Extracting Quantitative Information from Images Taken in the Wild: A Case Study of Two Vicariants of the Ophrys aveyronensis Species Complex
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study System
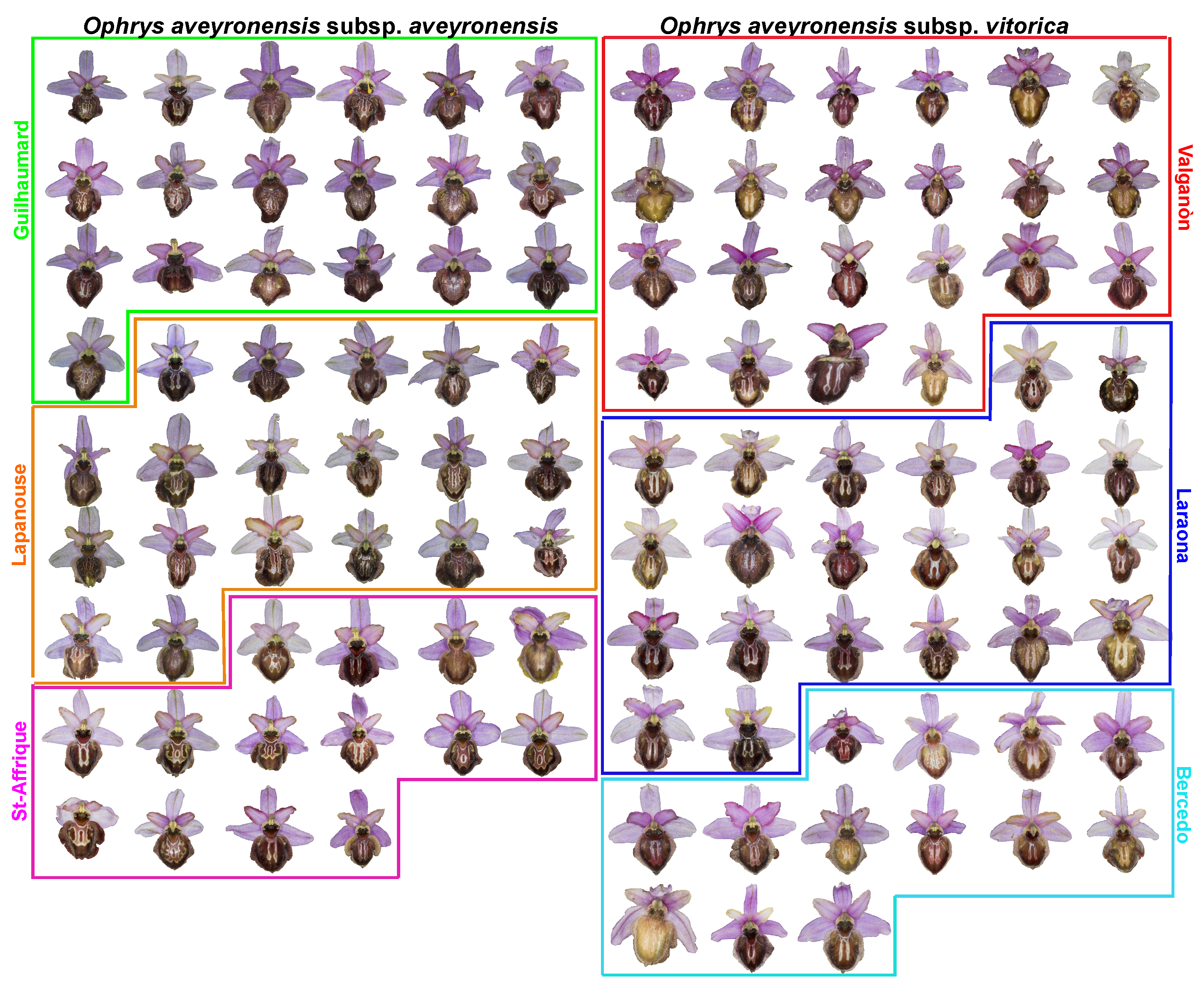
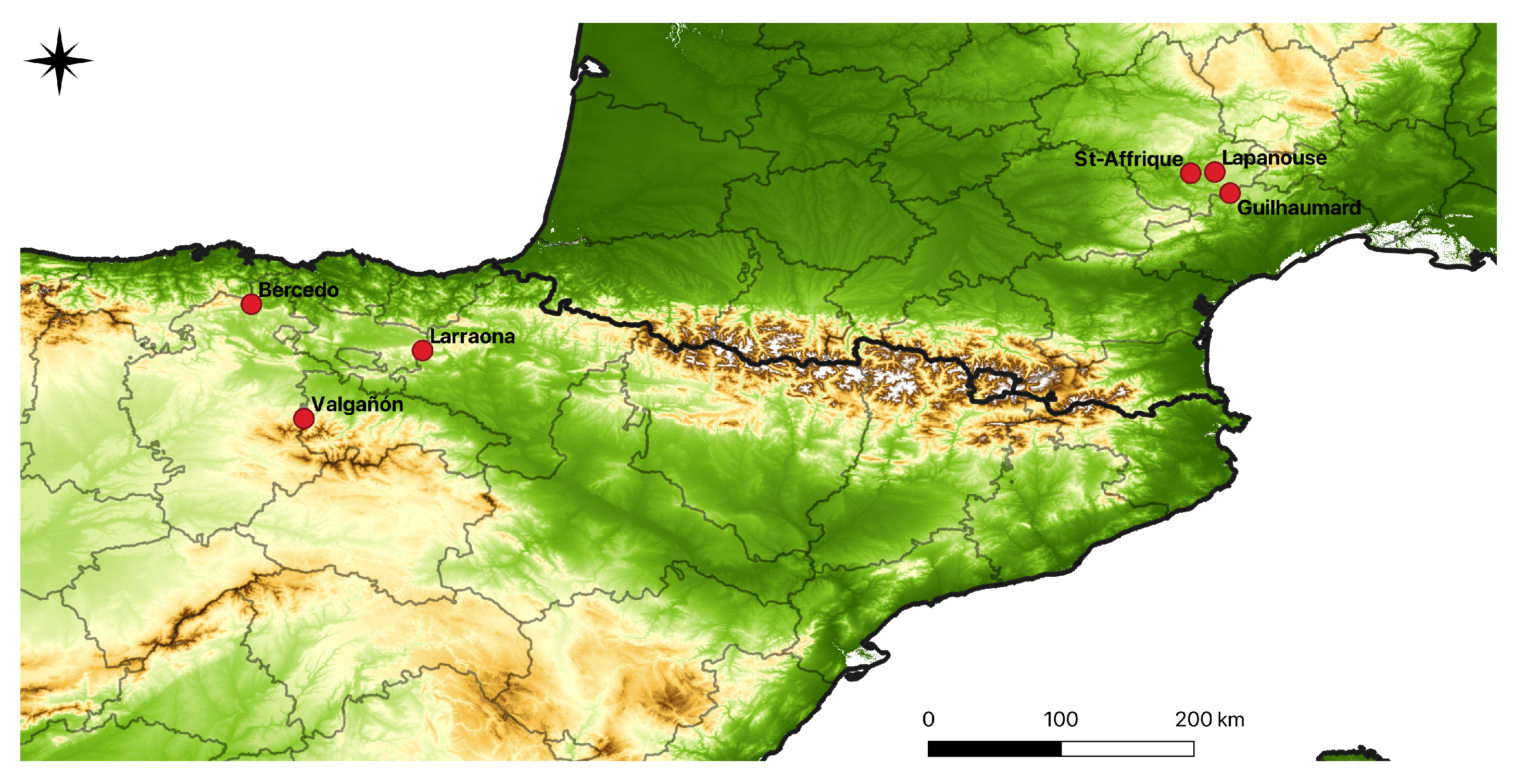
2.2. Collection Sites and Plant Material
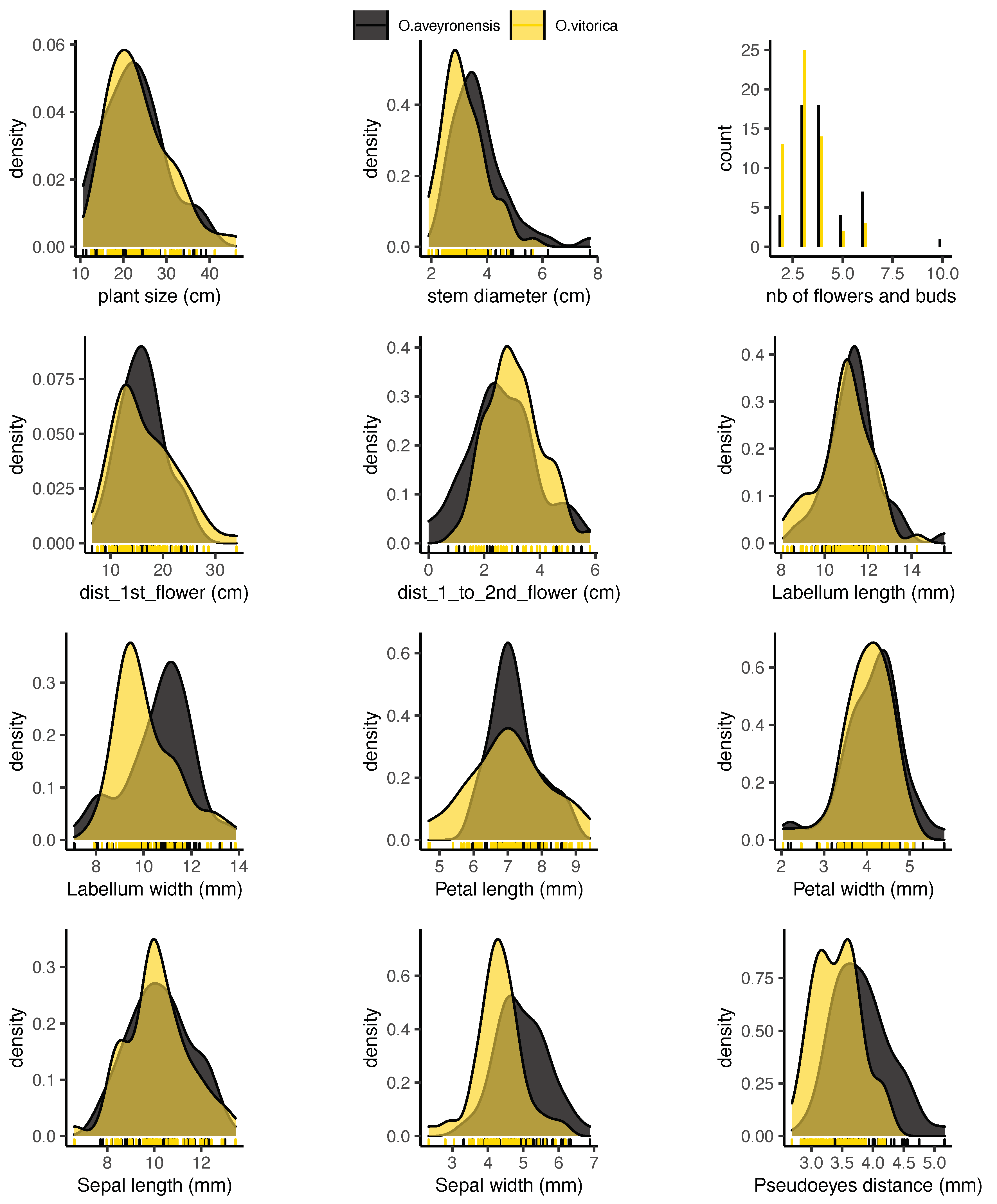
2.3. Morphological Traits Measured in the Field
2.4. Information Extracted from Images
2.4.1. Picture Acquisition
2.4.2. Post-Processing and Filtering of the Photographic Database Elements
2.4.3. Morphological Traits Extracted from Images
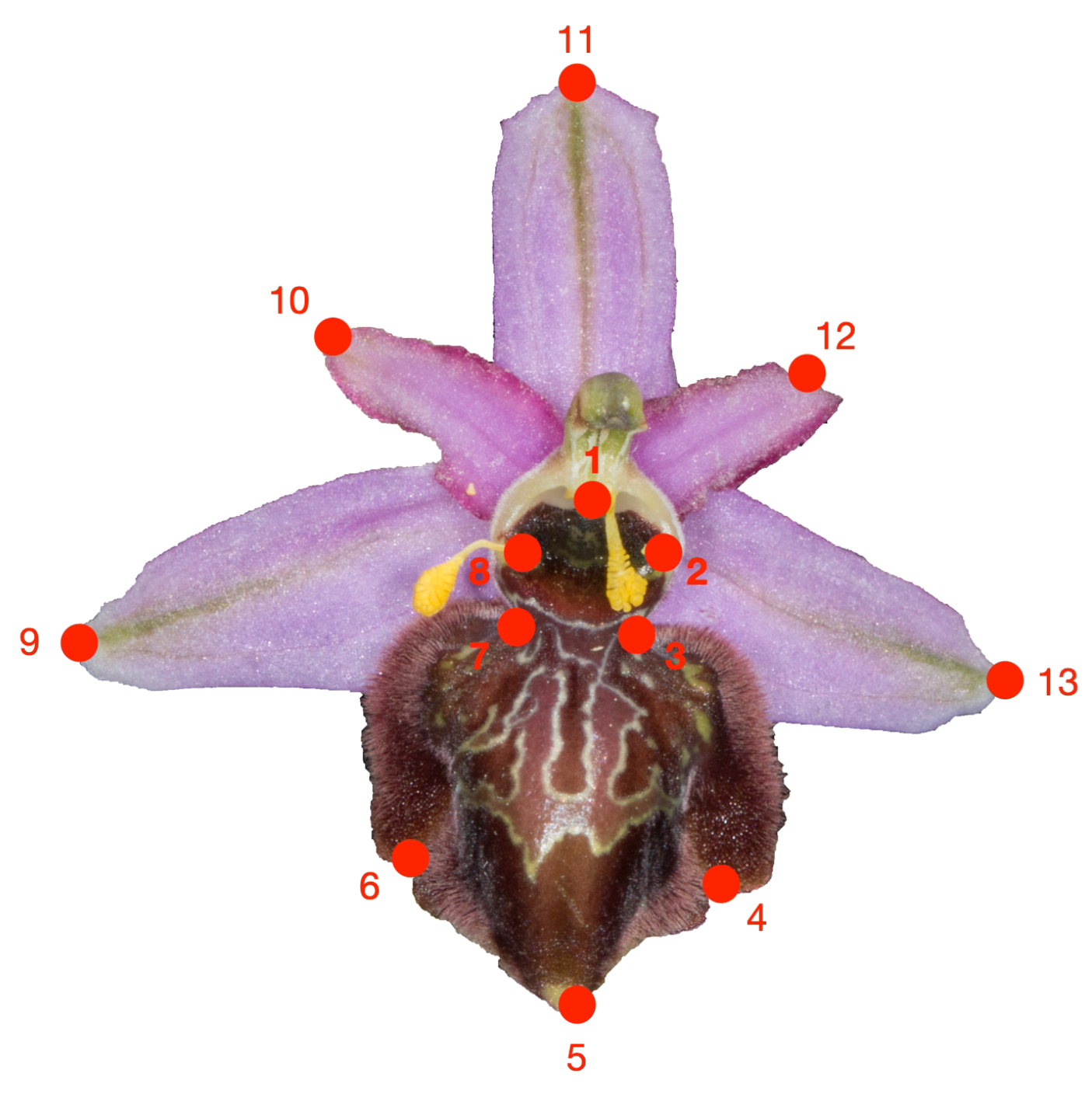
2.4.4. Flower Shape
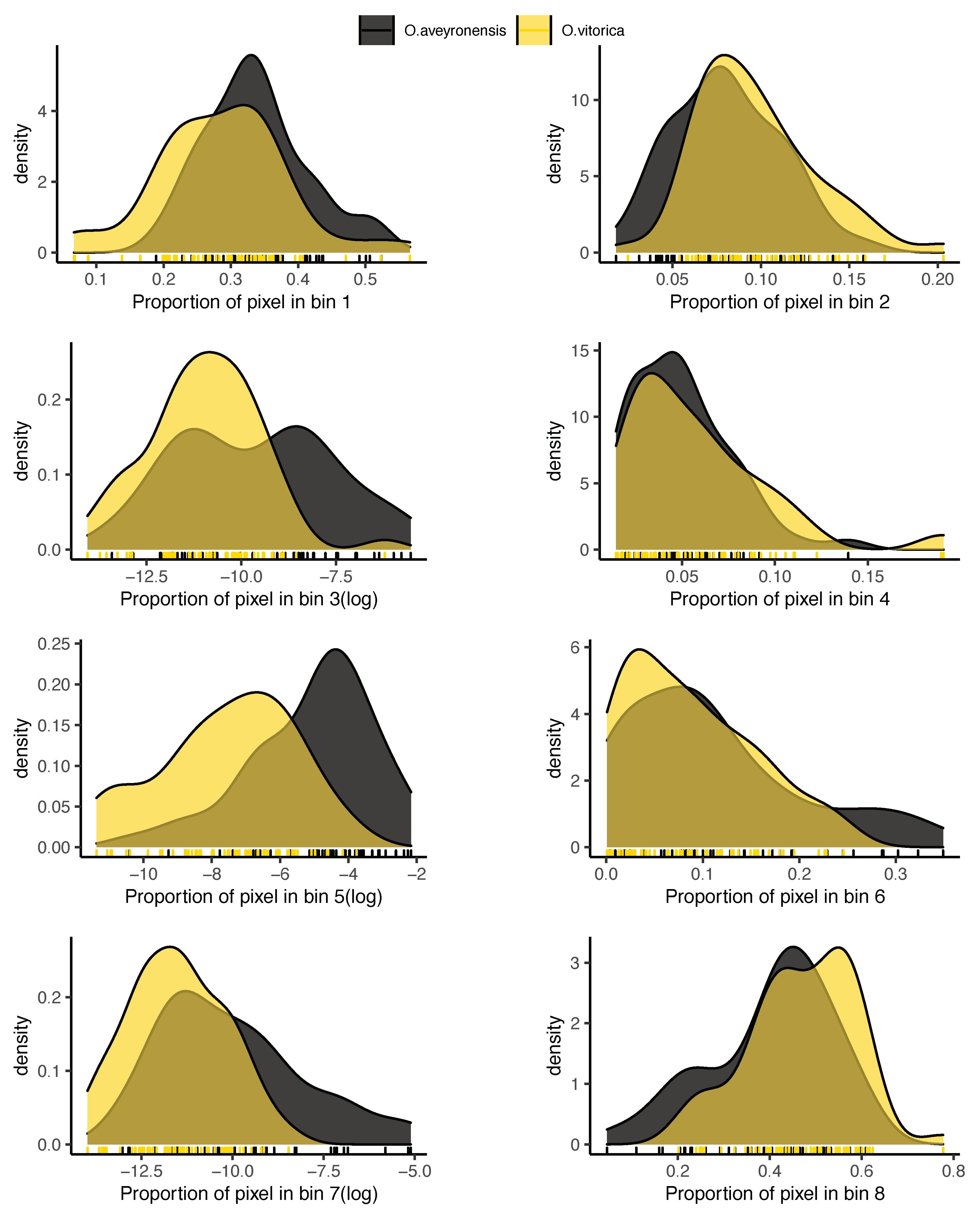
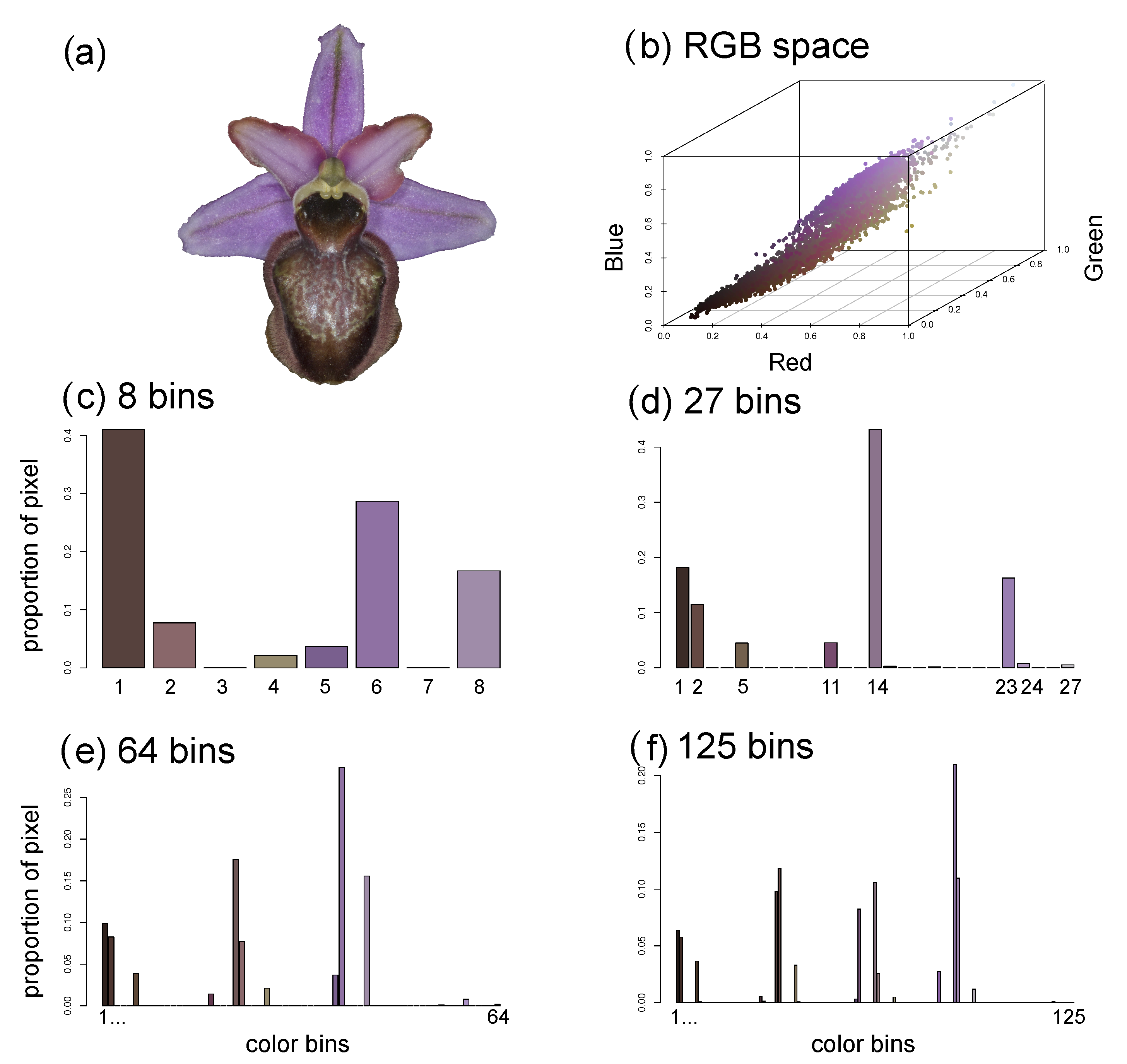
2.4.5. Flower Colour
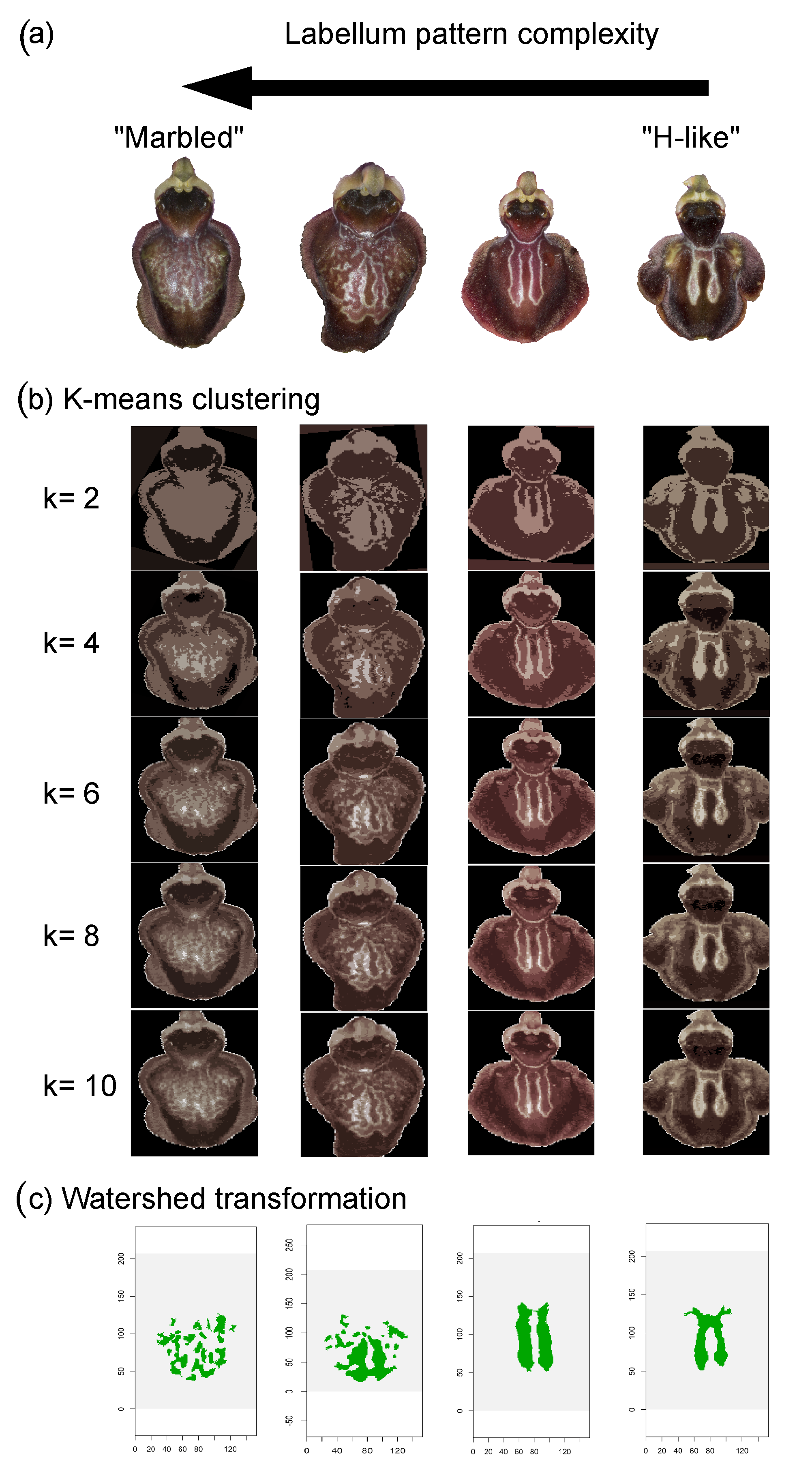
2.4.6. Labellum Colour Pattern
2.5. Statistical Analysis
2.5.1. Random Forest (RF)
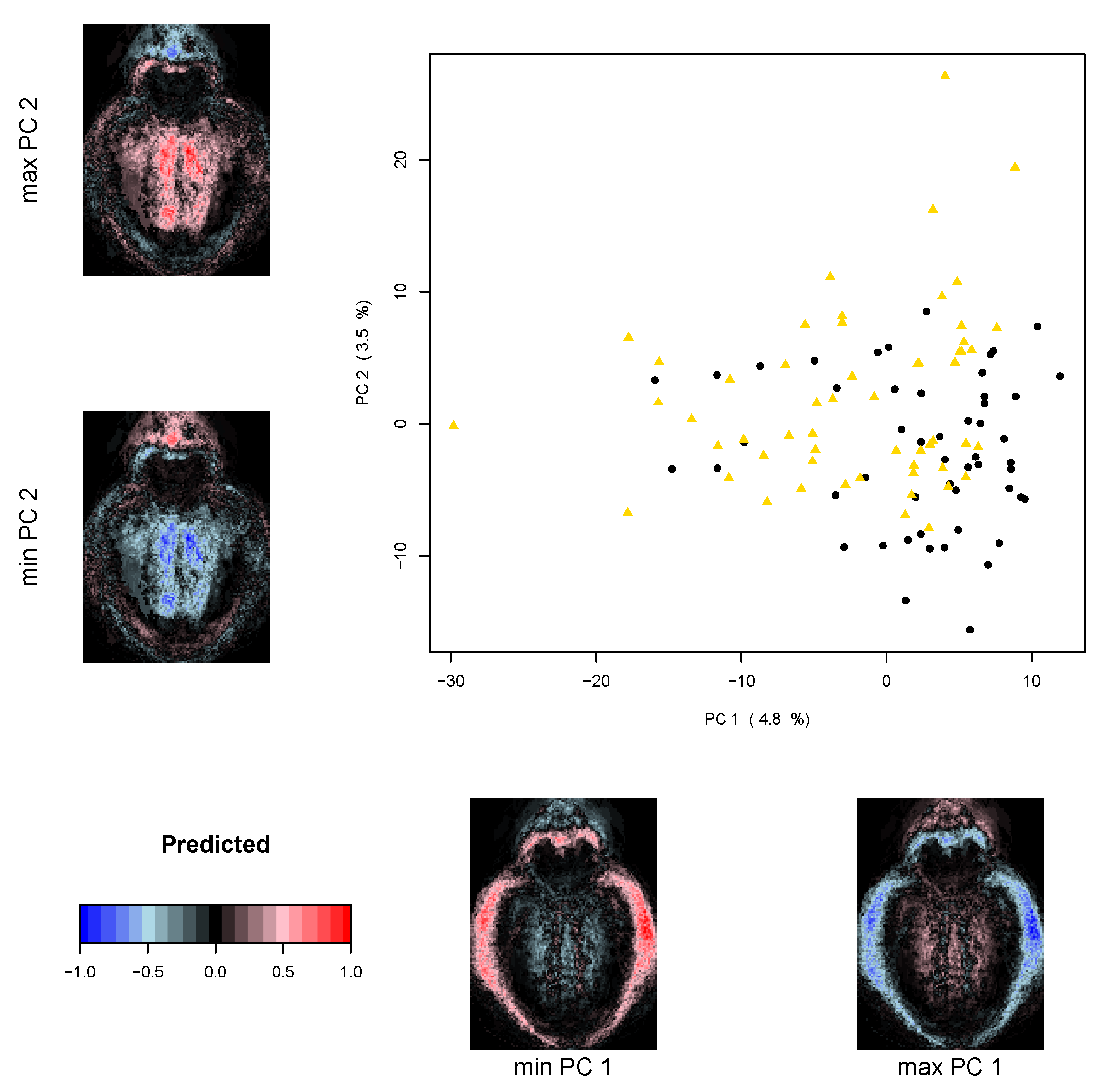
2.5.2. Multivariate Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| RF | Random forest |
Appendix A
Appendix A.1
| Subspecies | Population | Nb of Individuals | Latitude | Longitude |
|---|---|---|---|---|
| O. a. subsp. aveyronensis | St-Affrique | 14 | 43.98 | 2.93 |
| O. a. subsp. aveyronensis | Lapanouse | 19 | 43.98 | 3.09 |
| O. a. subsp. aveyronensis | Guilhaumard | 19 | 43.84 | 3.19 |
| O. a. subsp. vitorica | Bercedo | 13 | 43.09 | -3.43 |
| O. a. subsp. vitorica | Larraona | 22 | 42.78 | -2.27 |
| O. a. subsp. vitorica | Valgañón | 22 | 42.31 | -3.08 |
| Criteria | O. a. subsp. aveyronensis | O. a. subsp. vitorica |
|---|---|---|
| Number of flowers | 5–8 sometimes 3–12 (2–10) | 2–5 (2–6) |
| Sepal length | 10–16 mm (8–13) | 8–10.5 mm (7–13.5) |
| Sepal width | 6–8 mm (3.5–7) | 3–5 mm (2.5–6) |
| Petal length | 7–10 mm (6–9) | 4–6.5 mm (4.5–9.5) |
| Petal width | 4–5.5 mm (2–6) | 2–4 mm (2–5) |
| Labellum length | 11–15 mm (8.5–15.5) | 10–13.5 mm (8–14) |
| Labellum width | 12–18 mm (7–13) | 11–15 mm (8–14) |
