Abstract
The yellow grouper (Epinephelus awoara) is distributed in the West Pacific Ocean. Its genetic structure and demography were investigated using mitochondrial COI, Cyt b, the ND2 gene, the nuclear RyR3 gene, and 10 microsatellite DNA markers. A total of 120 individuals were collected from four locations along the coast of southeastern mainland China. High levels of haplotype diversity (0.968) were observed in mitochondrial DNA, and the average number of alleles ranged from 13.4 to 20.3 in microsatellite DNA data, which showed that all populations exhibited a high level of genetic diversity. Deficiency of heterozygosity was observed in all populations with positive FIS, showing that the characteristics of hermaphroditism might also be an underlying cause. The results of PCA, UPGMA clustering analysis and the significant genetic differentiation found in the Beibu Gulf population revealed the prevention of gene flow caused by the Qiongzhou Strait. The population of E. awoara also presented two major lineages, resulting in the appearance of the land bridge of the Taiwan Strait as a possible factor during the Pleistocene glaciation. Analysis of demographic history revealed that E. awoara underwent a reduction in effective population size in the past, followed by a single instantaneous increase in population size.
1. Introduction
Phylogeography focuses on the spatial distribution history and phylogenetic components of gene lineages between and within related species [1,2]. Genetic differentiation among populations within species is often attributed to historical climate change or physical barriers, as well as limited gene flow [3]. Due to the openness of the oceanic environment, there are few geographical barriers, and it is difficult for marine species to develop geographically genetic structures compared with inland species [4]. Marine fish have always been thought to be less genetically differentiated than freshwater fish due to the high degree of gene flow in marine environments. The dispersal potential of planktonic larvae and eggs depends on the lack of geographical barriers among populations, contributing to the low genetic differentiation of marine fish [5,6]. However, the interacting effects of biotic and abiotic factors in the marine environment, such as ocean currents, spawning aggregation, larval behavior, and pelagic larval duration, may play an important role in enhancing the long-distance dispersal of populations or limiting dispersal to distant populations and shaping the population genetic structure [7,8,9,10,11]. If these factors contribute to the isolation between adjacent seawater areas, then the potential for genetic differentiation between different geographical groups of the same species is increased. Some studies have even reported that certain populations of reef fish with high dispersal ability still show high genetic differentiation in the absence of obvious transmission barriers, such as Acanthurus triostegus [12], Decapterus maruadsi [13], Dascyllus trimaculatus [14].
In the continental margin of the western Pacific Ocean, there are many marginal basins, including the East China Sea (ECS), South China Sea (SCS), Yellow Sea, and Sea of Japan [15]. During Plio-Pleistocene glacial cycles, the environmental signals caused by sea level changes were amplified in the marginal waters of the western Pacific, which resulted in dramatic changes in the area and configuration of these waters and had a great impact on the distribution of local marine fish populations [16,17]. Therefore, the southeastern coastal area of China provides one of the best natural environments for studying how invasive events, population bottlenecks, prolonged isolation, and subsequent mixing affect the lineage structure and geographical differentiation of marine species [18]. Previous studies have shown low levels of genetic differentiation in some marine economic fishes on the southeastern coast of China, such as Epinephelus coiodes [19], Larimichthys polyactis [20] and Scomber japonicus [21], revealing that the strong gene flow caused by the lack of geographic isolation and the rapid spread of planktonic juveniles hindered the genetic differentiation between populations [22]. However, some studies have also demonstrated genetic differences between biogeographic groups. For example, Qiu et al. [23] and Ding et al. [24] showed that the exposure of the Taiwan Strait land bridge during the Pleistocene ice age introduced an ecological barrier for gene flow between populations in the East China Sea and the South China Sea, resulting in the differentiation of the southern and northern subpopulations in Bostrychus sinensis. Chen et al. [18] discovered genetic differentiation among the geographic populations of red grouper (E. akaara) and speculated that the differences in adult settlement habits, reproduction and sex changes of reef fish might prevent gene flow between populations.
The yellow grouper (Epinephelus awoara), belonging to the genus Epinephelus, is a warm water, euryhaline inshore bottom fish. They often inhabit areas within 100 m of estuaries and isobath lines [25]. E. awoara, with a wide distribution in the coastal waters of Vietnam, China, South Korea, and Japan, is mainly distributed from Jiangsu Province in the north to Hainan Province and Guangxi Province in southern China and is one of the main dominant species of grouper fishery in the southern provinces of China [26,27,28]. The wide geographical distribution of E. awoara makes it a suitable model species for studying biogeography and genetic subdivision. However, the wild population resources of E. awoara on the southeast coast of China have fluctuated significantly in recent decades. Since 2000, the resources of E. awoara have been decreasing, and the population size has become miniaturized under the influence of overfishing and habitat destruction [29,30,31]. Although the number of E. awoara caught in the south of the Taiwan Strait of China has recovered significantly in recent years, its geographical distribution has gradually retreated southward, and it has been difficult to find in the coastal areas of Jiangsu and Zhejiang Provinces in mainland China [32]. Previous studies on E. awoara have mainly focused on aquaculture, bait diseases, physiology and biochemistry, including the growth, nitrogen excretion and energy budget of juvenile E. awoara under different starvation levels [33]; the microbial community in the gastrointestinal tract [34]; and the genetic role of macrophage migration inhibitory factor (MIF) in vibrio parahaemolyticus infection of E. awoara [35]. However, the genetic diversity, genetic structure and other germplasm resources of the E. awoara population have rarely been investigated. Due to the sedentary nature of the adult stage, the population of E. awoara may have a unique genetic structure [32]. However, planktoniceggs and juveniles may promote gene flow between populations through the diffusion of ocean currents [36,37,38]. Therefore, the exact genetic difference among populations of E. awoara has not yet been identified. It is difficult to directly estimate the connectivity between populations due to the complex environment of the ocean, but molecular markers and correlation analysis techniques provide us with an indirect opportunity to estimate the interrelationships between populations [39]. Upadhyay et al. [40] compared the genetic diversity levels of Guangdong and Xiamen populations of E. awoara using the random amplified polymorphic DNA (RAPD) method and identified gene flow between the two populations that was mainly affected by the migration of larvae and young fishes during the floating period and the dispersal of adults with ocean currents. Zhao et al. [41] used the FIASCO (Fast Isolation by AFLP of Sequences Containing Repeats) method to construct a dinucleotide-enriched genomic library of E. awoara and developed 12 novel polymorphic microsatellite loci for E. awoara. However, it is worth mentioning that due to the shortcomings of the sampling groups and research methods, the previously obtained results may not exactly reveal the reality.
To conduct a more systematic and comprehensive assessment of the wild resources of E. awoara in offshore China, the geographical distribution of samples and the number of molecular markers analyzed were significantly increased in our study compared to the studies from Upadhyay et al. [40] and Zhao et al. [41]. We systematically analyzed the genetic diversity, genetic structure and population history dynamics for four wild populations of E. awoara distributed in the northern Taiwan Strait (MB), southern Taiwan Strait (MN), Guangdong Sea (DYW) and Beibu Gulf (BBW) of China using mitochondrial and nuclear gene sequence analysis techniques and microsatellite molecular markers. Our study addresses a comparative multimarker analysis of the phylogeographic structure and historical demography in E. awoara to test for common natural histories and environmental climatic factors (i.e., phylogeographic breaks), which may provide useful reference materials for the protection of the existing wild resources of E. awoara and the formulation of relevant fishery policies, as well as a theoretical basis and guidance for the conservation and breeding of parents of the green grouper in the aquaculture industry.
2. Materials and Methods
2.1. Sample Collection, Microsatellite Genotyping, and Mitochondrial and Nuclear Sequencing
In 2019, a total of 120 E. awoara individuals were collected from 4 different locations along the southeastern coast of mainland China, including 33 individuals in Ningde (the northern Fujian population (MB)), 36 individuals in Xiamen (the southern Fujian population (MN)), 34 individuals in Shenzhen (the Daya Bay population (DYW)) and 17 individuals in Weizhou Island (the Beibu Gulf population (BBW)) (Table 1; Figure 1). After morphological measurement of all samples, a piece of muscle from the base of the dorsal fin was removed, stored in anhydrous ethanol, and stored at −20 °C, until DNA extraction.

Table 1.
Summary statistics for genetic variation of E. awoara from four different locations.
