions of human activities with the ecosystems are dynamic and complex [20,21]. The rapidly increasing human population and the increasing ecological footprint per capita are placing further pressure on the natural landscapes [22]. Despite a growing awareness of the risks at which humans and other species are, and will be exposed as a result of anthropogenic activities, pressure continues to grow, whether to meet the vital needs of a positive demography or for leisure activities such as tourism, whose environmental footprint is also growing [13]. Biodiversity on the Earth is indeed highly affected by anthropic activities fundamentally causing alterations to the environment [23]. Biodiversity is known as a critical determinant of ecosystem functioning, starting from the ground level, i.e., the soil itself [24]. Therefore, understanding biodiversity changes is an important issue that promotes better knowledge and finer capability to estimate how soil biodiversity changes might impact ecosystem sustainability [25]. However, previous studies of anthropic effects on ecosystems have overwhelmingly focused on macroscopic plants and animals [22], both on land and in water. On a local scale, the microbial communities are more influenced by soil properties and microclimate changes than plant communities [26,27]. At the microscale, the effects on soil biodiversity are far less known, despite the importance of soil organisms (bacteria, fungi, arthropods, invertebrates, etc.), which are the major regulators of essential ecosystem functions and services, such as plant productivity, nutrient cycling, organic matter decomposition, and pollutant degradation [28,29,30,31,32,33].One of the important human activities, tourism, accounting for about 8% of global greenhouse gas emissions [13], is continuing to increase. At more local scales, the touristic–recreational use of territories may endanger their environmental value and that of the surrounding areas. An increase in the popularity of outdoor leisure activities leads to an increase in the number of visitors in the same tourist area. This increase may significantly affect belowground ecosystems, especially microbial communities [34]. Lucas-Borja et al. (2011) studied the microbiological properties of soil and vegetation in Mediterranean mountains, and the result indicated that increased tourist activity significantly impacted soil microbial processes and vegetal communities, mainly due to soil compaction [35]. A study in Finland also showed that continuous human trampling causes significant changes in soil microbial functions, even with light stepping [36]. Understanding the extent of such anthropic pressure can provide insights informing future environment management, restoration, and monitoring.In recent years, methods for studying microbial communities have progressed rapidly, and DNA sequencing has allowed a more thorough understanding of the key members of soil microbial communities and biodiversity patterns [37,38]. Changes in soil physical and chemical properties have been reported to shape the composition of a microbial community, in terms of density, diversity, and activity [39,40,41,42,43,44]. Moreover, soil microbes have a fundamental role in soil responses to human disturbances since they are tightly dependent on the surrounding abiotic and biotic environment [45,46,47]. The differences in physiology and ecology of bacteria and fungi suggest that they would be controlled by different environmental factors [48]. Previous studies have shown that fungi may be more sensitive than bacteria to changes in vegetation [49], and shifts in carbon pools may have different effects on bacteria and fungi [50,51].The importance of understanding community assembly processes is broadly recognized in microbial ecology [52,53], and the assembly of microbial communities is known to be influenced by both deterministic and stochastic processes [54,55]. Deterministic processes refer to habitat filtering or biotic interactions such as mutualism, commensalism, and parasitism, while stochastic processes refer to random demographic changes in mortality and passive dispersal [53,56,57]. Wang et al. (2019) pointed out that the mechanisms of soil bacterial and fungal community assembly are different [58]. Thus, elucidating the differential dynamics and factors that affect microbial community structuring can help in estimating how anthropogenic environmental changes ultimately impact the different types of microbial communities, which represent a vast and still largely obscure component of planetary biodiversity.