| Labellum shape and colour pattern | gibosity, hairiness, “marbled” and varied macula | “H-like” shape macula |
| Sepal colour | light to dark pink, rarely creamy white, exceptionally greenish | pink to pale purple |
Appendix A.2
References
- Bertrand, J.A.M.; Borsa, P.; Chen, W.J. Phylogeography of the sergeants Abudefduf sexfasciatus A. vaigiensis reveal complex introgression patterns of two widespread sympatric Indo-West Pacific reef fishes. Mol. Ecol. 2017, 9, 2527–2542. [Google Scholar] [CrossRef] [PubMed]
- Fišer, C.; Robinson, C.T.; Malard, F. Cryptic species as a window into the paradigm shift of the species concept. Mol. Ecol. 2016, 27, 613–635. [Google Scholar] [CrossRef] [PubMed]
- Sassone, A.B.; Arroyo, M.T.K.; Arroyo-Leuenberger, S.C.; García, N.; Román, M.J. One species with a disjunct distribution or two with convergent evolution? Taxon 2021, 4, 842–853. [Google Scholar] [CrossRef]
- Karanovic, T.; Djurakic, M.; Eberhard, S.M. Cryptic species or inadequate taxonomy? Implementation of 2D geometric morphometrics based on integumental organs as landmarks for delimitation and description of copepod taxa. Syst. Biol. 2016, 65, 304–327. [Google Scholar] [CrossRef] [Green Version]
- Korshunova, T.; Picton, B.; Furfaro, G.; Mariottini, P.; Pontes, M.; Prkić, J.; Fletcher, K.; Malmberg, K.; Lundin, K.; Martynov, A. multilevel fine-scale diversity challenges the ‘cryptic species’ concept. Sci. Rep. 2019, 9, 6732. [Google Scholar] [CrossRef]
- Lürig, M.D.; Donoughe, S.; Porto, A.; Tsuboi, M. Computer vision, machine learning, and the promise of phenomics in ecology and evolutionary biology. Front. Ecol. Evol. 2021, 9, 148. [Google Scholar] [CrossRef]
- Joly, A.; Bonnet, P.; Goëau, H.; Barbe, J.; Selmi, S.; Champ, J.; Dufour-Kowalski, S.; Affouard, A.; Carré, J.; Molino, J.F.; et al. A look inside the Pl@ntNet experience: The good, the bias and the hope. Multimed. Syst. 2016, 22, 751–766. [Google Scholar] [CrossRef] [Green Version]
- Wäldchen, J.; Rzanny, M.; Seeland, M.; Mäder, P.; Bucksch, A. Automated plant species identification—Trends and future directions. PLoS Comput. Biol. 2018, 14, e1005993. [Google Scholar] [CrossRef] [Green Version]