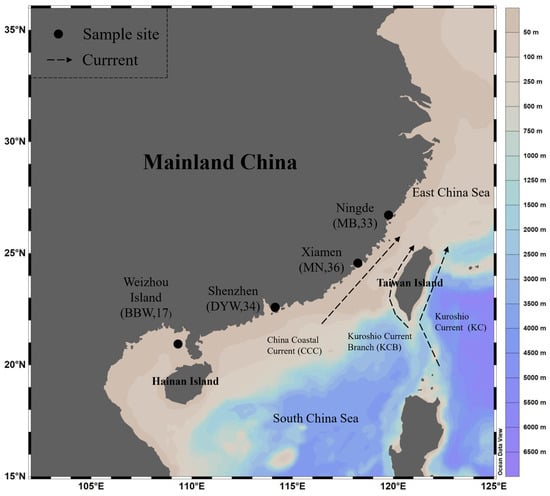
Figure 1.
Map of the southern coast of mainland China illustrating the sampling locations of Epinephelus awoara. KC: Kuroshio Current; KCB: Kuroshio Current Branch; CCC: China Coastal Current. Sampling site: Ningde, Xiamen, Shenzhen, Weizhou Island. Geographic population code: MB, MN, DYW, BBW. Numbers in the buckets are the number of individuals collected. The sampling site/geographic population code is described in Table 1.
Genomic DNA was extracted by the guanidine isothiocyanate method [42]. We used primers designed from the mitochondrial complete genome of E. awoara (GenBank Accession No MT269921): COI-F (5′-TTCTTCTCGACTAATCACAAAGACA-3′), COI-R (5′-CAGACTATACCCATGTAACCGAAC-3′), Cytb-F (5′-TCAACCAACCACAAAGACATTGGGAC-3′), Cytb-R (5′-TAGACTTCTGGGTGGCCAAAGAATCA-3′), ND2-F (5′-AAGGGCCACTTTGATAGAGTG-3′), ND2-R (5′-GTRAGTRYGGGGGYTTTTGCYCA-3′), RyR3-F (5′-TGACAGCTTCAGAGAGTATGACCCT-3′), and RyR3-R (5′-GCCAGTGAAGAGCATCCAGAAGAAG-3′). The mitochondrial cytochrome c oxidase subunit I gene (COI), cytochrome b oxidase subunit gene (Cyt b), NADH dehydrogenase subunit 2 gene (ND2) and nuclear ryanodine receptor gene (RyR3) were amplified by polymerase chain reaction (PCR). PCR was performed in 25 μL volume reactions containing 1 μL template DNA, 7.5 μL 10× buffer, 1.5 μL dNTPs (2.5 mmol·L−1), 1 μL primer (10 pmol·μL−1), 1.5 μL Taq enzyme (10 U·L−1) (TAKARA Products, Beijing, China), and 12.5 μL ddH2O. The PCR conditions were as follows: predenaturation for 5 min at 94 °C; 35 cycles of denaturation at 94 °C for 30 s, annealing at 52 °C (COI, Cyt b, ND2)/56 °C (RyR3) for 30 s, and extension at 72 °C for 1 min 30 s; and a final extension at 72 °C for 10 min. The amplified products were detected by 1% agarose gel electrophoresis. PCR products were recovered and purified by gel cutting and sequenced by Xiamen Borui Biotechnology Co. (Xiamen, China). A total of 10 microsatellite loci were redesigned from a previous study [43] (Table S1). The 5′ ends of all forward primers were labeled with FAM, HEX and ROX fluorescence groups. The PCR system was 15 μL, including approximately 0.8 μL template DNA, 1.5 μL 10× PCR buffer, 0.9 μL MgCl2 solution (25 mmol/μL), 1 μL dNTPs (2 mmol/μL), 1.5 μL upstream primer (10 pmol/μL), 1.5 μL downstream primer (10 pmol/μL), 0.2 μL rTaq enzyme (5 U/μL), and 7.6 μL sterilized deionized water. The reaction conditions were as follows: predenaturation for 5 min at 94 °C; 35 cycles of 30 s of denaturation at 94 °C, 30 s of renaturation at the respective annealing temperature, 1 min of extension at 72 °C, and 10 min of final extension at 72 °C after the cycle. The amplified products were detected by 1% agarose gel electrophoresis, and the qualified PCR products were sent to Qingdao Huada Bioengineering Co., Ltd. (Qingdao, China) for genotyping.
2.2. Data Analysis
2.2.1. Sequence Analysis
The COI, Cyt b and ND2 genes of the mtDNA sequences were edited and aligned using CLUSTRAL W, as implemented in MEGA X [44]. These three genes (COI, Cyt b, ND2) were concatenated for analysis. The haplotype number (Nh), haplotype diversity (h) and nucleotide diversity (θπ and θω) were calculated using DnaSP ver.5.0 [45]. The phylogenetic relationship was constructed with the maximum likelihood method (ML) in the PHYML ver. 3.0 network server [46], using MrBayes ver. 3.2.7 software [47] for Bayesian inference (BI) and MEGA X [44] for neighbor-joining (NJ). The Akaike information standard (AIC) and program PhyML 3.0 with Smart Model Selection (http://www.atgc-montpellier.fr/phyml-sms/ (accessed on 7 August 2021)) [48] were used for intelligent selection of the best fitting nucleotide substitution model. The haplotype network was inferred using the TCS algorithm as implemented in PopART v.1.7 software [49].
The levels of genetic differentiation between populations (pairwise FST values) and analysis of molecular variance (AMOVA) were conducted using Arlequin v. 3.5 [50], and 10,000 permutation steps were used for statistical significance analysis for each group comparison. In the AMOVA, the population was divided into five scenarios according to geographical barriers: (1) Scenario I: four geographic population (MB, MN, DYW, BBW) (2) Scenario II: the southern population (MN, DYW, BBW) and the northern population (MB) with the Taiwan Strait as the main dividing line; (3) Scenario III: the South China Sea Group (MB, MN) and the East China Sea Group (DYW, BBW); (4) Scenario IV: the southern group (BBW) and the northern population (MB, MN, DYW) separated by the Qiongzhou Strait; (5) Scenario V: the northern group (MB), the middle group (MN, DYW) and the southern group (BBW) divided by the Taiwan Strait and Qiongzhou Strait. In addition, isolation by distance (IBD) was tested using IBD 1.52 for Windows [51]. To test whether genetic differentiation increased with geographic distance, we used Google Earth combined with coordinate point information to measure the geographical distance between sampling locations.
Historical demographic scenarios were analyzed by neutral tests (Tajima’s D test [52] and Fu’s Fs test [53]) and mismatch distribution tests using DnaSP v. 5.0 [45]. The effective population size over time was evaluated by a Bayesian skyline plot (BSP) using BEAST v. 1.8.2 [54]. The tests were run for >200,000,000 MCMCS generations to ensure the convergence of all parameters (ESSs > 200), and the first 10% of samples from each chain were discarded as burn in. Plots for each analysis were generated using Tracer v.1.6 [55]. In this study, a mutation rate of 1.2% per million years by Bermingham (1997) and Kumazawa (2002) was calibrated for the mtDNA COI, Cyt b, and ND 2 genes [56,57].
The PHASE algorithm in DnaSP v. 5.0 software [45] was used to resolve the haplotype phases of the RyR3 sequences, and CLUSTRAL W software as implemented in MEGA X [44] was used to edit and align the RyR3 sequences. Genetic diversity parameters, phylogenetic clustering, population genetic differentiation and AMOVA molecular variance analysis were performed using the same software and procedures as used for the mtDNA data.
2.2.2. Microsatellite DNA Analysis
To assess possible genotyping errors, the presence of null alleles, and allelic dropout, all loci were examined using Micro-Checker [58]. The number of alleles (NA), observed heterozygosity (HO), expected heterozygosity (HE) and Hardy-Weinberg equilibrium (HWE) of each population were calculated using Arlequin v. 3.5. The allele richness (AR) and inbreeding coefficient (FIS) of each population were calculated using FSTAT v. 2.9.3 software [59]. Population genetic differentiation parameters (pairwise FST and RST values) and AMOVA molecular variance analysis were performed using the same software and program as used for the mtDNA data.
STRUCTURE ver. 2.2.3 [60] was used for Bayesian cluster analysis of microsatellite DNA data to determine the optimal number of genetically homogeneous groups. A burn-in of 50,000 in periods was followed by 100,000 iterations of the Markov Chain Monte Carlo (MCMC) simulations. The number of individual clusters (K) was set to 1–6, with 10 iterations for each particular value of k. Structure Harvester Web v. 0.6.94 [61] was used to determine the optimal values of k. In addition, principal component analysis (PCA) based on standardized covariance of genetic distance between populations was performed by GenAlEx v.6.503 [62] to reveal genetic variation patterns between populations and provide diversified graphic descriptions of allelic differences between populations. The method based on Nei’s DA Distance [63] was applied to calculate the genetic distances among all populations and create a tree using the arithmetic mean (UPGMA) method of unweighted pairwise grouping with 1000 repeated bootstrap analyses by using POPULATIONS v.1.2.28.