- To work on SOC and biodiversity, we capitalized on the occurrence of a naturally preserved peninsular area, just adjacent to a symmetrically distributed strip of land (see aerial view in Figure 1) that had the same original soils and vegetation, but that had been, for the last forty years, fully transformed into a tourist resort. The availability of these two neighboring sites—one of which is now exploited by human recreational business, and the other preserved by conservation law—allowed us to investigate the effects of human-driven ecosystem manipulation, in comparison to a more natural landscape evolution. Of course, it is important to note that there is no longer anything “natural” (in the sense intended by Clements, 1936; or Tansley, 1935) on our planet today. It is more correct to assume that the whole environment is now, to varying degrees “human-influenced”. Every place on the planet can be situated on a spectrum ranging from the completely urbanized, to what remains of areas that could be defined as “moderately altered by human action”, to those very rare that still appear as “wilderness”. Even so-called wilderness areas are increasingly affected by global atmospheric climatic and pollution drifts, meaning that they are far from pristine [59]. Here at the two sites, one more-natural and one more-man-made, we studied 10 ecosystems along a gradient of increasing complexity from the poorest in species near the shore to the rich and wooded hinterland. Referring to a concept of soil as the place where each ecosystem starts its formation and evolution [60,61,62,63], we delved into the organic resources available to these living beings, trying to link the total organic carbon of the soil, as if it were a source of nourishment, to its biodiversity.
- Islands are, moreover, ecologically isolated landmasses, further qualifying as useful model systems to address ecological questions (Warren in [64]). Here, we aimed at addressing the following questions: (i) How does soil microbial diversity change under human pressure such as habitat fragmentation and/or recreational tourism exploitation? (ii) What are the impacts of different land management practices on soil biodiversity? (iii) Are assembly mechanisms of soil microbial communities different in natural and anthropogenic ecosystems? (iv) Can ecological diversity indices be suitable to answer these questions? Finally, (v) is it possible to better define the concept of biodiversity itself?
2. Materials and Methods
2.1. Site Description and Sampling
- -
- Soil microorganisms: 3 replicates a few meters away from each other, using a brass cylinder 10 cm long and 1.3 cm in diameter, at a 0–10 cm depth, discarding the litter layer when present. We collected a total of 10 (points) × 2 (sites) × 3 (replicates) = 60 replicates to be submitted to DNA extraction; these 60 replicates belong to 20 sampling points;
- -
- Soil organic carbon: 6 cylindrical soil cores, 12 inches (=30.48 cm) long and 1 inch (=2.54 cm) in diameter, at 0–30 cm in the soil, with litter layer when present, scattered in the same areas investigated for microorganisms. We gathered the 6 soil cores (replicates) collected in each point in a single bag, obtaining a total of 10 (points) × 2 (sites) = 20 samples of soil.
2.2. DNA Extraction, Sequencing, and Bioinformatics
2.3. Soil Chemical Analysis
2.4. Statistical Analysis
3. Results: Are the Soil Microorganisms Different along the Vegetation Series and between Anthropized and Natural Sites?
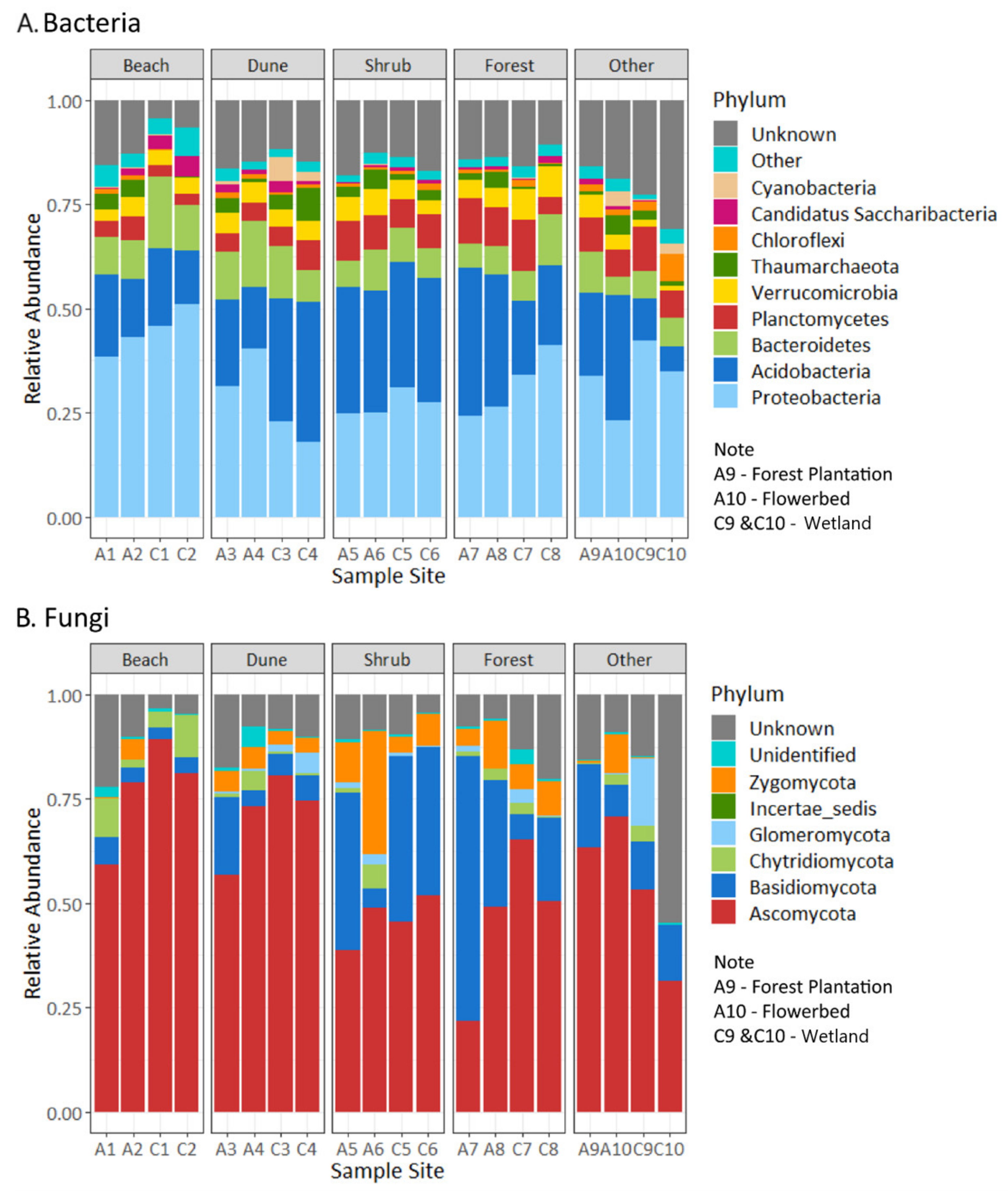
3.1. Soil Bacteria and Fungi
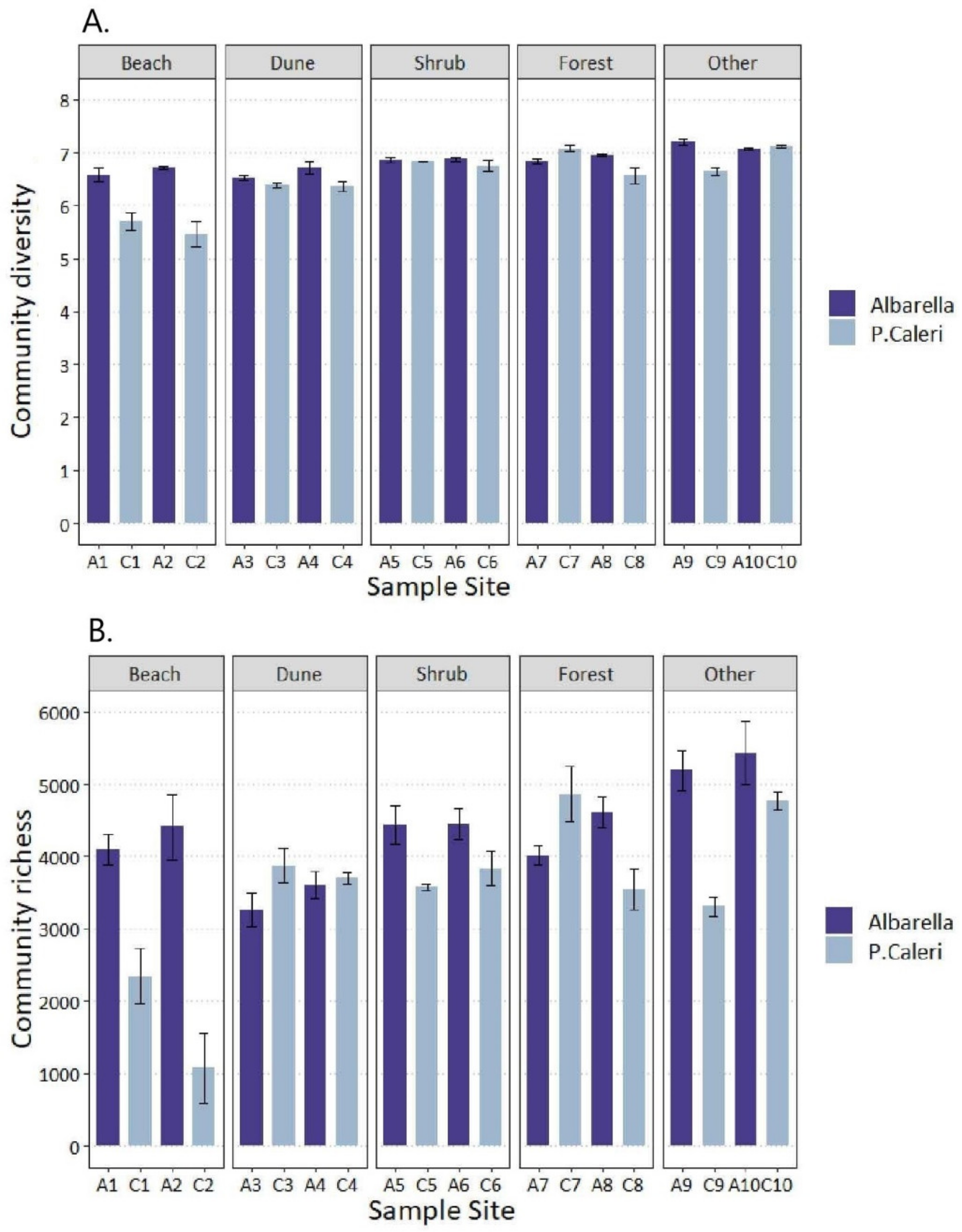
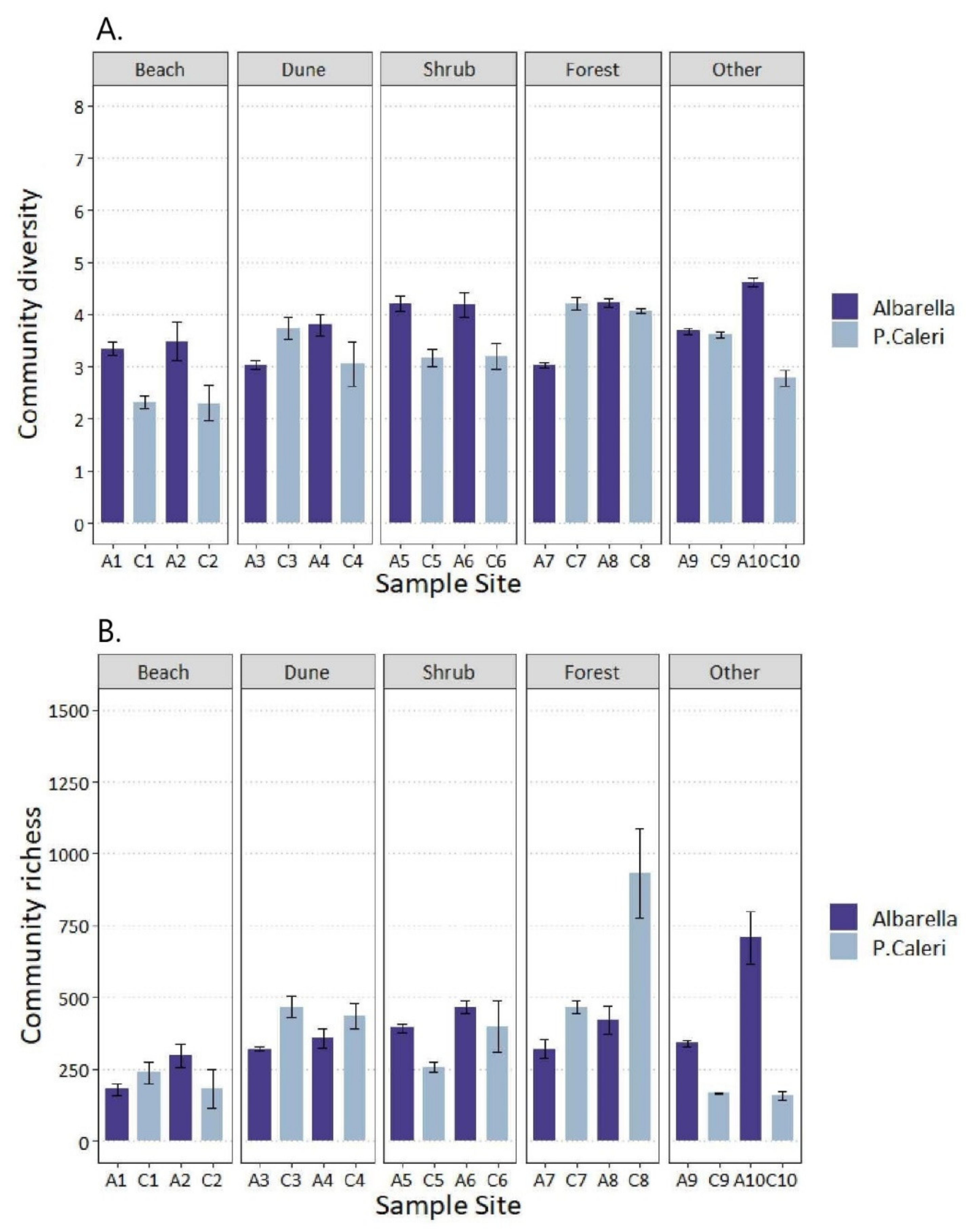
3.2. Soil Bacteria and Fungi Shannon’s and Chao1′s Diversities
- -
- The Shannon–Wiener index is the highest when individuals have the same frequency in each “species = sampled community” (high evenness); H’ is the lowest when all “species” but one is represented by a single individual, and one “species” cumulated all different individuals (low evenness).
- -
- The Chao1 index is high if the species represented by a single individual dominate in comparison to those represented by two individuals; the number of individuals (one or two) representing the species plays a great role (low evenness). The Chao1 index is low when the number of species represented by a single individual is similar to that of species represented by two individuals (high evenness).