- Bersweden, L.; Viruel, J.; Schatz, B.; Harland, J.; Gargiulo, R.; Cowan, R.S.; Calevo, J.; Juan, A.; Clarkson, J.J.; Leitch, A.R.; et al. Microsatellites and petal morphology reveal new patterns of admixture in Orchis Hybrid Zones. Am. J. Bot. 2021, 8, 1388–1404. [Google Scholar] [CrossRef]
- Yang, W.; Feng, H.; Zhang, X.; Zhang, J.; Doonan, J.H.; Batchelor, W.D.; Xiong, L.; Yan, J. Crop phenomics and high-throughput phenotyping: Past decades, current challenges, and future perspectives. Mol. Plant 2020, 13, 187–214. [Google Scholar] [CrossRef] [Green Version]
- Trunschke, J.; Lunau, K.; Pyke, G.H.; Ren, Z.X.; Wang, H. Color evolution and the evidence of pollinator-mediated selection. Front. Plant Sci. 2021, 12, 1–20. [Google Scholar]
- Joffard, N.; Le Roncé, I.; Langlois, A.; Renoult, J.; Buatois, B.; Dormont, L.; Schatz, B. Floral trait differentiation in Anacamptis coriophora: Phenotypic selection on scents, but not on colour. J. Evol. Biol. 2020, 33, 1028–1038. [Google Scholar] [CrossRef] [PubMed]
- Troscianko, J.; Stevens, M. Image calibration and analysis toolbox—A free software suite for objectively measuring reflectance, colour and pattern. Methods Ecol. Evol. 2015, 11, 1320–1331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, I.Z.W.; Stevens, M.; Todd, P.A. PAT-GEOM: A software package for the analysis of animal patterns. Methods Ecol. Evol. 2015, 4, 591–600. [Google Scholar] [CrossRef] [Green Version]
- Porto, A.; Voje, K.L. ML-morph: A fast, accurate and general approach for automated detection and landmarking of biological structures in images. Methods Ecol. Evol. 2020, 11, 500–512. [Google Scholar] [CrossRef] [Green Version]
- van den Berg, C.P.; Troscianko, J.; Endler, J.A.; Marshall, N.J.; Cheney, K.L. Quantitative Colour Pattern Analysis (QCPA): A comprehensive framework for the analysis of colour patterns in nature. Methods Ecol. Evol. 2020, 2, 316–332. [Google Scholar] [CrossRef]