We used Bottleneck v.1.2.02 [64] with the infinite allele model (IAM), stepwise mutation model (SMM), and two-phase model (TPM) to investigate whether any population had recently experienced a reduction in effective population size. The use of the two-phase model (TPM) included a 70% stepwise mutation model (SMM) and a 30% infinite allele model (IAM).
2.2.3. DIY-ABC Analysis
Because of the demographic history complexity, we used an approximate Bayesian computation (ABC) method to investigate the possible historical demographic scenarios of E. awoara using the program DIYABC v.2.0.4 [65]. We followed the recommendation by Cabrera and Palsboll [66] to define the five possible demographic scenarios of E. awoara (Figure 2). In Scenario 1 (CON model), a constant population size was maintained over time. In Scenario 2 (INC model), the population experienced a recent bottleneck event. In Scenario 3 (DEC model), the population expanded recently. In Scenario 4 (DECINC model), the populations expanded and then experienced a single instantaneous decrease in population size. In Scenario 5 (INCDEC model), the populations experienced a bottleneck event followed by a single instantaneous increase in population size. We simulated three million datasets for each scenario and included all summary statistics computed by DIYABC software for mtDNA, nuclear DNA and microsatellite markers [65]. The posterior probability of each scenario was obtained by the logistic regression approach.
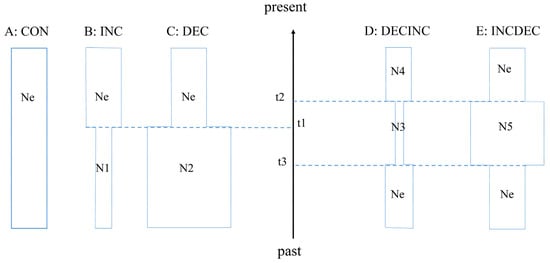
Figure 2.
Schematic representation of five demographic scenarios for Epinephelus awoara tested by approximate Bayesian computation (ABC). Time and effective population size are not to scale. Graphical representation of the five scenarios: (A) scenario 1 (CON model); (B) scenario 2 (INC model); (C) scenario 3 (DEC model); (D) scenario 4 (DECINC model); (E) scenario 5 (INCDEC model).
3. Result
3.1. Mitochondrial DNA
A total of 82 haplotypes were obtained from 2295-bp concatenated mtDNA (COI + Cyt b + ND2) sequences of 120 E. awoara. Eight haplotypes were shared, of which two haplotypes were shared by four populations (Hap13, Hap23), one haplotype was shared by three populations (Hap07), and five haplotypes were shared by two populations (Hap08, Hap22, Hap35, Hap41, Hap49) (Table S2). The haplotype diversity (h) of each population ranged from 0.926 (BBW) to 0.994 (MB), and the average haplotype diversity value was 0.968. The nucleotide diversity (θπ) of each population ranged from 0.0033 (DYW) to 0.0083 (MB), and the average value of nucleotide diversity was 0.0037 (Table 1). The historical nucleotide diversity was higher than the current genetic diversity (θω (0.0060) > θπ (0.0037)), indicating that the population of E. awoara showed a pattern of decline [67].
The pairwise FST values between populations ranged from −0.0203 (between MN and BBW) to 0.0121 (between DYW and BBW), with a mean value of 0.0135, and the FST values were low and nonsignificant, indicating that there was no significant genetic differentiation among populations (Table 2). The IBD analysis of all populations showed no significant correlation between geographical distance and population genetic differentiation (R = −0.3416, p = 0.709). Five possible geographically separated scenarios were tested using analysis of molecular variance (AMOVA). The results showed that the genetic variation among groups was low in five scenarios: Scenario I (−0.57%), Scenario II (−0.14%), Scenario III (0.42%), Scenario IV (0.11%) and Scenario V (−0.45%), indicating the absence of a potential barrier hindering the gene flow among groups. In addition, the variation among populations also showed a low level (−0.53%~−0.22%) (Table 3).

Table 2.
Matrix of pairwise FST among four populations based on microsatellite DNA (below diagonal) and mitochondrial gene (above diagonal) in E. awoara.

Table 3.
Analysis of molecular variance (AMOVA) for E. awoara populations based on mtDNA, nuDNA and microsatellite DNA.
The phylogenetic clustering tree constructed based on three algorithms (NJ, ML and Bayesian) supported the formation of two major lineages (I and II) (Figure S1), but no clustering corresponding to the sampling location was detected. Eighty-two haplotypes were roughly divided into two branches, with each haplotype having no obvious geographical distribution pattern and each population haplotype clustering across it. In addition, the star-like structure of a haplotype network indicated a sudden population expansion of E. awoara (Figure 3). Significant negative Tajima’s D and Fu’s Fs values (D = −2.096, P = 0.000; Fs = −24.585, P = 0.000) indicate that the E.awoara populations experienced a recent demographic expansion. The unimodal mismatch distribution showed similar results (Figure S2), and the Bayesian skyline plots revealed that the population of E. awoara remained stable for a period of time, with a continuous growth trend beginning approximately 100,000 years ago and rapid expansion beginning approximately 50,000 years ago (Figure 4).
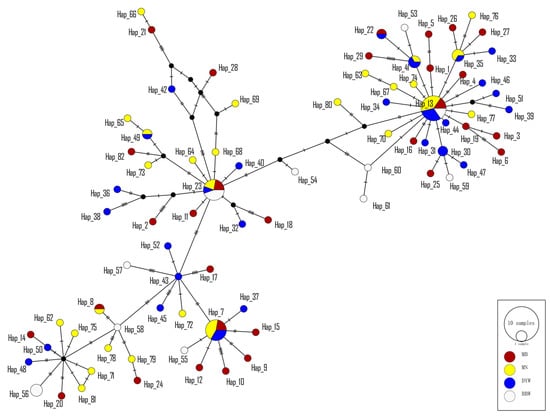
Figure 3.
The haplotype network using the TCS algorithm inferred from mitochondrial COI, Cyt b and ND2 genes of Epinephelus awoara from four geographic samples in mainland China. Each circle indicated each haplotype, and the size of each circle is related to its haplotype distribution frequency. Each color in haplotype network circles represents each sampling location.
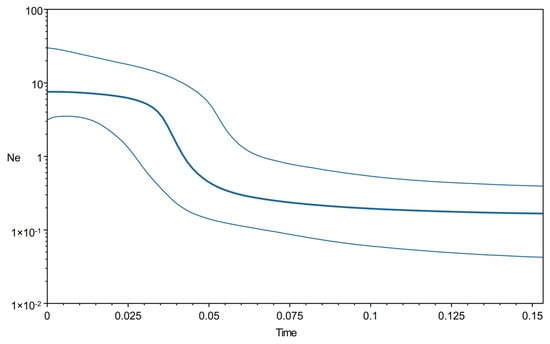
Figure 4.
Bayesian skyline plot of the effective population sizes through time for Epinephelus awoara. The y-axis is the product of effective population size (Ne) and generation length in a log scale while the x-axis is the time scale before present in units of million years ago.
3.2. Nuclear DNA
A total of 240 RyR3 sequences were obtained from 120 E. awoara RyR3 individuals for genetic analysis. The total sequence length was 649 bp, and 9 haplotypes were detected, among which 3 haplotypes (Hap01, Hap02, Hap03) were found in all populations. One haplotype (Hap04) was found in three populations, and two haplotypes (Hap06 and Hap07) appeared in two populations (Table S3). The haplotype diversity of the RyR3 sequence ranged from 0.169 (DYW) to 0.410 (BBW), and the average haplotype diversity (h = 0.280) was lower than that of mitochondria (h = 0.968). The nucleotide diversity ranged from 0.00031 (DYW) to 0.00084 (BBW), and the mean nucleotide diversity θω = 0.00052 (Table 1).
The pairwise FST values between populations ranged from −0.0015 (between DYW and MB) to 0.0460 (between DYW and BBW) in the RyR3 gene, with a mean value of 0.017 (Table S4). There was no significant genetic differentiation between populations (p > 0.05), except for the DYW and BBW populations. Although the DYW and BBW populations were geographically adjacent, they still showed higher genetic differentiation. Molecular analysis of variance (AMOVA) was used to test the five possible geographically separated scenarios, and the genetic differences among each geographically separated group were as follows: Scenario I (1.37%), Scenario II (−1.61%), Scenario III (−1.27%), Scenario IV (4.07%) and Scenario V (1.67%). In Scenarios I to V, only −0.01%~–2.26% genetic differences between populations were detected (Table 3).