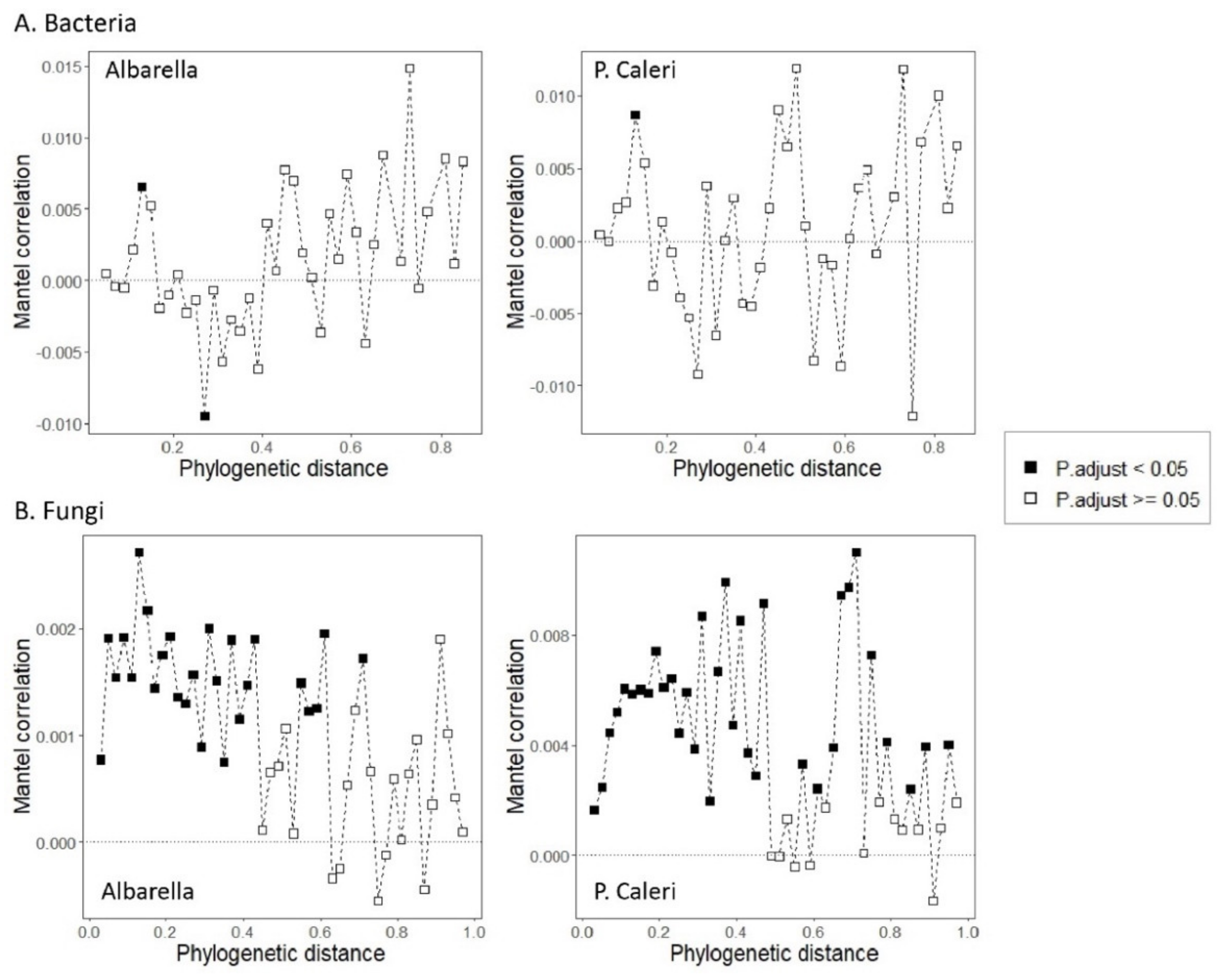
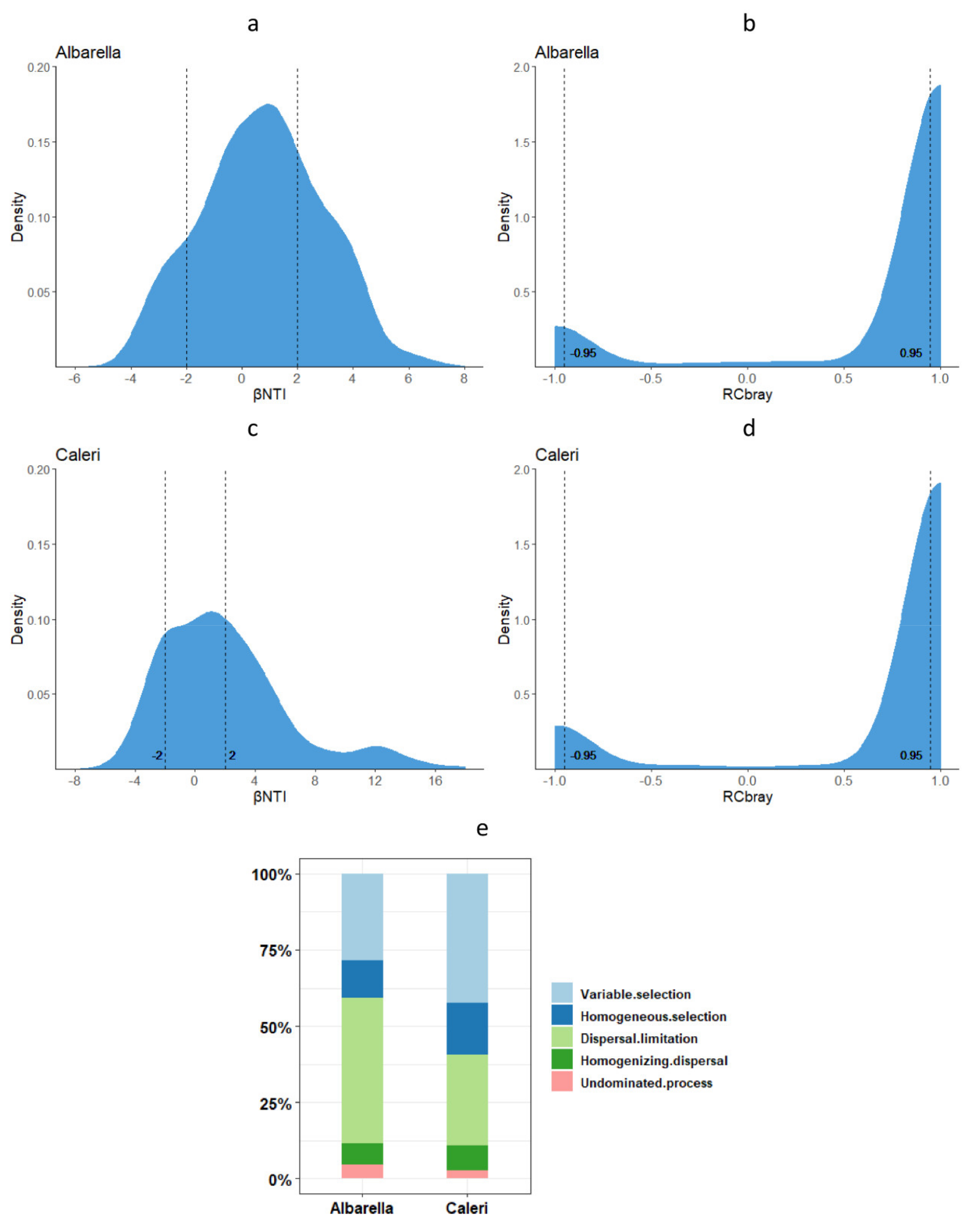
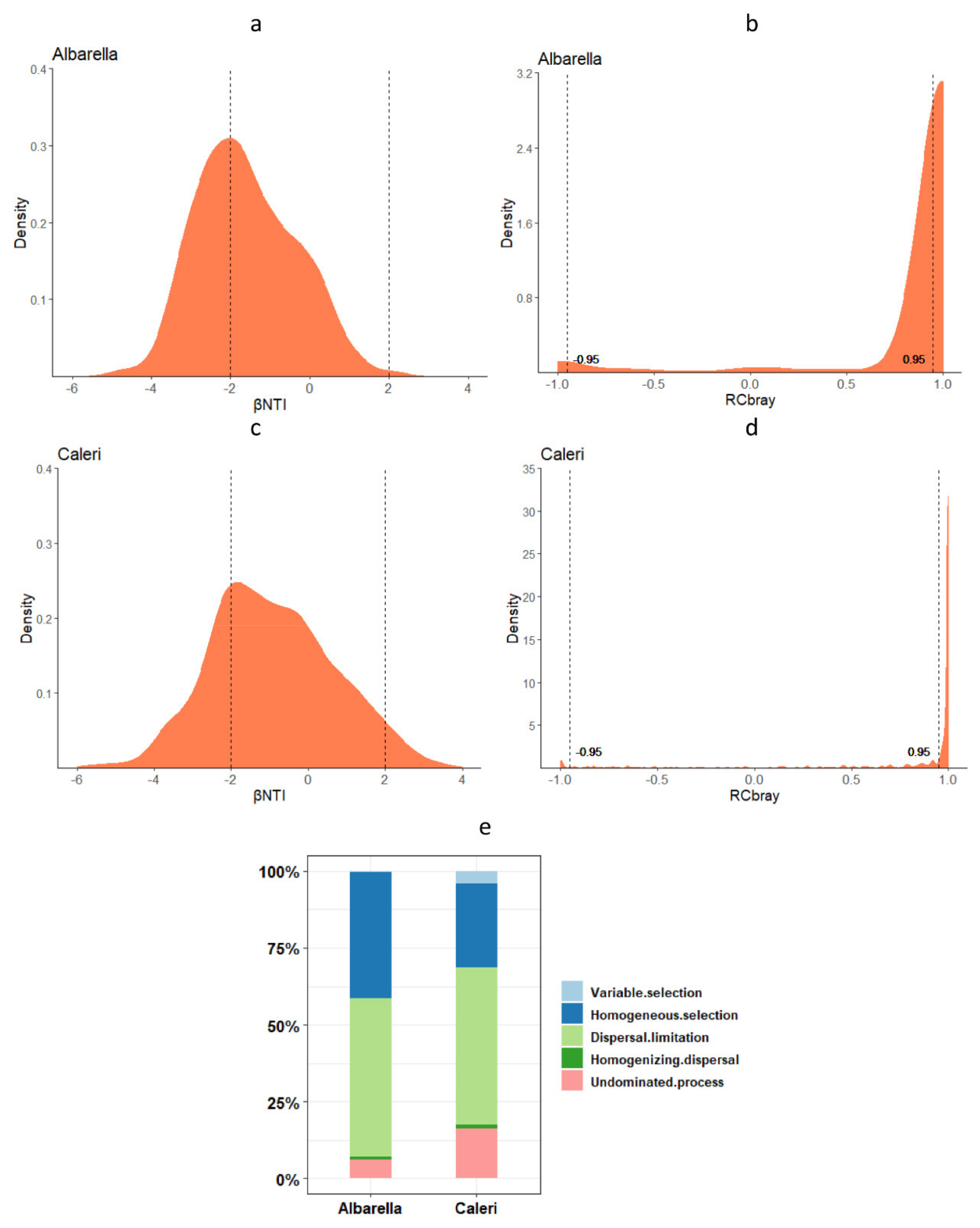
3.3. Ecological Processes Governing Bacterial and Fungal Community Assembly
- (1)
- Naturally assembled microorganisms living in nearby sampling points could have more affinity with each other and be better coordinated by necessity in the exploitation of resources;
- (2)
- That these natural assemblages of microorganisms were better adapted to live together than microorganisms grouped by chance following human intervention;
- (3)
- That the affinity of natural groupings had a strong probability of being the expression of an underlying phylogenesis.