- Porto, A.; Rolfe, S.; Maga, A.M. ALPACA: A fast and accurate computer vision approach for automated landmarking of three-dimensional biological structures. Methods Ecol. Evol. 2021, 12, 2129–2144. [Google Scholar] [CrossRef]
- Ott, T.; Lautenschlager, U. GinJinn2: Object detection and segmentation for ecology and evolution. Methods Ecol. Evol. 2021, 13, 603–610. [Google Scholar] [CrossRef]
- Schwartz, S.T.; Alfaro, M.E. Sashimi: A toolkit for facilitating high-throughput organismal image segmentation using deep learning. Methods Ecol. Evol. 2020, 12, 2341–2354. [Google Scholar] [CrossRef]
- Valvo, J.J.; Aponte, J.D.; Daniel, M.J.; Dwinell, K.; Rodd, H.; Houle, D.; Hughes, K.A. Using Delaunay triangulation to sample whole-specimen color from digital images. Ecol. Evol. 2021, 11, 12468–12484. [Google Scholar] [CrossRef]
- Adams, D.C.; Otárola-Castillo, E. Geomorph: An package for the collection and analysis of geometric morphometric shape data. Methods Ecol. Evol. 2013, 4, 393–399. [Google Scholar] [CrossRef]
- Bonhomme, V.; Picq, S.; Gaucherel, S.; Claude, J. Momocs: Outline Analysis Using. R. J. Stat. Softw. 2014, 13, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Van Belleghem, S.M.; Papa, R.; Ortiz-Zuazaga, H.; Hendrickx, F.; Jiggins, C.D.; Owen McMillan, W.; Counterman, B.A. Patternize: An R package for quantifying colour pattern variation. Methods Ecol. Evol. 2018, 13, 390–398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maia, R.; Gruson, H.; Endler, J.A.; White, T.E. Pavo2: New tools for the spectral and spatial analysis of colour in R. Methods Ecol. Evol. 2019, 7, 1097–1107. [Google Scholar] [CrossRef] [Green Version]
- Weller, H.I.; Westneat, M.W. Quantitative color profiling of digital images with earth mover’s distance using the R package colordistance. PeerJ 2019, 7, e6398. [Google Scholar] [CrossRef] [Green Version]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Fernandez-Delgado, M.; Papa, R.; Cernadas, E.; Barro, S.; Amorim, D. Do we need hundreds of classifiers to solve real world classification problems? J. Mach. Learn. Res. 2014, 15, 3133–3181. [Google Scholar]