3.3. Microsatellite DNA
No evidence of null alleles, large allele dropout, or stuttering was found using Micro-Checker v. 2.2.3. A total of 272 alleles were detected at 10 loci of 120 E. awoara individuals, of which 49 were only present in a single population. The average number of alleles ranged from 13.40 (BBW) to 20.30 (MB), with an average of 18.10. The average allele richness (AR) ranged from 14.71 (MB) to 15.80 (DYW), with an average of 15.40. The average expected heterozygosity was 0.849, ranging from 0.826 (MN) to 0.876 (BBW), and the observed heterozygosity was 0.870 (BBW) to 0.917 (MB), with an average of 0.900. The heterozygote deficiency of all populations was reflected in the positive FIS, ranging from 0.023 (BBW) to 0.104 (MN) (Table 1).
All 10 microsatellite loci that were detected were polymorphic and characterized by the number of alleles (N) ranging from 18 (Qsby-72) to 34 (Qsby-11, Qsby-70), with an average of 27. The average allele richness (AR) ranged from 11.647 (Qsby72) to 18.762 (Qsby-65), with an average of 15.003. The average observed heterozygosity (HE) ranged from 0.879 (Qsby-60) to 0.953 (Qsby-65), with an average of 0.924. The average expected heterozygosity (HO) ranged from 0.653 (Qsby-11) to 0.924 (Qsby-70), with an average of 0.845. Four microsatellite marker loci (Qsby9, Qsby-24, Qsby-60, and Qsby-71) showed heterozygous excess (FIS < 0), and the rest showed a deficiency of heterozygotes (Table S5).
The pairwise FST values ranged from 0.0039 (MB and DYW) to 0.0235 (MB and BBW), in which the FST values between the BBW population and the other populations were larger than those of other populations, and the differentiation was significant (p < 0.001) (Table 2). The AMOVA results showed that 91% of the variation occurred within individuals, i.e., one group (Scenario I, 90.79), two groups (Scenario II, 90.89%), two groups (Scenario III, 90.90%), three groups (Scenario IV, 89.97%) and three groups (Scenario V, 90.66%) (Table 3). In addition, a relatively low proportion of genetic variability was found among the groups, Scenario I (0.97%), Scenario II (−0.24%), Scenario III (−0.37%), and Scenario IV (1.35%), Scenario V (0.64%), indicating that there was no geographical barrier among the groups. Only a low proportion of genetic variation partitioning was found among populations within groups (0.48%~1.22%), which indicated that the genetic differences were mainly concentrated within populations, and there was almost no variation between populations. These results were also consistent with the findings obtained for mitochondrial and nuclear genes (Table 3).
STRUCTURE clustering analysis was conducted using microsatellite DNA data to assign individuals to the population based on the estimated individual mixing ratio and to determine the most likely population number (i.e., k value) required to interpret the observed genotypes. The results of STRUCTURE showed that the four populations were divided into two different genetic groups (K = 1, LnP(K) = −6960.600; Stdev LnP(K) = 1.308; K = 2, LnP(K) = −6971.330, Stdev LnP(K) = 14.258, Delta K = 3.0937; K = 3, LnP(K) = −7026.170, Stdev LnP(K) = 289.217, Delta K = 0.3221), and all the sampled individuals showed mixing between the two groups (Figure 5). The PCA plot showed that the four populations were divided into two clusters (Figure 6), among which MB, DYW and MN were one group and BBW was another group. The variance of the first principal component and the second principal component of microsatellite DNA was 71.01% and 18.38%, respectively, making up a total of 89.39%, using the genetic distance between populations to study the population pattern. The cluster-based method algorithms were employed to build a phylogenetic tree with UPGMA for the microsatellite DNA markers and showed differences between the BBW population and the other three populations (Figure 7).
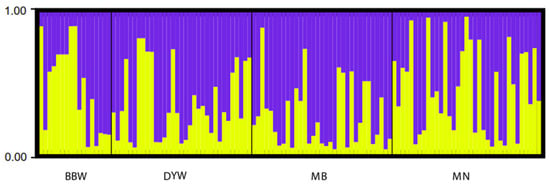
Figure 5.
Clustering of individuals by structure at K = 2. Individuals are represented by vertical bars. Each vertical column represents one individual, and the separation of the column into two colors represents the estimated probability of belonging to one population or the other. Different colors in the same individual indicate the percentage of the genome shared with each cluster according to the admixture proportions. The y-axis represents the probability to belong to a certain cluster, while on the x-axis is reported each population (code name given in Table 1) delimited by a black solid vertical line.
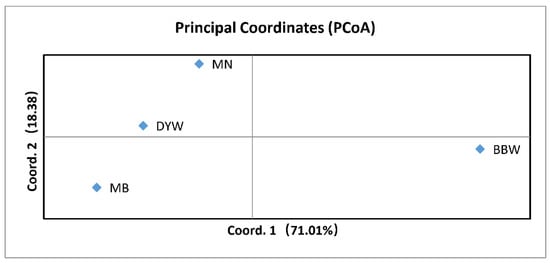
Figure 6.
Plot of principal coordinate (PCA) axes for six populations of Epinephelus awoara based on population genetic distance. Percentage values represent variation justified by each axis. MN: Southern Fujian population; MB: Northern Fujian population; DYW: Daya Bay population; BBW: Beibu Gulf population. See Table 1 for the sampling sites/population codes.
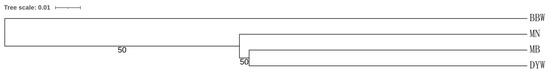
Figure 7.
A UPGMA tree reconstructed based on Nei’s DA Distance among four populations in Epinephelus awoara. MN: Southern Fujian population; MB: Northern Fujian population; DYW: Daya Bay population; BBW: Beibu Gulf population.
Bottelneck software results showed that all four populations had a typical L-type distribution of allelic frequencies in the mode-shift indicator, which suggests a stable population. Under the IAM model, all populations showed significant loss of heterozygosity, indicating that genetic bottlenecks were detected in E. awoara due to mutation-drift equilibrium. Under the TPM, only the MN population showed a significant loss of heterozygosity, and under the SMM, none of the populations showed a significant loss of heterozygosity. Therefore, we suggest that the studied populations of E. awoara experienced a recent genetic bottleneck (Table S6).
We used the five scenarios of ABC analysis to determine the possible demographic history of E. awoara. In the ABC modeling, Scenario 5 (INCDEC model) was highly favored (posterior probability = 0.6023 [0.5445, 0.6601]) over Scenario 2 (INC model) (posterior probability = 0.3370 [0.2486, 0.4254]) and Scenario 1 (CON model) (posterior probability = 0.0606 [0.0000, 0.1930]). This scenario indicated that E. awoara experienced a reduction in the effective population size in the past, followed by a single instantaneous increase in population size.
4. Discussion
4.1. Genetic Diversity
Protecting the population genetic diversity of marine fish is an important part of marine fishery management and policy-making [68]. Genetic diversity plays an important role in the ability of a species to respond to environmental changes via genetic or phenotypic variation [69]. High genetic diversity increases or maintains the adaptability of a population to an unpredictable environment, which is the basis for the stability of each population and the entire ecosystem [70].
In our study, three molecular markers were used to analyze the genetic diversity of four populations of E. awoara from the southeast coast of China. Mitochondrial DNA has many attributes that make it suitable for population genetic studies, including its rapid rate of evolution, lack of recombination, and maternal inheritance [71]. Investigating the key determinants of genetic diversity, as well as phylogeographic patterns, would help to design comprehensive conservation schemes when protecting biodiversity in marine fish. Analysis of mtDNA showed that, compared with other common marine economic species in coastal China (Mugil cephalus (h = 0.7398, π = 0.0449) [72]; Salanx ariakensis (h = 0.9475, π = 0.0075) [73]; B. sinensis (h = 0.7707, π = 0.0013) [23]; E. japonicus (h = 0.958, π = 0.0064) [74]), the population of E. awoara exhibited higher haplotype diversity (h = 0.968) and lower nucleotide diversity (π = 0.0037). The high haplotype diversity and low nucleotide diversity of the E. awoara population were similar to those observed for other marine fish in the South China Sea, suggesting that the population may have experienced a rapid demographic expansion from a small effective population in a short period of time in history.