3.4. Plastics
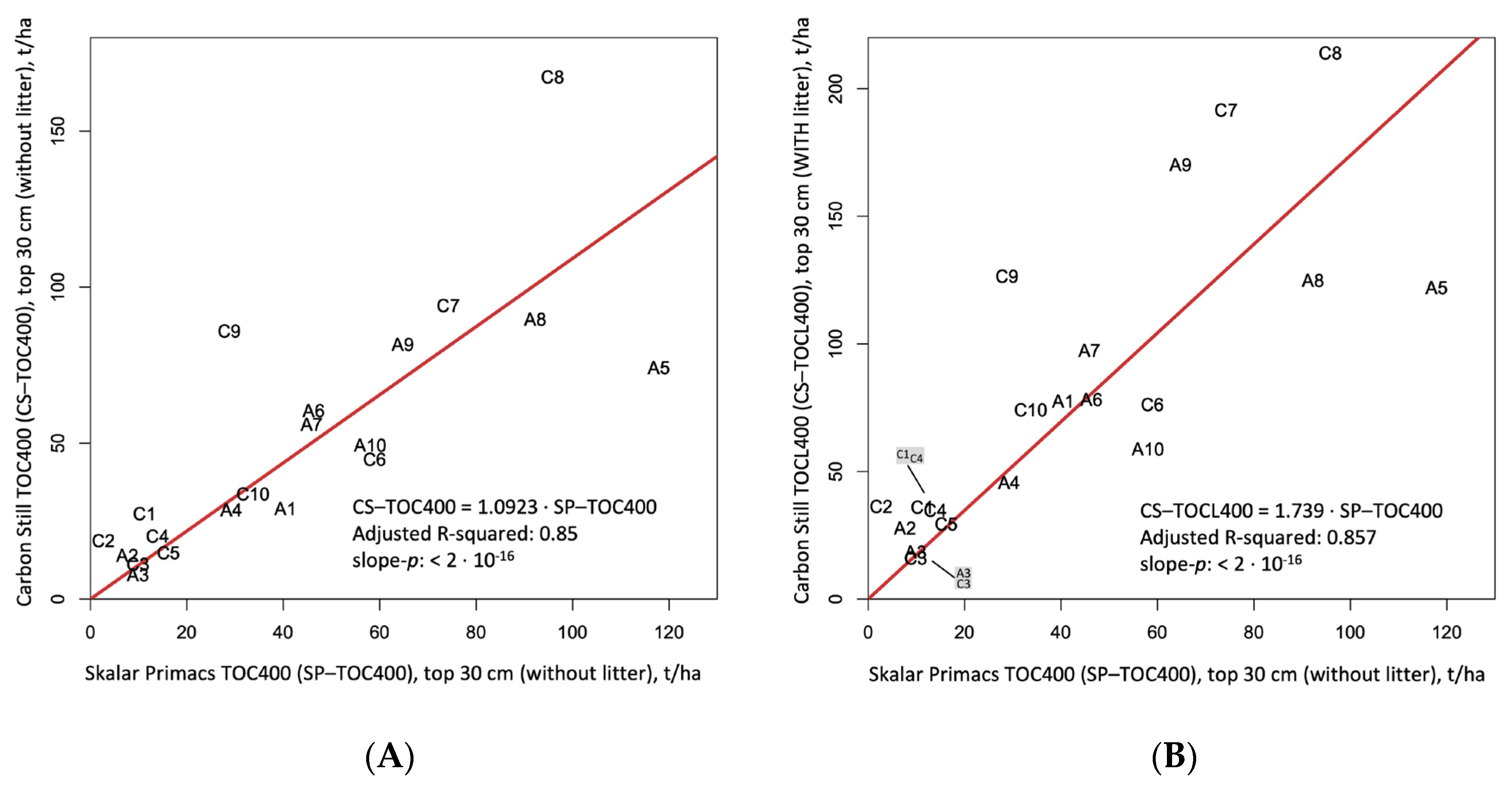
3.5. Carbon Still or Skalar Primacs Soil Organic Carbon Measurements?
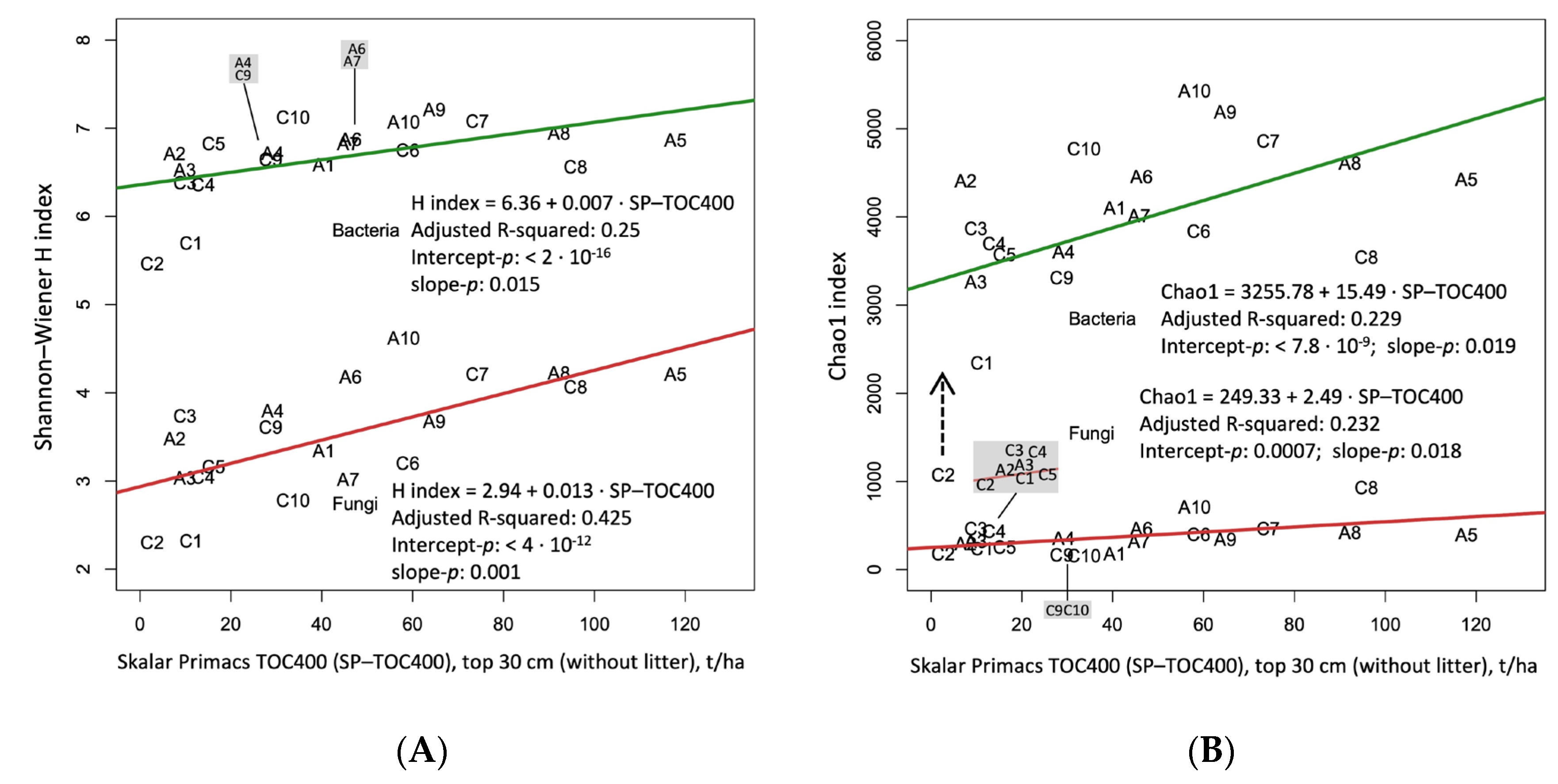
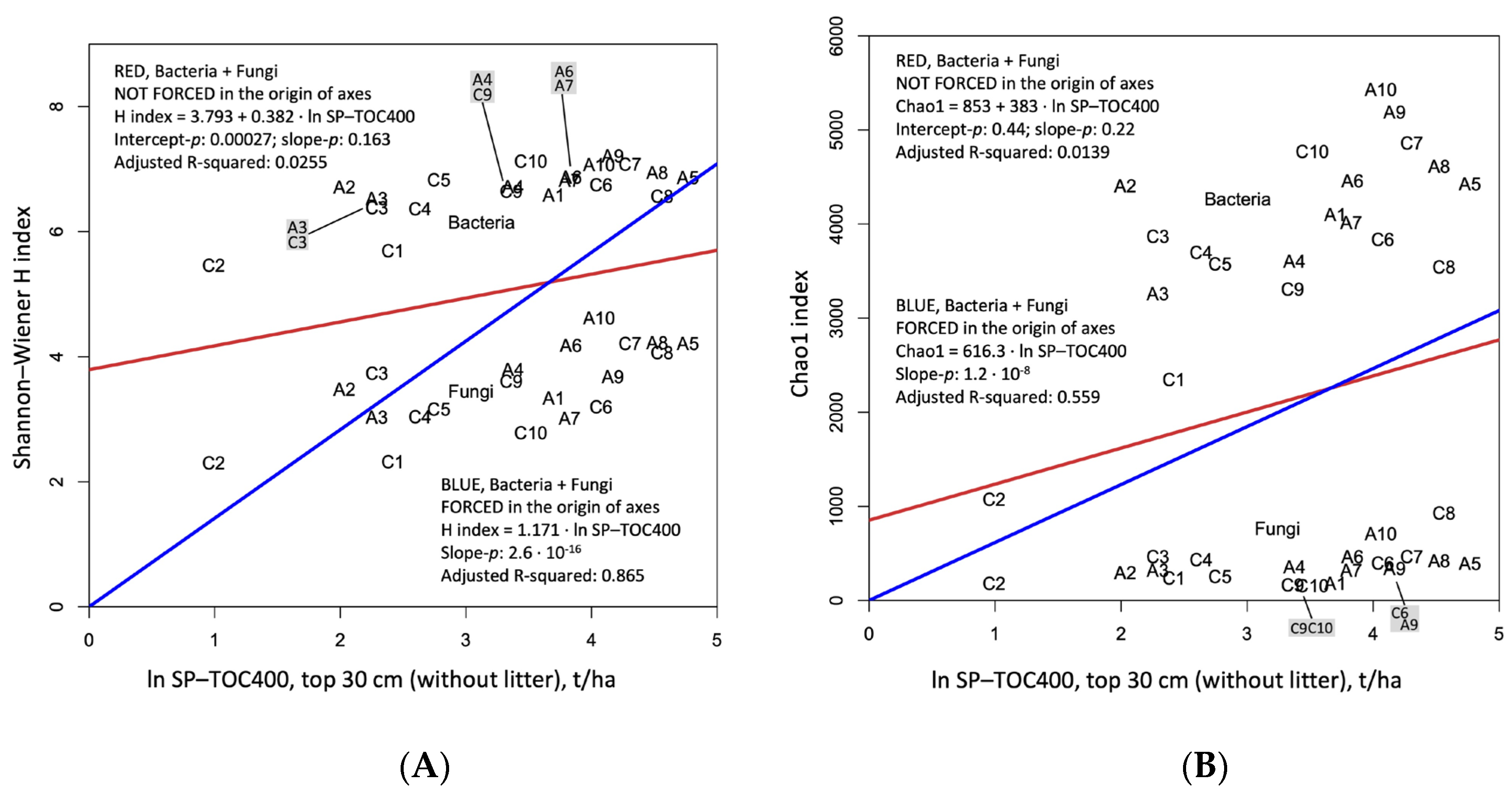
3.6. Biodiversity and Soil Organic Carbon Gradient 1
4. Discussion
4.1. Biodiversity and Soil Organic Carbon Gradient 2
- (1)
- Expressed by the Shannon and Chao1 indices, biodiversity grows from the left to the right of the graphs, and corresponds to an increasing organic carbon in the first 30 cm of soils;
- (2)
- Bacteria and fungi show the same trend, even if bacterial OTUs return indices almost double those of fungi;
- (3)
- The regression line that best summarizes the plotted points is only the one that is forced into the origin;
- (4)
- To improve the regression, the abscissa axis reports the organic carbon on a logarithmic (ln) scale, and value 1 corresponds to e = 2.718... (about 2.7 t/ha) and 5 to about 150 t/ha of SOC (Figure 10). Shannon–Wiener is a logarithmic (ln) index, so the straight line is a good compromise that allows one to understand the growth of biodiversity with the soil carbon content expressed in a logarithmic way. We can say that Shannon–Wiener = 1.42 • ln (TOC400).
- (5)
- The Chao1 scale is not logarithmic, but the new regression line explains more than 50% of the variance. There is a big difference between fungi and bacteria for this index that gives a lot of importance to the OTUs which are represented by one or two fragments of identification genetic code:
4.2. Soil Biodiversity and Anthropic Soil Use
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Resource Identification Initiative
References
- Margulis, L. Symbiotic Planet: A New Look at Evolution; Science Ma; Perseus Books Group: New York, NY, USA, 1998. [Google Scholar]
- Lovelock, J.E.; Margulis, L. Atmospheric homeostasis by and for the biosphere: The gaia hypothesis. Tellus 1974, 26, 2–10. [Google Scholar] [CrossRef]
- Darwin, C.M.A. On the Origin of Species by Means of Natural Selection or the Preservation of Favoured Races in the Struggle for Life; By Charles Darwin, M.A., fellow of the Royal, Geological, Linnean, etc., Societies; Author of ’Journal of researches during H. M. S. Bea; John Murray: London, UK, 1859. [Google Scholar]
- Phillips, J. The Biotic Community. J. Ecol. 1931, 19, 1–24. [Google Scholar] [CrossRef]
- Clements, F.E. Nature and Structure of the Climax. J. Ecol. 1936, 24, 252. [Google Scholar] [CrossRef]
- Tansley, A.G. The Use and Abuse of Vegetational Concepts and Terms. Ecology 1935, 16, 284–307. Available online: https://www.jstor.org/stable/1930070 (accessed on 8 August 2022). [CrossRef]
- Van der Valk, A.G. From Formation to Ecosystem: Tansley’s Response to Clements’ Climax. J. Hist. Biol. 2013, 47, 293–321. [Google Scholar] [CrossRef]
- Richter, D.D.; Billings, S.A. ‘One physical system’: Tansley’s ecosystem as Earth’s critical zone. New Phytol. 2015, 206, 900–912. [Google Scholar] [CrossRef]
- Benton, M.J. Origins of Biodiversity. PLoS Biol. 2016, 14, e2000724. [Google Scholar] [CrossRef]
- Raup, D.M. Taxonomic Diversity during the Phanerozoic. Science 1972, 177, 1065–1071. [Google Scholar] [CrossRef]
- Miller, S.L.; Urey, H.C. Organic Compound Synthes on the Primitive Eart. Science 1959, 130, 245–251. [Google Scholar] [CrossRef]
- Banerjee, S.; Walder, F.; Büchi, L.; Meyer, M.; Held, A.Y.; Gattinger, A.; Keller, T.; Charles, R.; van der Heijden, M.G.A. Agricultural intensification reduces microbial network complexity and the abundance of keystone taxa in roots. ISME J. 2019, 13, 1722–1736. [Google Scholar] [CrossRef]
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Vergès, M.-C.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H.; et al. Microbiome definition re-visited: Old concepts and new challenges. Microbiome 2020, 8, 103. [Google Scholar] [CrossRef] [PubMed]
- Narango, D.L.; Tallamy, D.W.; Shropshire, K.J. Few keystone plant genera support the majority of Lepidoptera species. Nat. Commun. 2020, 11, 5751. [Google Scholar] [CrossRef] [PubMed]
- Zanella, A.; Bolzonella, C.; Lowenfels, J.; Ponge, J.-F.; Bouché, M.; Saha, D.; Kukal, S.S.; Fritz, I.; Savory, A.; Blouin, M.; et al. Humusica 2, article 19: Techno humus systems and global change–conservation agriculture and 4/1000 proposal. Appl. Soil Ecol. 2018, 122, 271–296. [Google Scholar] [CrossRef]
- Zanella, A. Humans, humus, and universe. Appl. Soil Ecol. 2018, 123, 561–567. [Google Scholar] [CrossRef]
- Soto-Navarro, C.; Ravilious, C.; Arnell, A.; de Lamo, X.; Harfoot, M.; Hill, S.L.L.; Wearn, O.R.; Santoro, M.; Bouvet, A.; Mermoz, S.; et al. Mapping co-benefits for carbon storage and biodiversity to inform conservation policy and action. Philos. Trans. R. Soc. B Biol. Sci. 2020, 375, 20190128. [Google Scholar] [CrossRef]
- Edlinger, A.; Saghaï, A.; Herzog, C.; Degrune, F.; Garland, G. Towards a multidimensional view of biodiversity and ecosystem functioning in a changing world. New Phytol. 2020, 228, 820–822. [Google Scholar] [CrossRef] [PubMed]
- Phillips, H.R.P.; Guerra, C.A.; Bartz, M.L.C.; Briones, M.J.I.; Brown, G.; Crowther, T.W.; Ferlian, O.; Gongalsky, K.B.; Hoogen, J.V.D.; Krebs, J.; et al. Global distribution of earthworm diversity. Science 2019, 366, 480–485. [Google Scholar] [CrossRef]
- DeFries, R.S.; Foley, J.A.; Asner, G.P. Land-Use Choices: Balancing Human Needs and Ecosystem Function. Front. Ecol. Environ. 2004, 2, 249. [Google Scholar] [CrossRef]