- Breitkopf, H.; Onstein, R.E.; Cafasso, D.; Schlüter, P.M.; Cozzolino, S. Multiple shifts to different pollinators fueled rapid diversification in sexually deceptive Ophrys orchids. New Phytol. 2015, 207, 377–389. [Google Scholar] [CrossRef]
- Baguette, M.; Bertrand, J.A.M.; Stevens, V.M.; Schatz, B. Why are there so many bee-orchid species? Adaptive radiation by intra-specific competition for mnesic pollinators. Biol. Rev. 2020, 95, 1630–1663. [Google Scholar] [CrossRef]
- Triponez, Y.; Arrigo, N.; Pellissier, L.; Schatz, B.; Alvarez, N. Morphological, ecological and genetic aspects associated with endemism in the Fly Orchid group. Mol. Ecol. 2006, 22, 1431–1446. [Google Scholar] [CrossRef]
- Paulus, H.F. Deceived males—Pollination biology of the Mediterranean orchid genus Ophrys (Orchidaceae). J. Eur. Orch. 2006, 38, 303–353. [Google Scholar]
- Rakosy, D.; Cuervo, M.; Paulus, H.F.; Ayasse, M. Looks matter: Changes in flower form affect pollination effectiveness in a sexually deceptive orchid. J. Evol. Biol. 2017, 30, 1978–1993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spaethe, J.; Moser, W.H.; Paulus, H.F. Increase of pollinator attraction by means of a visual signal in the sexually deceptive orchid, Ophrys heldreichii (Orchidaceae). Plant Syst. Evol. 2007, 264, 31–40. [Google Scholar] [CrossRef]
- Spaethe, J.; Streinzer, M.; Paulus, H.F. Why sexually deceptive orchids have colored flowers. Commun. Integr. Biol. 2010, 3, 139–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Streinzer, M.; Ellis, T.; Paulus, H.F.; Spaethe, J. Visual discrimination between two sexually deceptive Ophrys species by a bee pollinator. Arthropod-Plant Interact. 2010, 4, 141–148. [Google Scholar] [CrossRef] [Green Version]
- Streinzer, M.; Ellis, T.; Paulus, H.F.; Spaethe, J. Floral shape mimicry and variation in sexually deceptive orchids with a shared pollinator: Floral shape, mirmicry, and sexual deception. Biol. J. Linn. Soc. 2012, 106, 469–481. [Google Scholar]