Microsatellite DNA is an allelic genotype pattern and has a high mutation rate, so it is more suitable for monitoring population structure at present. Our study showed that the average number of alleles (Na) in four populations of E. awoara ranged from 13.4 (BBW) to 20.3 (MB), which was generally higher than that of other marine fishes in the coastal areas of mainland China (Lepturacanthus savala (Na = 10.3~14.0) [75]; Scombridae (Na = 7.455~8.818) [76]; L. polyactis (Na = 10.43~13.71) [20]; Acanthopagrus schlegelii (Na = 5.43~8.00) [77]; Sebastiscus marmoratus (Na = 2~12) [78]), which showed high genetic diversity. We also found that the expected heterozygosity of the four populations was higher than the observed heterozygosity, and the FIS value was positive, showing a deficiency of heterozygotes. But our results of Hardy–Weinberg equilibrium test is not significant, so we can not confirm the existence of inbreeding or the deficiency of heterozygotes in E. awoara’s populations. In some previous studies, we found that marine fish populations off the coast of mainland China generally showed a slight heterozygote deficiency [20,76,77], which might be related to the reduction in effective population size caused by overfishing and the imbalance in the sex ratio [79]. The larvae of E. awoara show the characteristics of hermaphroditism, and the first sexual maturity occurs at approximately 2~3 years of age, showing the female phase. The body length ranges from 210 to 240 mm, and more than 50% of mature females are 6 years old, with a body length of 24 cm [80]. Female E. awoara in the northern sea of Zhejiang, China, can be sexually translatable when the body length reaches 250–400 mm, and some of them can be transformed into males; when the body length is greater than 420 mm, almost all of them are male [81]. As a result, large male groupers are more likely to be caught. In Hong Kong, the size of E. awoara has decreased from 20 to 38 cm in the 1960s to 17 to 27 cm in the 2000s, showing a decrease of 16 to 29 percent, according to a survey of wild caught samples. A market survey conducted in Hong Kong between November 2004 and January 2006 recorded a total of 401 wild catch samples, almost all of which (97%) were immature, with an average length of only 22 cm [82]. These findings suggest that large males of E. awoara experience greater fishing pressure. Therefore, the lack of males leads to an imbalance in the sex ratio in the population of E. awoara and the overfishing still have large possibility to cause inbreeding leading to the deviation from the Hardy–Weinberg equilibrium.
Previous studies have indicated that fluctuations in population size caused by biological or abiotic environmental changes would affect the level of diversity of most organisms [83,84]. For example, negative effects of climate change, diseases, and human activities on population size would lead to a rapid decline in heterozygosity [85]. Because of the increase in fishing pressure and the destruction of the ecological environment in recent decades, the biomass of E. awoara has decreased sharply. However, our results showed that the genetic diversity of the E. awoara population remained at a high level, indicating that the fluctuation in population size was not the main factor affecting the diversity of E. awoara. Romiguier et al. [86] investigated genome-wide genetic diversity patterns of 90 different animals from different taxa and found that any variables that might be related to the population history of a species did not significantly affect their genetic diversity, while closely related species tended to have similar levels of genetic diversity. In fact, there is a strong correlation between genetic diversity and the life history characteristics of species, such as body weight, longevity and reproductive strategies [87]. The influence of life history traits on genetic diversity was mediated by the effective population size, NE [86]. One theory suggests that the minimum population size is the most important factor for the long-term mean (NE) [88]. Species with a selection breeding strategy, such as E. awoara, can produce many offspring in a productive event, which makes it easier for them to maintain a minimum sustainable sizable species density under the high strength of environmental stress or human influence.
4.2. Population Structure
Molecular variance analysis (AMOVA) of the five scenarios showed that most of the genetic variation resided within the population, and there was no significant genetic differentiation among the groups. In addition, we were unable to find obvious geographic clusters from the “star-like” haplotype network, indicating a lack of population structure. However, the results of the pairwise population FST showed higher significant differentiation between the BBW population and the other populations in microsatellite DNA data. The pairwise FST test revealed no genetic differences among populations except the BBW and DYW populations in mtDNA. AMOVA revealed that 4.07% of the total genetic variance was found among groups (Scenario IV) in microsatellite DNA, suggesting that the Qiongzhou Strait might be a possible physical barrier. The results of PCA and UPGMA also revealed that the BBW population was separated from the other populations by the Qiongzhou strait. Considering the biological characteristics of the adult E. awoara settling in deep water without long-distance migration, the migration of the individual mainly depends on the larval and young fish at the plankton stage or living in shallow water [89]. For juvenile E. awoara lacking autonomous swimming ability, ocean current is the most important factor affecting its migration. The eastern entrance of the Qiongzhou Strait was mainly influenced by semidiurnal tide waves from the South China Sea, while the western entrance was influenced by diurnal tide waves from the Beibu Gulf [90]. The tidal current between the two independent tidal wave systems was strong, forming eddies at the entrance of the Qiongzhou Strait [91] and, thus, causing a certain degree of gene flow barrier for E. awoara, which mainly depended on the individual diffusion driven by the water flow during the plankton period. However, due to the interaction between the east and west sides of the Qiongzhou Strait, the direction of the residual current is from east to west throughout the year, except for the flow into the South China Sea from the Beibu Gulf in summer under the influence of the southwest monsoon [90]. Thus, the degree of differentiation is relatively low. In addition, regardless of the stage of the life history of E. awoara, we should not overlook its ability to swim through the Qiongzhou Strait to a certain extent, which further limits its genetic differentiation to a low level. However, in the eastern Qiongzhou Strait, due to the lack of an obvious geographical barrier and the transportation effect of the Kuroshio branch and South China Sea surface current [92], suitable conditions existed for extensive gene flow among the three geographical populations, so no genetic differentiation was found. Similarly, restricted gene flow between the Beibu Gulf and the South China Sea has been reported in other marine organisms, such as Monodonta labio [93], Gobiopterus lacustris [94], P. argenteus [95], and L. savala [75]. We found a large difference in genetic differentiation between populations on both sides of the Qiongzhou Strait among species that lacked autonomous swimming ability and relied on ocean current diffusion, which confirmed that ocean currents hindered organisms from crossing the Qiongzhou Strait to some extent.
Our study suggested that the population of E. awoara also presented two major lineages, and the existing populations were all mixed populations of two clusters. During the ice age from the Eocene to the end of the Pleistocene, the decline in sea level connected the two sides of the Taiwan Strait and formed a land bridge [96], which geographically blocked gene exchange between the marine communities north and south of the strait. Previous studies have reported the populations of many species isolated by the land bridge of the Taiwan Strait throughout history, such as Chelon haematocheilus [97], Terapon jarbua [98], B. sinensis [23,24], and L. savala [75]. Because of the distribution of E. awoara, we can consider the appearance of the land bridge of the Taiwan Strait as a possible factor in the historical divergence between the southern population (MN, DYW, BBW) and the northern population (MB) of the Taiwan Strait.
4.3. Demographic History and DIY-ABC
In our study, Tajima’s D and Fu’s Fs neutral tests and mismatch distribution tests showed that the population of E. awoara experienced population expansion. The pattern of the haplotype network is star-like, which also fits the typical pattern of population expansion. A Bayesian skyline plot (BSP) showed that the population size of E. awoara remained stable for a period of time and a continuous growth trend occurred from approximately 100,000 years ago to a rapid expansion from approximately 50,000 years ago. The above results suggest that the late Pleistocene may have had an important influence on shaping the population history of E. awoara. Over the past million years, periodic fluctuations in the global climate have occurred. It is dominated by a series of large glacial and interglacial periods [99]. The rise and fall of sea level and climate change are considered to have a great impact on the geographical distribution and population size of marine species, thereby leading to great variation in the genetic structure and historical dynamics of marine biological communities [100], especially over the edge of the northwestern Pacific Ocean [16]. In addition, changes in nutrient concentrations, water temperature and the availability of a suitable habitat could all participate as factors regulating the population numbers of marine species [101]. These factors may contribute to the fluctuation in the size of the E. awoara population. Upon their departure from harsh conditions, rapid expansion might occur. Several previous studies on marine organisms on the southeastern coast of China have reported similar population expansion events, such as those occurring in L. savala [75], B. sinensis [23,24], Tridacna maxima [102], and Scatophagus argus [103]. However, the historical nucleotide diversity was higher than the current genetic diversity based on mtDNA data, indicating that the population of E. awoara showed a pattern of decline [67]. According to a bottleneck analysis based on microsatellite DNA, all E. awoara populations experienced a recent bottleneck. Thus, the present results showed that E. awoara presented a complex demographic history. In our approximate Bayesian computation (ABC) analysis, the demographic history of all E. awoara populations underwent a decline in population size following a recent population expansion. Following the late Pleistocene, we postulate that the sea level fluctuations of various scales within glacial periods promoted the bottleneck and recent population expansion in E. awoara. Today, E. awoara populations are threatened by severe climate change, habitat destruction and overfishing. Although their populations have historically expanded, like many other marine creatures, they have shrunk in recent times.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d14060439/s1, The values above the branches are bootstrap values for the NJ, ML, and BI analyses; Table S1: Repeat motifs and primers of 10 microsatellites of E. awoara; Table S2: The distribution information of the shared haplotypes of mitochondrial COI, Cyt b and ND2 sequence (Hap01–Hap82); Table S3: The distribution information of the shared haplotypes of RyR3 sequence (Hap01–Hap82); Table S4: Matrix of pairwise FST among four populations based on RyR3 gene; Table S5: Characteristics and genetic diversity indices for ten microsatellite loci in Epinephelus awoara; Table S6: Bottleneck results for four populations based on Wilcoxon’s signed-rank test; Figure S1: Phylogenetic trees reconstructed from mitochondrial sequences of COI Cyt b and ND2 gene in Epinephelus awoara; Figure S2: Mismatch distribution plot from mitochondrial sequences of COI Cyt b and ND2 gene in Epinephelus awoara.