- Rindfuss, R.R.; Walsh, S.J.; Turner, B.L.; Fox, J.; Mishra, V. Developing a science of land change: Challenges and methodological issues. Proc. Natl. Acad. Sci. USA 2004, 101, 13976–13981. [Google Scholar] [CrossRef]
- Geisen, S.; Wall, D.H.; van der Putten, W.H. Challenges and Opportunities for Soil Biodiversity in the Anthropocene. Curr. Biol. 2019, 29, R1036–R1044. [Google Scholar] [CrossRef]
- Lenzen, M.; Sun, Y.-Y.; Faturay, F.; Ting, Y.-P.; Geschke, A.; Malik, A. The carbon footprint of global tourism. Nat. Clim. Chang. 2018, 8, 522–528. [Google Scholar] [CrossRef]
- Ellis, E.C.; Ramankutty, N. Putting people in the map: Anthropogenic biomes of the world. Front. Ecol. Environ. 2008, 6, 439–447. [Google Scholar] [CrossRef]
- Cardinale, B.J. Biodiversity improves water quality through niche partitioning. Nature 2011, 472, 86–89. [Google Scholar] [CrossRef] [PubMed]
- Maynard, D.S.; Crowther, T.W.; Bradford, M.A. Competitive network determines the direction of the diversity–function relationship. Proc. Natl. Acad. Sci. USA 2017, 114, 11464–11469. [Google Scholar] [CrossRef] [PubMed]
- Dreesens, L.L.; Lee, C.K.; Cary, S.C. The Distribution and Identity of Edaphic Fungi in the McMurdo Dry Valleys. Biology 2014, 3, 466–483. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, I.R.; de Mello, C.M.A.; Neto, R.A.F.; da Silva, D.K.A.; de Melo, A.L.; Oehl, F.; Maia, L.C. Diversity of arbuscular mycorrhizal fungi along an environmental gradient in the Brazilian semiarid. Appl. Soil Ecol. 2014, 84, 166–175. [Google Scholar] [CrossRef]
- Dirzo, R.; Young, H.S.; Galetti, M.; Ceballos, G.; Isaac, N.J.B.; Collen, B. Defaunation in the Anthropocene. Science 2014, 345, 401–406. [Google Scholar] [CrossRef]
- Young, H.S.; McCauley, D.J.; Galetti, M.; Dirzo, R. Patterns, Causes, and Consequences of Anthropocene Defaunation. Annu. Rev. Ecol. Evol. Syst. 2016, 47, 333–358. [Google Scholar] [CrossRef]
- Bardgett, R. The Biology of Soil: A Community and Ecosystem Approach; Oxford University Press: Oxford, UK, 2005. [Google Scholar]
- Zhang, T.; Chen, H.Y.H.; Ruan, H. Global negative effects of nitrogen deposition on soil microbes. ISME J. 2018, 12, 1817–1825. [Google Scholar] [CrossRef]
- Bar-On, Y.M.; Phillips, R.; Milo, R. The biomass distribution on Earth. Proc. Natl. Acad. Sci. USA 2018, 115, 6506–6511. [Google Scholar] [CrossRef]
- Bastida, F.; García, C.; Fierer, N.; Eldridge, D.J.; Bowker, M.A.; Abades, S.; Alfaro, F.D.; Berhe, A.A.; Cutler, N.A.; Gallardo, A.; et al. Global ecological predictors of the soil priming effect. Nat. Commun. 2019, 10, 3481. [Google Scholar] [CrossRef] [PubMed]
- Levine, J.M.; Adler, P.B.; Yelenik, S. A meta-analysis of biotic resistance to exotic plant invasions. Ecol. Lett. 2004, 7, 975–989. [Google Scholar] [CrossRef]
- Lucas-Borja, M.E.; Bastida, F.; Moreno-Ortego, J.L.; Nicolás, C.; Andres, M.; López-Serrano, F.R.; Del Cerro, A. The effects of human trampling on the microbiological properties of soil and vegetation in mediterranean mountain areas. Land Degrad. Dev. 2010, 22, 383–394. [Google Scholar] [CrossRef]
- Sherman, C.; Unc, A.; Doniger, T.; Ehrlich, R.; Steinberger, Y. The effect of human trampling activity on a soil microbial community at the Oulanka Natural Reserve, Finland. Appl. Soil Ecol. 2018, 135, 104–112. [Google Scholar] [CrossRef]
- Chodak, M.; Gołębiewski, M.; Morawska-Płoskonka, J.; Kuduk, K.; Niklińska, M. Diversity of microorganisms from forest soils differently polluted with heavy metals. Appl. Soil Ecol. 2012, 64, 7–14. [Google Scholar] [CrossRef]
- Cristescu, M.E. From barcoding single individuals to metabarcoding biological communities: Towards an integrative approach to the study of global biodiversity. Trends Ecol. Evol. 2014, 29, 566–571. [Google Scholar] [CrossRef] [PubMed]
- Shentu, J.-L.; He, Z.-L.; Zeng, Y.-Y.; He, S.-Y.; Du, S.-T.; Shen, D.-S. Microbial Biomass and PLFA Profile Changes in Rhizosphere of Pakchoi (Brassica chinensis L.) as Affected by External Cadmium Loading. Pedosphere 2014, 24, 553–562. [Google Scholar] [CrossRef]
- Deslippe, J.R.; Hartmann, M.; Simard, S.W.; Mohn, W.W. Long-term warming alters the composition of Arctic soil microbial communities. FEMS Microbiol. Ecol. 2012, 82, 303–315. [Google Scholar] [CrossRef]
- Karhu, K.; Auffret, M.; Dungait, J.; Hopkins, D.; Prosser, J.; Singh, B.; Subke, J.-A.; Wookey, P.; Ågren, G.I.; Sebastià, M.T.; et al. Temperature sensitivity of soil respiration rates enhanced by microbial community response. Nature 2014, 513, 81–84. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, G.; Xue, S.; Wang, G. Soil bacterial community dynamics reflect changes in plant community and soil properties during the secondary succession of abandoned farmland in the Loess Plateau. Soil Biol. Biochem. 2016, 97, 40–49. [Google Scholar] [CrossRef]
- Zhang, X.; Johnston, E.; Liu, W.; Li, L.; Han, X. Environmental changes affect the assembly of soil bacterial community primarily by mediating stochastic processes. Glob. Chang. Biol. 2015, 22, 198–207. [Google Scholar] [CrossRef] [PubMed]
- De Vries, F.; Liiri, M.E.; Bjørnlund, L.; Bowker, M.A.; Christensen, S.; Setälä, H.M.; Bardgett, R.D. Land use alters the resistance and resilience of soil food webs to drought. Nat. Clim. Chang. 2012, 2, 276–280. [Google Scholar] [CrossRef]