- Stejskal, K.; Streinzer, M.; Dyer, A.; Paulus, H.F.; Spaethe, J. Functional significance of labellum pattern variation in a sexually deceptive Orchid (Ophrys heldreichii): Evidence of individual signature learning effects. PLoS ONE 2015, 10, e0142971. [Google Scholar] [CrossRef] [Green Version]
- Vereecken, N.J.; Schiestl, F.P. On the roles of colour and scent in a specialized floral mimicry system. Ann. Bot. 2009, 104, 1077–1084. [Google Scholar] [CrossRef] [Green Version]
- Delforge, P. Orchidées d’Europe. d Afrique du Nord et du Proche-Orient. Delachaux Et Niestlé 2016, 15, 1–544. [Google Scholar]
- Wood, J.J. Two New Combinations in Ophrys (Orchidaceae). Kew Bull. 1983, 38, 135–137. [Google Scholar] [CrossRef]
- Delforge, P. L’Ophrys de l’Aveyron. L’Orchidophile 1984, 15, 177–183. [Google Scholar]
- Hermosilla, C.E.; Sabando, J. Notas sobre orquídeas (V). Estud. Mus. Cienc. Nat. Álava 1999, 13, 123–156. [Google Scholar]
- Hermosilla, C.E.; Soca, R. Distribution of Ophrys aveyronensis (J.J.Wood) Delforge and survey of its hybrids. Caesiana 1999, 13, 31–38. [Google Scholar]
- Benito Ayuso, J. Estudios sobre polinizacion en el genero Ophrys (Orchidaceae). Flora Montiberica 2019, 74, 32–37. [Google Scholar]
- Kreutz, C.A.J. Beitrag zur Taxonomie und Nomenklatur europäischer, mediterraner, nordafrikanischer und vorderasiatischer Orchideen. Ber. Arbeitskrs. Heim. Orchid. 2007, 24, 77–141. [Google Scholar]
- Paulus, H.F. Zur Bestäubungsbiologie der Gattung Ophrys in Nordspanien: Freilandstudien an Ophrys aveyronensis, O. subinsectifera, O. riojana, O. vasconica und O. forestieri. J. Eur. Orch. 2017, 49, 427–471. [Google Scholar]
- Rohlf, F.J. tpsDig, Digitize Landmarks and Outlines, Version 2.0; Department of Ecology and Evolution, State University of New York at Stony Brook: Stony Brook, NY, USA, 2006. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: https://www.R-project.org/ (accessed on 1 February 2021).