Author Contributions
S.D. and H.L. designed research; K.Y. and R.L. performed research; K.Y. and H.L. analysed the data; K.Y., H.L. and S.D. wrote the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Key R & D Program of China (2018YFD0900803) and the National Natural Science Foundation of China (41976093).
Institutional Review Board Statement
This study was approved by the animal ethics committee of College of Ocean and Earth Sciences, Xiamen University.
Informed Consent Statement
Not applicable.
Data Availability Statement
The raw data which support this study are available in GenBank with accession numbers from ON324572 to ON324826.
Acknowledgments
We thank Jigui Yuan, Lina Wu, Sui Gu and other colleagues for their assistance during the fieldwork and data analysis.
Conflicts of Interest
All authors declare no conflict of interest.
References
- Avise, J.C.; Arnold, J.; Ball, R.M.; Bermingham, E.; Lamb, T.; Neigel, J.E.; Reeb, C.A.; Saunders, N.C. Intraspecific Phylogeography: The Mitochondrial DNA Bridge Between Population Genetics and Systematics. Annu. Rev. Ecol. Evol. Syst. 1987, 18, 489–522. [Google Scholar] [CrossRef]
- Hewitt, G.M. Genetic consequences of climatic oscillations in the Quaternary. Philos. Trans. R. Soc. B—Biol. Sci. 2004, 359, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Teske, P.R.; von der Heyden, S.; McQuaid, C.D.; Barker, N.P. A review of marine phylogeography in southern Africa. S. Afr. J. Sci. 2011, 107, 43–53. [Google Scholar] [CrossRef]
- Ibrahim, K.M.; Nichols, R.A.; Hewitt, G.M. Spatial patterns of genetic variation generated by different forms of dispersal during range expansion. Heredity 1996, 77, 282–291. [Google Scholar] [CrossRef]
- Bohonak, A.J. Dispersal, gene flow, and population structure. Q. Rev. Biol. 1999, 74, 21–45. [Google Scholar] [CrossRef]
- Liu, B.J.; Zhang, B.D.; Xue, D.X.; Gao, T.X.; Liu, J.X. Population Structure and Adaptive Divergence in a High Gene Flow Marine Fish: The Small Yellow Croaker (Larimichthys polyactis). PLoS ONE 2016, 11, e0144637. [Google Scholar] [CrossRef]
- Barber, P.H.; Palumbi, S.R.; Erdmann, M.V.; Moosa, M.K. Sharp genetic breaks among populations of Haptosquilla pulchella (Stomatopoda) indicate limits to larval transport: Patterns, causes, and consequences. Mol. Ecol. 2002, 11, 659–674. [Google Scholar] [CrossRef]
- Benzie, J.A.H. Major genetic differences between crown-of-thorns starfish (Acanthaster planci) populations in the Indian and Pacific Oceans. Evolution 1999, 53, 1782–1795. [Google Scholar] [CrossRef]
- Selkoe, K.A.; Toonen, R.J. Marine connectivity: A new look at pelagic larval duration and genetic metrics of dispersal. Mar. Ecol. Prog. Ser. 2011, 436, 291–305. [Google Scholar] [CrossRef]
- White, C.; Selkoe, K.A.; Watson, J.; Siegel, D.A.; Zacherl, D.C.; Toonen, R.J. Ocean currents help explain population genetic structure. Proc. R. Soc. B—Biol. Sci. 2010, 277, 1685–1694. [Google Scholar] [CrossRef]
- Kingsford, M.J.; Leis, J.M.; Shanks, A.; Lindeman, K.C.; Morgan, S.G.; Pineda, J. Sensory environments, larval abilities and local self-recruitment. Bull. Mar. Sci. 2002, 70, 309–340. [Google Scholar]
- Planes, S.; Fauvelot, C. Isolation by distance and vicariance drive genetic structure of a coral reef fish in the Pacific Ocean. Evolution 2002, 56, 378–399. [Google Scholar] [CrossRef] [PubMed]
- Jamaludin, N.A.; Mohd-Arshaad, W.; Akib, N.A.M.; Abidin, D.H.Z.; Nghia, N.V.; Nor, S.M. Phylogeography of the Japanese scad, Decapterus maruadsi (Teleostei; Carangidae) across the Central Indo-West Pacific: Evidence of strong regional structure and cryptic diversity. Mitochondrial DNA Part A 2020, 31, 298–310. [Google Scholar] [CrossRef] [PubMed]
- Salas, E.; Bernardi, G.; Berumen, M.L.; Gaither, M.; Rocha, L. RADseq analyses reveal concordant Indian Ocean biogeographic and phylogeographic boundaries in the reef fish Dascyllus trimaculatus. R. Soc. Open Sci. 2019, 6, 172413. [Google Scholar] [CrossRef]
- Tamaki, K.; Honza, E. Global tectonics and formation of marginal basins—Role of the western pacific. Episodes 1991, 14, 224–230. [Google Scholar] [CrossRef]
- Liu, J.X.; Gao, T.X.; Yokogawa, K.; Zhang, Y.P. Differential population structuring and demographic history of two closely related fish species, Japanese sea bass (Lateolabrax japonicus) and spotted sea bass (Lateolabrax maculatus) in Northwestern Pacific. Mol. Phylogenet. Evol. 2006, 39, 799–811. [Google Scholar] [CrossRef]
- Wang, P. Response of Western Pacific marginal seas to glacial cycles: Paleoceanographic and sedimentological features. Mar. Geol. 1999, 156, 5–39. [Google Scholar] [CrossRef]
- Chen, S.P.; Liu, T.; Li, Z.F.; Gao, T.X. Genetic population structuring and demographic history of red spotted grouper (Epinephelus akaara) in South and East China Sea. Afr. J. Biotechnol. 2008, 7, 3554–3562. [Google Scholar]
- Antoro, S.; Na-Nakorn, U.; Koedprang, W. Study of genetic diversity of orange-spotted grouper, Epinephelus coioides, from Thailand and Indonesia using microsatellite markers. Mar. Biotechnol. 2006, 8, 17–26. [Google Scholar] [CrossRef]
- Li, Y.; Han, Z.; Song, N.; Gao, T.X. New evidence to genetic analysis of small yellow croaker (Larimichthys polyactis) with continuous distribution in China. Biochem. Syst. Ecol. 2013, 50, 331–338. [Google Scholar] [CrossRef]
- Yan, S.; Catanese, G.; Brown, C.L.; Wang, M.; Yang, C.; Yang, T. Phylogeographic study on the chub mackerel (Scomber japonicus) in the Northwestern Pacific indicates the late Pleistocene population isolation. Mar. Ecol. 2015, 36, 753–765. [Google Scholar] [CrossRef]
- Palumbi, S. Molecular biogeography of the Pacific. Coral Reefs 1997, 16, S47–S52. [Google Scholar] [CrossRef]
- Qiu, F.; Li, H.; Lin, H.; Ding, S.; Miyamoto, M.M. Phylogeography of the inshore fish, Bostrychus sinensis, along the Pacific coastline of China. Mol. Phylogenet. Evol. 2016, 96, 112–117. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.; Mishra, M.; Wu, H.; Liang, S.; Miyamoto, M.M. Characterization of hybridization within a secondary contact region of the inshore fish, Bostrychus sinensis, in the East China Sea. Heredity 2018, 120, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Xu, H.; Chen, Z.; Wang, Y. Marine fish cage culture in China. The Future of Mariculture: A Regional Approach for Responsible Development in the Asia-Pacific Region. 2008; p. 285. Available online: https://www.fao.org/3/i0202e/i0202e14.pdf (accessed on 19 August 2021).