- Griffiths, B.S.; Philippot, L. Insights into the resistance and resilience of the soil microbial community. FEMS Microbiol. Rev. 2013, 37, 112–129. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Liang, Y.; Jiang, Y.; Yang, Y.; Xue, K.; Xiong, J.; Zhou, J.; Sun, B. Planting increases the abundance and structure complexity of soil core functional genes relevant to carbon and nitrogen cycling. Sci. Rep. 2015, 5, 14345. [Google Scholar] [CrossRef] [PubMed]
- Van der Wal, A.; van Veen, J.A.; Smant, W.; Boschker, H.T.; Bloem, J.; Kardol, P.; van der Putten, W.H.; de Boer, W. Fungal biomass development in a chronosequence of land abandonment. Soil Biol. Biochem. 2006, 38, 51–60. [Google Scholar] [CrossRef]
- Heinemeyer, A.; Ridgway, K.P.; Edwards, E.J.; Benham, D.G.; Young, J.P.W.; Fitter, A.H. Impact of soil warming and shading on colonization and community structure of arbuscular mycorrhizal fungi in roots of a native grassland community. Glob. Chang. Biol. 2003, 10, 52–64. [Google Scholar] [CrossRef]
- Bossuyt, H.; Denef, K.; Six, J.; Frey, S.; Merckx, R.; Paustian, K. Influence of microbial populations and residue quality on aggregate stability. Appl. Soil Ecol. 2001, 16, 195–208. [Google Scholar] [CrossRef]
- Six, J.; Frey, S.D.; Thiet, R.K.; Batten, K.M. Bacterial and Fungal Contributions to Carbon Sequestration in Agroecosystems. Soil Sci. Soc. Am. J. 2006, 70, 555–569. [Google Scholar] [CrossRef]
- Dumbrell, A.J.; Nelson, M.; Helgason, T.; Dytham, C.; Fitter, A.H. Relative roles of niche and neutral processes in structuring a soil microbial community. ISME J. 2009, 4, 337–345. [Google Scholar] [CrossRef]
- Nemergut, D.R.; Schmidt, S.K.; Fukami, T.; O’Neill, S.P.; Bilinski, T.M.; Stanish, L.F.; Knelman, J.E.; Darcy, J.L.; Lynch, R.C.; Wickey, P.; et al. Patterns and Processes of Microbial Community Assembly. Microbiol. Mol. Biol. Rev. 2013, 77, 342–356. [Google Scholar] [CrossRef]
- Caruso, T.; Chan, Y.; Lacap, D.C.; Lau, M.C.Y.; McKay, C.P.; Pointing, S.B. Stochastic and deterministic processes interact in the assembly of desert microbial communities on a global scale. ISME J. 2011, 5, 1406–1413. [Google Scholar] [CrossRef] [PubMed]
- Stomeo, F.; Valverde, A.; Pointing, S.B.; McKay, C.P.; Warren-Rhodes, K.A.; Tuffin, M.I.; Seely, M.; Cowan, D.A. Hypolithic and soil microbial community assembly along an aridity gradient in the Namib Desert. Extremophiles 2013, 17, 329–337. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Ning, D. Stochastic Community Assembly: Does It Matter in Microbial Ecology? Microbiol. Mol. Biol. Rev. 2017, 81, e00002-17. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Li, S.; Yang, X.; Zhou, J.; Shu, W.; Jiang, L. Mechanisms of soil bacterial and fungal community assembly differ among and within islands. Environ. Microbiol. 2019, 22, 1559–1571. [Google Scholar] [CrossRef]
- Wang, Q.; Ma, M.; Jiang, X.; Zhou, B.; Guan, D.; Cao, F.; Chen, S.; Li, J. Long-term N fertilization altered 13C-labeled fungal community composition but not diversity in wheat rhizosphere of Chinese black soil. Soil Biol. Biochem. 2019, 135, 117–126. [Google Scholar] [CrossRef]
- Coleman, D.C.; Crossley, D.; Hendrix, P.F. Future Developments in Soil Ecology. In Fundamentals of Soil Ecology; Elsevier: Amsterdam, The Netherlands, 2004; pp. 271–298. [Google Scholar] [CrossRef]
- Zanella, A.; Ponge, J.-F.; Fritz, I.; Pietrasiak, N.; Matteodo, M.; Nadporozhskaya, M.; Juilleret, J.; Tatti, D.; Le Bayon, C.; Rothschild, L.; et al. Humusica 2, article 13: Para humus systems and forms. Appl. Soil Ecol. 2018, 122, 181–199. [Google Scholar] [CrossRef]
- Miller, M.; Dick, R.P. Dynamics of soil C and microbial biomass in whole soil and aggregates in two cropping systems. Appl. Soil Ecol. 1995, 2, 253–261. [Google Scholar] [CrossRef]
- St. Martin, C.C.G.; Rouse-Miller, J.; Barry, G.T.; Vilpigue, P. Compost and Compost Tea Microbiology: The ‘-Omics’ Era; Springer: Cham, Switzerland, 2020; pp. 3–30. [Google Scholar]
- Jenny, H. Factors of Soil Formation: A System of Quantitative Pedology; Dover Publications Inc.: New York, NY, USA, 1941. [Google Scholar]
- Allmon, W.D. Species, lineages, splitting, and divergence: Why we still need ‘anagenesis’ and ‘cladogenesis’. Biol. J. Linn. Soc. 2016, 120, 474–479. [Google Scholar] [CrossRef]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes—Application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols; Elsevier: Amsterdam, The Netherlands, 1990; pp. 315–322. [Google Scholar]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108 (Supp. S1), 4516–4522. [Google Scholar] [CrossRef]
- Apprill, A.; McNally, S.; Parsons, R.; Weber, L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 2015, 75, 129–137. [Google Scholar] [CrossRef]
- Parada, A.E.; Needham, D.M.; Fuhrman, J.A. Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 2016, 18, 1403–1414. [Google Scholar] [CrossRef] [PubMed]
- Coller, E.; Cestaro, A.; Zanzotti, R.; Bertoldi, D.; Pindo, M.; Larger, S.; Albanese, D.; Mescalchin, E.; Donati, C. Microbiome of vineyard soils is shaped by geography and management. Microbiome 2019, 7, 140. [Google Scholar] [CrossRef]
- Albanese, D.; Fontana, P.; De Filippo, C.; Cavalieri, D.; Donati, C. MICCA: A complete and accurate software for taxonomic profiling of metagenomic data. Sci. Rep. 2015, 5, 9743. [Google Scholar] [CrossRef] [PubMed]
- Quéméré, E.; Hibert, F.; Miquel, C.; Lhuillier, E.; Rasolondraibe, E.; Champeau, J.; Rabarivola, C.; Nusbaumer, L.; Chatelain, C.; Gautier, L.; et al. A DNA Metabarcoding Study of a Primate Dietary Diversity and Plasticity across Its Entire Fragmented Range. PLoS ONE 2013, 8, e58971. [Google Scholar] [CrossRef]