- Liaw, A.; Wiener, M.R. Classification and regression by random forest. R News 2002, 2, 18–22. [Google Scholar]
- Anderson, M.J. Permutational Multivariate Analysis of Variance (PERMANOVA). Wiley StatsRef Stat. Ref. Online 2017, 1, 1–15. [Google Scholar]
- Scaccabarozzi, D.; Guzzetti, L.; Phillips, R.D.; Milne, L.; Tommasi, N.; Cozzolino, S.; Dixon, K.W. Ecological factors driving pollination success in an orchid that mimics a range of Fabaceae. Bot. J. Linn. Soc. 2020, 2, 253–269. [Google Scholar] [CrossRef]
- Oksanen, J.; Kindt, R.; Legendre, P.; O’Hara, B.; Simpson, G.; Solymos, P.; Stevens, M.H.H.; Wagner, H. The VEGAN Package: Community Ecology Package. CRAN R PACKAGES. 2009. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 1 March 2021).
- Schatz, B.; Genoud, D.; Claessens, J.; Kleynen, J. Orchid-pollinator network in Euro-Mediterranean region: What we know, what we think we know, and what remains to be done. Acta Oecol. 2019, 107, 103605. [Google Scholar] [CrossRef]
- Joffard, N.; Massol, F.; Grenié, M.; Montgelard, C.; Schatz, B. Effect of pollination strategy, phylogeny and distribution on pollination niches of Euro-Mediterranean orchids. J. Ecol. 2019, 107, 478–490. [Google Scholar] [CrossRef] [Green Version]
- Michez, D.; Rasmont, P.; Terzo, M.; Vereecken, N.J. Bees of Europe. Hymenoptera of Europe 1; NAP Edition: Paris, France, 2019; pp. 1–547. [Google Scholar]
- Fadzly, N.; Zuharah, W.F.; Jenn, N.; Jenny, W. Can plants fool artificial intelligence? Using machine learning to compare between bee orchids and bees. Plant Signal. Behav. 2021, 16, 1935605. [Google Scholar] [CrossRef] [PubMed]
- Lunau, K.; Scaccabarozzi, D.; Willing, L.; Dixon, K. A bee’s eye view of remarkable floral colour patterns in the south-west Australian biodiversity hotspot revealed by false colour photography. Ann. Bot. 2021, 128, 821–824. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, J.A.M.; Baguette, M.; Joffard, N.; Schatz, B. Challenges Inherent in the Systematics and Taxonomy of Genera that have Recently Experienced Explosive Radiation: The Case of Orchids of the Genus Ophrys. In Systematics and the Exploration of Life; Wiley: Hoboken, NJ, USA, 2021; pp. 113–134. [Google Scholar]