- Coker, D.J.; DiBattista, J.D.; Sinclair-Taylor, T.H.; Berumen, M.L. Spatial patterns of cryptobenthic coral-reef fishes in the Red Sea. Coral Reefs 2018, 37, 193–199. [Google Scholar] [CrossRef]
- Dooley, J. A Checklist of Fishes of the South China Sea: Malacanthidae. In A Checklist of Fishes of the South China Sea; National University of Singapore: Singapore, 2000; pp. 569–667. [Google Scholar]
- Lindberg, G.U.; Krasyukova, Z.V. Fishes of the Sea of Japan and the Adjacent Areas of the Sea of Okhotsk and the Yellow Sea; CRC Press: Boca Raton, FL, USA, 2020. [Google Scholar]
- To, A.W.; Shea, S.K. Rapid Reef Fish Survey at Daya Bay, Guangdong Province, China; City University of Hong Kong: Hong Kong, 2015. [Google Scholar]
- Hodgkiss, I. Fifty common marine food fishes of Hong Kong. Mem. Hong Kong Nat. Hist. Soc. 1988, 18, 19–33. [Google Scholar]
- Cornish, A.S. The reef fish species of the Cape Daguilar Marine Reserve, Hoi Ha Wan Marine Park, Yan Chau Tong Marine Park and Ping Chau, Hong Kong. Mar. Flora Fauna Hong Kong S. China V 2000, 5, 369. [Google Scholar]
- Heemstra, P.C.; Randall, J.E. Groupers of the world. FAO Fish. Synop. 1993, 16, 1. [Google Scholar]
- Sun, L.; Chen, H.; Huang, L. Growth, faecal production, nitrogenous excretion and energy budget of juvenile yellow grouper (Epinephelus awoara) relative to ration level. Aquaculture 2007, 264, 228–235. [Google Scholar] [CrossRef]
- Zhou, Z.G.; Liu, Y.C.; Shi, P.J.; He, S.X.; Yao, B.; Ringo, E. Molecular characterization of the autochthonous microbiota in the gastrointestinal tract of adult yellow grouper (Epinephelus awoara) cultured in cages. Aquaculture 2009, 286, 184–189. [Google Scholar] [CrossRef]
- Xu, X.J.; Sang, B.H.; Luo, G. A genetic role for macrophage migration inhibitory factor (MIF) in Epinephelus awoara infected with Vibrio parahaemolyticus. Eur. J. Inflamm. 2016, 14, 184–189. [Google Scholar] [CrossRef]
- Doyle, M.; Morse, W.; Jr, A. A Comparison of Larval Fish Assemblages in the Temperate Zone of the Northeast Pacific and Northwest Atlantic Oceans. Bull. Mar. Sci. 1993, 53, 588–644. [Google Scholar]
- Kim, W.J.; Kim, K.K.; Han, H.S.; Nam, B.H.; Kim, Y.O.; Kong, H.J.; Noh, J.K.; Yoon, M. Population structure of the olive flounder (Paralichthys olivaceus) in Korea inferred from microsatellite marker analysis. J. Fish Biol. 2010, 76, 1958–1971. [Google Scholar] [CrossRef] [PubMed]
- Umino, T.; Kajihara, T.; Shiozaki, H.; Ohkawa, T.; Jeong, D.-S.; Ohara, K. Wild stock structure of Girella punctata in Japan revealed shallow genetic differentiation but subtle substructure in subsidiary distributions. Fish. Sci. 2009, 75, 909–919. [Google Scholar] [CrossRef]
- Buchholz-Sørensen, M.; Vella, A. Population structure, genetic diversity, effective population size, demographic history and regional connectivity patterns of the endangered dusky grouper, Epinephelus marginatus (Teleostei: Serranidae), within Malta’s fisheries management zone. PLoS ONE 2016, 11, e0159864. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, S.K.; Jun, W.; Yong-Ouan, S.; Shao-Xiong, D.; Chaturvedi, S. Genetic diversity of yellow grouper (Epinephelus awoara) determined by random amplified polymorphic DNA (RAPD) analysis. Fish. Bull. 2006, 104, 638–642. [Google Scholar]
- Zhao, L.; Shao, C.; Liao, X.; Ma, H.; Zhu, X.; Chen, S. Twelve novel polymorphic microsatellite loci for the Yellow grouper (Epinephelus awoara) and cross-species amplifications. Conserv. Genet. 2009, 10, 743–745. [Google Scholar] [CrossRef]
- Karim, A.A.; Norziah, M.H.; Seow, C.C. Methods for the study of starch retrogradation. Food Chem. 2000, 71, 9–36. [Google Scholar] [CrossRef]
- Arias, M.C.; Aulagnier, S.; Baerwald, E.F.; Barclay, R.M.; Batista, J.S.; Beasley, R.R.; Bezerra, R.A.; Blanc, F.; Bridge, E.S.; Cabria, M.T. Microsatellite records for volume 8, issue 1. Conserv. Genet. Resour. 2016, 8, 43–81. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Lefort, V.; Longueville, J.-E.; Gascuel, O. SMS: Smart model selection in PhyML. Mol. Biol. Evol. 2017, 34, 2422–2424. [Google Scholar] [CrossRef] [PubMed]
- Leigh, J.; Bryant, D. PopART: Full-Feature Software for Haplotype Network Construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Bohonak, A.J. IBD (isolation by distance): A program for analyses of isolation by distance. J. Hered. 2002, 93, 153–154. [Google Scholar] [CrossRef] [PubMed]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Fu, Y.-X. Statistical Tests of Neutrality of Mutations Against Population Growth, Hitchhiking and Background Selection. Genetics 1997, 147, 915–925. [Google Scholar] [CrossRef]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian Phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef]
- Bouckaert, R.; Heled, J.; Kühnert, D.; Vaughan, T.; Wu, C.-H.; Xie, D.; Suchard, M.A.; Rambaut, A.; Drummond, A.J. BEAST 2: A software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2014, 10, e1003537. [Google Scholar] [CrossRef] [PubMed]
- Kumazawa, Y. Molecular clock estimation in fishes and its application to biogeographical studies. Fish. Sci. 2002, 68, 357–360. [Google Scholar] [CrossRef][Green Version]
- Bermingham, E.; McCafferty, S.S.; Martin, A. Fish biogeography and molecular clocks: Perspectives from the Panamanian Isthmus. Mol. Syst. Fishes 1997, 113–128. [Google Scholar]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. Micro-Checker: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Goudet, J. FSTAT (Version 2.9.3): A Program to Estimate and Test Gene Diversities and Fixation Indices. 2001. Available online: www.unil.ch/izea/softwares/fstat.html (accessed on 19 August 2021).
- Weir, B.S.; Cockerham, C.C. estimating f-statistics for the analysis of population-structure. Evolution 1984, 38, 1358–1370. [Google Scholar] [CrossRef]
- Earl, D.A.; Vonholdt, B.M. Structure Harvester: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Nei, M. Genetic Distance between Populations. Am. Nat. 1972, 106, 283–292. [Google Scholar] [CrossRef]
- Piry, S.; Luikart, G.; Cornuet, J.M. Bottleneck: A computer program for detecting recent reductions in the effective population size using allele frequency data. J. Hered. 1999, 90, 502–503. [Google Scholar] [CrossRef]
- Cornuet, J.-M.; Pudlo, P.; Veyssier, J.; Dehne-Garcia, A.; Gautier, M.; Leblois, R.; Marin, J.-M.; Estoup, A. DIYABC v2. 0: A software to make approximate Bayesian computation inferences about population history using single nucleotide polymorphism, DNA sequence and microsatellite data. Bioinformatics 2014, 30, 1187–1189. [Google Scholar] [CrossRef]
- Cabrera, A.A.; Palsbøll, P.J. Inferring past demographic changes from contemporary genetic data: A simulation-based evaluation of the ABC methods implemented in DIYABC. Mol. Ecol. Resour. 2017, 17, e94–e110. [Google Scholar] [CrossRef] [PubMed]
- Templeton, A.R.; Sing, C. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping. IV. Nested analyses with cladogram uncertainty and recombination. Genetics 1993, 134, 659–669. [Google Scholar] [CrossRef] [PubMed]
- Billington, N.; Hebert, P. Mitochondrial DNA diversity in fishes and its implications for introductions. Can. J. Fish. Aquat. Sci. 1991, 48, 80–94. [Google Scholar] [CrossRef]
- Booy, G.; Hendriks, R.J.J.; Smulders, M.J.M.; Van Groenendael, J.M.; Vosman, B. Genetic diversity and the survival of populations. Plant Biol. 2000, 2, 379–395. [Google Scholar] [CrossRef]