- IUSS Working Group WRB; FAO. World Reference Base for Soil Resources 2014 INTERNATIONAL Soil Classification System; Food and Agriculture Organization of the United Nations: Rome, Italy, 2015. [Google Scholar]
- Malms, S.; Krahnstöver, T.; Linnemann, V.; Wintgens, T.; Montag, D.; Pinnekamp, J.; Benstoem, F. Project BePAK—Evaluation of Methods for Quantifying Activated Carbon in Effluents of Wastewater Treatment Plants. In Proceedings of the 10th IWA Micropol & Ecohazard Conference 2017, Vienna, Austria, 17–20 September 2017; Available online: https://www.researchgate.net/publication/319955284_Evaluation_of_Methods_for_Quantifying_Activated_Carbon_in_Effluents_of_Wastewater_Treatment_Plants (accessed on 8 August 2022).
- Yeomans, A.J. Priority One: Together We Can Beat Global Warming; Keyline Pub. Co.: Arundel, Qld, Australia, 2005; 492p. [Google Scholar]
- Meixner, K.; Kubiczek, M.; Fritz, I. Microplastic in soil–current status in Europe with special focus on method tests with Austrian samples. AIMS Environ. Sci. 2020, 7, 174–191. [Google Scholar] [CrossRef]
- Stegen, J.C.; Lin, X.; Fredrickson, J.K.; Konopka, A.E. Estimating and mapping ecological processes influencing microbial community assembly. Front. Microbiol. 2015, 6, 370. [Google Scholar] [CrossRef]
- Stegen, J.C.; Lin, X.; Fredrickson, J.K.; Chen, X.; Kennedy, D.W.; Murray, C.J.; Rockhold, M.L.; Konopka, A. Quantifying community assembly processes and identifying features that impose them. ISME J. 2013, 7, 2069–2079. [Google Scholar] [CrossRef]
- Wang, B.; Brewer, P.E.; Shugart, H.H.; Lerdau, M.T.; Allison, S.D. Soil aggregates as biogeochemical reactors and implications for soil–atmosphere exchange of greenhouse gases—A concept. Glob. Chang. Biol. 2018, 25, 373–385. [Google Scholar] [CrossRef]
- Feng, Y.; Chen, R.; Stegen, J.C.; Guo, Z.; Zhang, J.; Li, Z.; Lin, X.; Feng, Y.; Chen, R.; Stegen, J.C.; et al. Two key features influencing community assembly processes at regional scale: Initial state and degree of change in environmental conditions. Mol. Ecol. 2018, 27, 5238–5251. [Google Scholar] [CrossRef]
- Clark, M.A.; Moran, N.A.; Baumann, P. Sequence evolution in bacterial endosymbionts having extreme base compositions. Mol. Biol. Evol. 1999, 16, 1586–1598. [Google Scholar] [CrossRef] [PubMed]
- Lovelock, J.E. Hands up for the Gaia hypothesis. Nature 1990, 344, 100–102. [Google Scholar] [CrossRef]
- Casarotto, D. Quantità di carbonio organico nei primi 30 cm di suolo dei principali ecosistemi naturali del giardino botanico litoraneo di Porto Caleri (VE). Prova finale. Corso di Laurea triennale in Tecnologie Forestali e Ambientali; Padova, Italy. Unpublished work. 2021. [Google Scholar]
- Longo, M. Quantità di carbonio organico nei primi 30 cm di suolo dei principali ecosistemi di Albarella (Rovigo, Italia). Prova finale. Corso di Laurea triennale in Tecnologie Forestali e Ambientali. Padova, Italy. Unpublished work. 2021. [Google Scholar]
- Zanella, A.; Ponge, J.-F.; Jabiol, B.; Sartori, G.; Kolb, E.; Le Bayon, R.-C.; Gobat, J.-M.; Aubert, M.; De Waal, R.; Van Delft, B.; et al. Humusica 1, article 5: Terrestrial humus systems and forms—Keys of classification of humus systems and forms. Appl. Soil Ecol. 2018, 122, 75–86. [Google Scholar] [CrossRef]
- Prescott, C.; Grayston, S. Tree species influence on microbial communities in litter and soil: Current knowledge and research needs. For. Ecol. Manag. 2013, 309, 19–27. [Google Scholar] [CrossRef]
- Urbanová, M.; Šnajdr, J.; Baldrian, P. Composition of fungal and bacterial communities in forest litter and soil is largely determined by dominant trees. Soil Biol. Biochem. 2015, 84, 53–64. [Google Scholar] [CrossRef]
- Hu, P.-L.; Liu, S.-J.; Ye, Y.-Y.; Zhang, W.; Wang, K.-L.; Su, Y.-R. Effects of environmental factors on soil organic carbon under natural or managed vegetation restoration. Land Degrad. Dev. 2018, 29, 387–397. [Google Scholar] [CrossRef]
- Yang, Y.; Dou, Y.; An, S. Testing association between soil bacterial diversity and soil carbon storage on the Loess Plateau. Sci. Total Environ. 2018, 626, 48–58. [Google Scholar] [CrossRef]
- Coller, E.; Longa, C.M.O.; Morelli, R.; Zanoni, S.; Ippolito, M.C.C.; Pindo, M.; Cappelletti, C.; Ciutti, F.; Menta, C.; Zanzotti, R.; et al. Soil Communities: Who Responds and How Quickly to a Change in Agricultural System? Sustainability 2021, 14, 383. [Google Scholar] [CrossRef]
- Kuypers, M.M.M.; Marchant, H.K.; Kartal, B. The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 2018, 16, 263–276. [Google Scholar] [CrossRef]
- Poole, P.; Ramachandran, V.; Terpolilli, J. Rhizobia: From saprophytes to endosymbionts. Nat. Rev. Genet. 2018, 16, 291–303. [Google Scholar] [CrossRef]
- Yuan, H.; Ge, T.; Chen, X.; Liu, S.; Zhu, Z.; Wu, X.; Wei, W.; Whiteley, A.; Wu, J. Abundance and Diversity of CO2-Assimilating Bacteria and Algae Within Red Agricultural Soils Are Modulated by Changing Management Practice. Microb. Ecol. 2015, 70, 971–980. [Google Scholar] [CrossRef] [PubMed]
- Brabcová, V.; Štursová, M.; Baldrian, P. Nutrient content affects the turnover of fungal biomass in forest topsoil and the composition of associated microbial communities. Soil Biol. Biochem. 2018, 118, 187–198. [Google Scholar] [CrossRef]