- Searle, J.B. Phylogeography—The History and Formation of Species. Heredity 2000, 85, 201. [Google Scholar] [CrossRef]
- Hoelzel, A.; Hancock, J.; Dover, G. Evolution of the cetacean mitochondrial D-loop region. Mol. Biol. Evol. 1991, 8, 475–493. [Google Scholar]
- Sun, P.; Shi, Z.-h.; Yin, F.; Peng, S.-m. Genetic Variation Analysis of Mugil cephalus in China Sea Based on Mitochondrial COI Gene Sequences. Biochem. Genet. 2012, 50, 180–191. [Google Scholar] [CrossRef]
- Hua, X.; Wang, W.; Yin, W.; He, Q.; Jin, B.; Li, J.; Chen, J.; Fu, C. Phylogeographical analysis of an estuarine fish, Salanx ariakensis (Osmeridae: Salanginae) in the north-western Pacific. J. Fish Biol. 2009, 75, 354–367. [Google Scholar] [CrossRef]
- Yu, Z.N.; Kong, X.Y.; Guo, T.H.; Jiang, Y.-y.; Zhuang, Z.-m.; Jin, X.-s. Mitochondrial DNA sequence variation of Japanese anchovy Engraulis japonicus from the Yellow Sea and East China Sea. Fish. Sci. 2005, 71, 299. [Google Scholar] [CrossRef]
- Gu, S.; Yi, M.-R.; He, X.-B.; Lin, P.-S.; Liu, W.-H.; Luo, Z.-S.; Lin, H.-D.; Yan, Y.-R. Genetic diversity and population structure of cutlassfish (Lepturacanthus savala) along the coast of mainland China, as inferred by mitochondrial and microsatellite DNA markers. Reg. Stud. Mar. Sci. 2021, 43, 101702. [Google Scholar] [CrossRef]
- Zeng, L.; Cheng, Q.; Chen, X. Microsatellite analysis reveals the population structure and migration patterns of Scomber japonicus (Scombridae) with continuous distribution in the East and South China Seas. Biochem. Syst. Ecol. 2012, 42, 83–93. [Google Scholar] [CrossRef]
- Wang, X.; Weng, Z.; Yang, Y.; Hua, S.; Zhang, H.; Meng, Z. Genetic Evaluation of Black Sea Bream (Acanthopagrus schlegelii) Stock Enhancement in the South China Sea Based on Microsatellite DNA Markers. Fishes 2021, 6, 47. [Google Scholar] [CrossRef]
- Sun, D.-Q.; Shi, G.E.; Liu, X.-Z.; Wang, R.-X.; Xu, T.-J. Genetic diversity and population structure of the marbled rockfish, Sebastiscus marmoratus, revealed by SSR markers. J. Genet. 2013, 92, 21–24. [Google Scholar] [CrossRef]
- Falconer, D.S.; Mackay, T.F. Quantitative Genetics; Longman: London, UK, 1983. [Google Scholar]
- To, A.W.; De Mitcheson, Y.S. Shrinking baseline: The growth in juvenile fisheries, with the Hong Kong grouper fishery as a case study. Fish Fish. 2009, 10, 396–407. [Google Scholar] [CrossRef]
- Chen, B.; Luo, H.Z.; Fu, R.B. Biology and hatchery of Epinephelus awoara. Hebei Fish. 2006, 2, 29–31. [Google Scholar]
- To, W.L. The Biology, Fisheries of Groupers (Family: Serranidae) in Hong Kong and Adjacent Waters, and Implications for Management. Ph.D Thesis, University of Hong Kong, Hong Kong, 2009. [Google Scholar]
- Bjørnstad, O.N.; Grenfell, B.T. Noisy clockwork: Time series analysis of population fluctuations in animals. Science 2001, 293, 638–643. [Google Scholar] [CrossRef]
- Sun, J.; Cornelius, S.P.; Janssen, J.; Gray, K.A.; Motter, A.E. Regularity underlies erratic population abundances in marine ecosystems. J. R. Soc. Interface 2015, 12, 20150235. [Google Scholar] [CrossRef]
- Banks, S.C.; Cary, G.J.; Smith, A.L.; Davies, I.D.; Driscoll, D.A.; Gill, A.M.; Lindenmayer, D.B.; Peakall, R. How does ecological disturbance influence genetic diversity? Trends Ecol. Evol. 2013, 28, 670–679. [Google Scholar] [CrossRef]
- Romiguier, J.; Gayral, P.; Ballenghien, M.; Bernard, A.; Cahais, V.; Chenuil, A.; Chiari, Y.; Dernat, R.; Duret, L.; Faivre, N. Comparative population genomics in animals uncovers the determinants of genetic diversity. Nature 2014, 515, 261–263. [Google Scholar] [CrossRef]
- Ellegren, H.; Galtier, N. Determinants of genetic diversity. Nat. Rev. Genet. 2016, 17, 422–433. [Google Scholar] [CrossRef]
- Wright, S. Size of population and breeding structure in relation to evolution. Science 1938, 87, 430–431. [Google Scholar]
- Palumbi, S.R. Genetic divergence, reproductive isolation, and marine speciation. Annu. Rev. Ecol. Syst. 1994, 25, 547–572. [Google Scholar] [CrossRef]
- Shi, M.; Chen, C.; Xu, Q.; Lin, H.; Liu, G.; Wang, H.; Wang, F.; Yan, J. The role of Qiongzhou Strait in the seasonal variation of the South China Sea circulation. J. Phys. Oceanogr. 2002, 32, 103–121. [Google Scholar] [CrossRef]
- Chen, C.; Li, P.; Shi, M.; Zuo, J.; Chen, M.; Sun, H. Numerical study of the tides and residual currents in the Qiongzhou Strait. Chin. J. Oceanol. Limnol. 2009, 27, 931–942. [Google Scholar] [CrossRef]
- Qiu, B.; Imasato, N. A numerical study on the formation of the Kuroshio Counter Current and the Kuroshio Branch Current in the East China Sea. Cont. Shelf Res. 1990, 10, 165–184. [Google Scholar] [CrossRef]
- Zhao, D.; Li, Q.; Kong, L.; Yu, H. Cryptic diversity of marine gastropod Monodonta labio (Trochidae): Did the early Pleistocene glacial isolation and sea surface temperature gradient jointly drive diversification of sister species and/or subspecies in the Northwestern Pacific? Mar. Ecol. 2017, 38, e12443. [Google Scholar] [CrossRef]
- Wang, Z.-D.; Liao, J.; Huang, C.-Q.; Long, S.-S.; Zhang, S.; Guo, Y.-S.; Liu, L.; Liu, C.-W. Significant genetic differentiation of Gobiopterus lacustris, a newly recorded transparent goby in China. Mitochondrial DNA Part A 2018, 29, 785–791. [Google Scholar] [CrossRef]
- Peng, S.; Shi, Z.; Hou, J.; Wang, W.; Zhao, F.; Zhang, H. Genetic diversity of silver pomfret (Pampus argenteus) populations from the China Sea based on mitochondrial DNA control region sequences. Biochem. Syst. Ecol. 2009, 37, 626–632. [Google Scholar] [CrossRef]
- Voris, H.K. Maps of Pleistocene sea levels in Southeast Asia: Shorelines, river systems and time durations. J. Biogeogr. 2000, 27, 1153–1167. [Google Scholar] [CrossRef]
- Liu, J.X.; Gao, T.X.; Wu, S.F.; Zhang, Y.P. Pleistocene isolation in the Northwestern Pacific marginal seas and limited dispersal in a marine fish, Chelon haematocheilus (Temminck & Schlegel, 1845). Mol. Ecol. 2007, 16, 275–288. [Google Scholar]
- Liu, S.-Y.V.; Huang, I.-H.; Liu, M.-Y.; Lin, H.-D.; Wang, F.-Y.; Liao, T.-Y. Genetic stock structure of Terapon jarbua in Taiwanese waters. Mar. Coast. Fish. 2015, 7, 464–473. [Google Scholar] [CrossRef]
- Imbrie, J.; Boyle, E.; Clemens, S.; Duffy, A.; Howard, W.; Kukla, G.; Kutzbach, J.; Martinson, D.; McIntyre, A.; Mix, A. On the structure and origin of major glaciation cycles 1. Linear responses to Milankovitch forcing. Paleoceanography 1992, 7, 701–738. [Google Scholar] [CrossRef]
- Hewitt, G. The genetic legacy of the Quaternary ice ages. Nature 2000, 405, 907–913. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.Q.; Gao, T.X.; Yanagimoto, T.; Sakurai, Y. Genetic population structure of Nibea albiflora in yellow sea and east china sea. Fish. Sci. 2008, 74, 544–552. [Google Scholar] [CrossRef]
- Neo, M.L.; Liu, L.-L.; Huang, D.; Soong, K. Thriving populations with low genetic diversity in giant clam species, Tridacna maxima and Tridacna noae, at Dongsha Atoll, South China Sea. Reg. Stud. Mar. Sci. 2018, 24, 278–287. [Google Scholar] [CrossRef]
- Yan, Y.-R.; Hsu, K.-C.; Yi, M.-R.; Li, B.; Wang, W.-K.; Kang, B.; Lin, H.-D. Cryptic diversity of the spotted scat Scatophagus argus (Perciformes: Scatophagidae) in the South China Sea: Pre-or post-production isolation. Mar. Freshw. Res. 2020, 71, 1640–1650. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).