- Zheng, H.; Vesterdal, L.; Schmidt, I.K.; Rousk, J. Ecoenzymatic stoichiometry can reflect microbial resource limitation, substrate quality, or both in forest soils. Soil Biol. Biochem. 2022, 167, 108613. [Google Scholar] [CrossRef]
- Fanin, N.; Mooshammer, M.; Sauvadet, M.; Meng, C.; Alvarez, G.; Bernard, L.; Bertrand, I.; Blagodatskaya, E.; Bon, L.; Fontaine, S.; et al. Soil enzymes in response to climate warming: Mechanisms and feedbacks. Funct. Ecol. 2022, 36, 1378–1395. [Google Scholar] [CrossRef]
- Bisschop, K.; Kortenbosch, H.H.; van Eldijk, T.J.B.; Mallon, C.A.; Salles, J.F.; Bonte, D.; Etienne, R.S. Microbiome Heritability and Its Role in Adaptation of Hosts to Novel Resources. Front. Microbiol. 2022, 13, 703183. [Google Scholar] [CrossRef]
- Shade, A.; Caporaso, J.G.; Handelsman, J.; Knight, R.; Fierer, N. A meta-analysis of changes in bacterial and archaeal communities with time. ISME J. 2013, 7, 1493–1506. [Google Scholar] [CrossRef]
- Tecon, R.; Or, D. Biophysical processes supporting the diversity of microbial life in soil. FEMS Microbiol. Rev. 2017, 41, 599–623. [Google Scholar] [CrossRef]
- Yan, Y.; Klinkhamer, P.G.; van Veen, J.A.; Kuramae, E.E. Environmental filtering: A case of bacterial community assembly in soil. Soil Biol. Biochem. 2019, 136, 107531. [Google Scholar] [CrossRef]
- Holyoak, M.; Leibold, M.A.; Holt, R.D. Metacommunities: Spatial Dynamics and Ecological Communities; University of Chicago Press: Chicago, IL, USA, 2005. [Google Scholar]
- Leibold, M.A.; Holyoak, M.; Mouquet, N.; Amarasekare, P.; Chase, J.M.; Hoopes, M.F.; Holt, R.D.; Shurin, J.B.; Law, R.; Tilman, D.; et al. The metacommunity concept: A framework for multi-scale community ecology. Ecol. Lett. 2004, 7, 601–613. [Google Scholar] [CrossRef]
- Ewers, R.M.; Thorpe, S.; Didham, R.K. Synergistic interactions between edge and area effects in a heavily fragmented landscape. Ecology 2007, 88, 96–106. [Google Scholar] [CrossRef]
- Ferrenberg, S.; O’Neill, S.P.; E Knelman, J.; Todd, B.; Duggan, S.; Bradley, D.; Robinson, T.; Schmidt, S.K.; Townsend, A.R.; Williams, M.W.; et al. Changes in assembly processes in soil bacterial communities following a wildfire disturbance. ISME J. 2013, 7, 1102–1111. [Google Scholar] [CrossRef] [PubMed]
- Langenheder, S.; Lindström, E.S. Factors influencing aquatic and terrestrial bacterial community assembly. Environ. Microbiol. Rep. 2019, 11, 306–315. [Google Scholar] [CrossRef] [PubMed]
- Fukami, T. Historical Contingency in Community Assembly: Integrating Niches, Species Pools, and Priority Effects. Annu. Rev. Ecol. Evol. Syst. 2015, 46, 1–23. [Google Scholar] [CrossRef]
- Wagg, C.; Bender, S.F.; Widmer, F.; van der Heijden, M.G.A. Soil biodiversity and soil community composition determine ecosystem multifunctionality. Proc. Natl. Acad. Sci. USA 2014, 111, 5266–5270. [Google Scholar] [CrossRef] [PubMed]



| Variable | Source | Sig. (p-Value) |
|---|---|---|
| Bacteria | ||
| Proteobacteria | Site | 0.654 |
| Sample | 0.001 | |
| Acidobacteria | Site | 0.116 |
| Sample | 0.000 | |
| Bacteroidetes | Site | 0.687 |
| Sample | 0.004 | |
| Planctomycetes | Site | 0.286 |
| Sample | 0.000 | |
| Verrucomicrobia | Site | 0.034 |
| Sample | 0.000 | |
| Thaumarchaeota | Site | 0.020 |
| Sample | 0.001 | |
| Chloroflexi | Site | 0.314 |
| Sample | 0.000 | |
| Candidatus Saccharibacteria | Site | 0.432 |
| Sample | 0.000 | |
| Cyanobacteria/Chloroplast | Site | 0.449 |
| Sample | 0.001 | |
| Fungi | ||
| Ascomycota | Site | 0.602 |
| Sample | 0.001 | |
| Basidiomycota | Site | 0.633 |
| Sample | 0.002 | |
| Chytridiomycota | Site | 0.007 |
| Sample | 0.011 | |
| Glomeromycota | Site | 0.375 |
| Sample | 0.000 | |
| Zygomycota | Site | 0.003 |
| Sample | 0.000 |
| Variable | Source | df | Mean Square | F-Value | Sig. (p-Value) |
|---|---|---|---|---|---|
| Bacteria | |||||
| Shannon–Wiener | Site | 1 | 1.7576 | 9.944 | 0.002 |
| Sample | 19 | 0.5776 | 22.35 | 0.000 | |
| Chao1 | Site | 1 | 11,121,957 | 11.95 | 0.001 |
| Sample | 19 | 2,935,996 | 12.61 | 0.000 | |
| Fungi | |||||
| Shannon–Wiener | Site | 1 | 3.970 | 9.32 | 0.003 |
| Sample | 19 | 1.2612 | 10.7 | 0.000 | |
| Chao1 | Site | 1 | 1780 | 0.044 | 0.834 |
| Sample | 19 | 104,903 | 12 | 0.000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mo, L.; Zanella, A.; Bolzonella, C.; Squartini, A.; Xu, G.-L.; Banas, D.; Rosatti, M.; Longo, E.; Pindo, M.; Concheri, G.; et al. Land Use, Microorganisms, and Soil Organic Carbon: Putting the Pieces Together. Diversity 2022, 14, 638. https://doi.org/10.3390/d14080638
Mo L, Zanella A, Bolzonella C, Squartini A, Xu G-L, Banas D, Rosatti M, Longo E, Pindo M, Concheri G, et al. Land Use, Microorganisms, and Soil Organic Carbon: Putting the Pieces Together. Diversity. 2022; 14(8):638. https://doi.org/10.3390/d14080638
Chicago/Turabian StyleMo, Lingzi, Augusto Zanella, Cristian Bolzonella, Andrea Squartini, Guo-Liang Xu, Damien Banas, Mauro Rosatti, Enrico Longo, Massimo Pindo, Giuseppe Concheri, and et al. 2022. "Land Use, Microorganisms, and Soil Organic Carbon: Putting the Pieces Together" Diversity 14, no. 8: 638. https://doi.org/10.3390/d14080638
APA StyleMo, L., Zanella, A., Bolzonella, C., Squartini, A., Xu, G.-L., Banas, D., Rosatti, M., Longo, E., Pindo, M., Concheri, G., Fritz, I., Ranzani, G., Bellonzi, M., Campagnolo, M., Casarotto, D., Longo, M., Linnyk, V., Ihlein, L., & Yeomans, A. J. (2022). Land Use, Microorganisms, and Soil Organic Carbon: Putting the Pieces Together. Diversity, 14(8), 638. https://doi.org/10.3390/d14080638