- Bertrand, J.A.M.; Gibert, A.; Llauro, C.; Panaud, O. Characterization of the complete plastome of Ophrys aveyronensis, a Euro-Mediterranean orchid with an intriguing disjunct geographic distribution. Mitochondrial DNA Part B 2019, 4, 3256–3257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
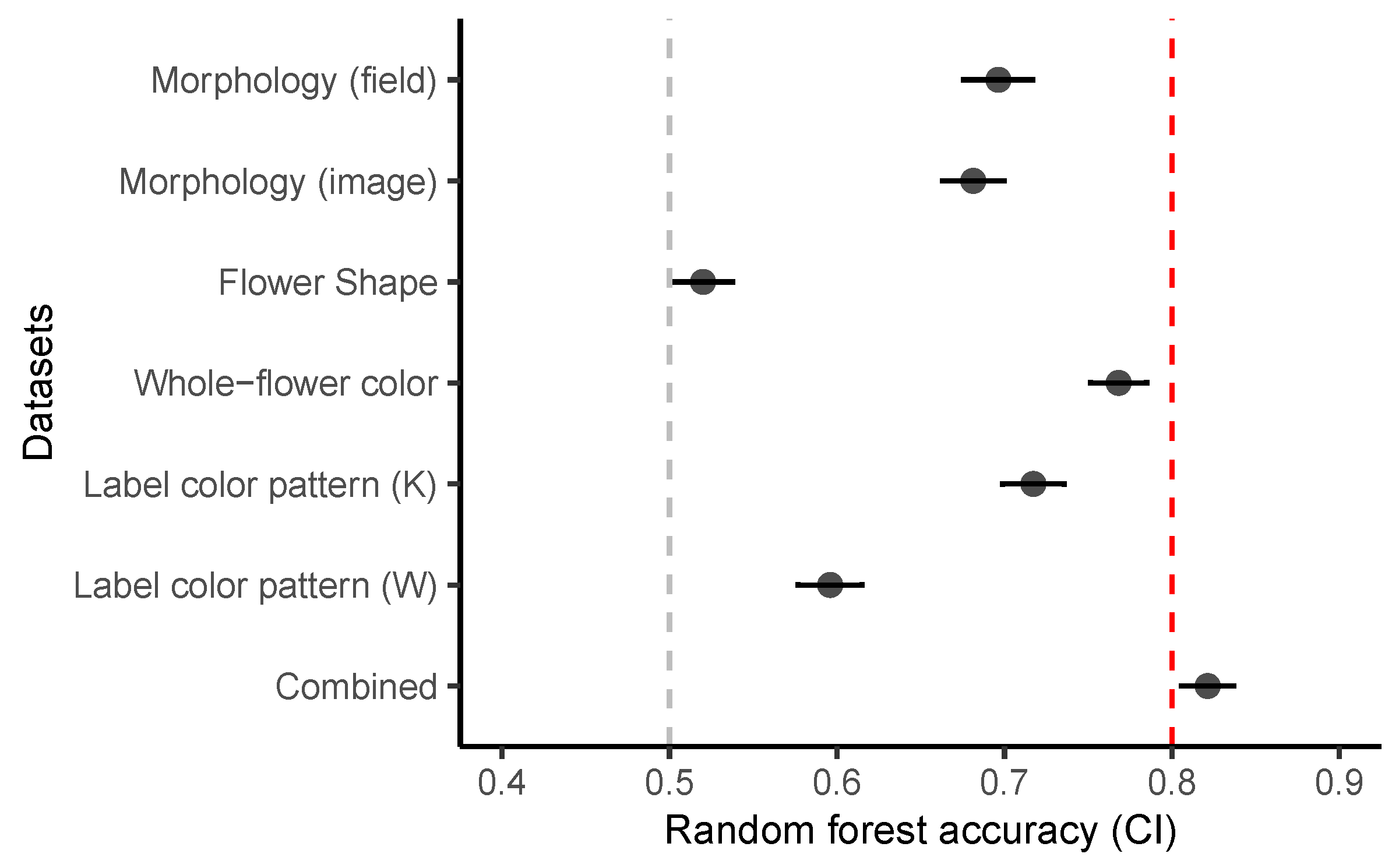
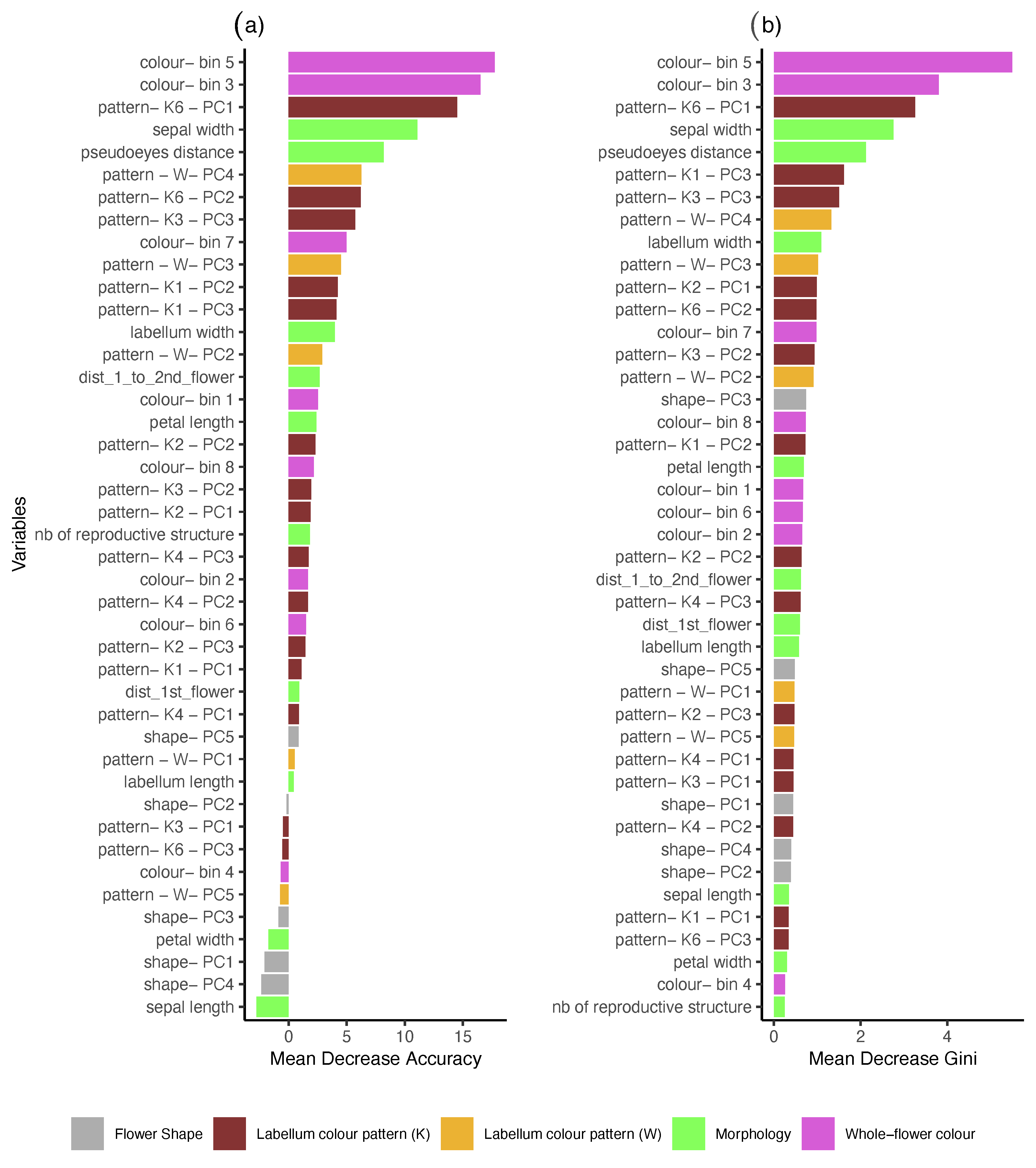
| Dataset | mtry | Accuracy | Sensitivity (TPR) | Specificity (TNR) | OOBerror | AUC |
|---|---|---|---|---|---|---|
| Morphology (field) | 3 | 0.73 | 0.64 | 0.82 | 33 | 0.78 |
| Morphology (image) | 2 | 0.59 | 0.5 | 0.7 | 33 | 0.72 |
| Flower shape | 2 | 0.59 | 0.5 | 0.64 | 51 | 0.51 |
| Whole-flower colour | 3 | 0.82 | 0.78 | 0.85 | 25 | 0.80 |
| Labellum colour pattern (K) | 4 | 0.68 | 0.62 | 0.71 | 24 | 0.80 |
| Labellum colour pattern (W) | 2 | 0.64 | 0.6 | 0.65 | 39 | 0.66 |
| Combined | 9 | 0.95 | 0.9 | 1 | 15 | 0.94 |
| Phenotype | Effect | Df | F | p |
|---|---|---|---|---|
| Morphology (field) | subspecies | 1 | 1.236 | 0.277 |
| populations | 4 | 8.861 | 0.001 | |
| Morphology (image) | subspecies | 1 | 12.655 | 0.001 |
| populations | 4 | 1.135 | 0.327 | |
| Flower shape | subspecies | 1 | 1.413 | 0.206 |
| populations | 4 | 1.117 | 0.3 | |
| Whole-flower colour | subspecies | 1 | 22.992 | 0.001 |
| populations | 4 | 11.425 | 0.001 | |
| Labellum colour pattern (K) | subspecies | 1 | 1.628 | 0.001 |
| populations | 4 | 1.266 | 0.001 | |
| Labellum colour pattern (W) | subspecies | 1 | 2.222 | 0.001 |
| populations | 4 | 1.267 | 0.016 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gibert, A.; Louty, F.; Buscail, R.; Baguette, M.; Schatz, B.; Bertrand, J.A.M. Extracting Quantitative Information from Images Taken in the Wild: A Case Study of Two Vicariants of the Ophrys aveyronensis Species Complex. Diversity 2022, 14, 400. https://doi.org/10.3390/d14050400
Gibert A, Louty F, Buscail R, Baguette M, Schatz B, Bertrand JAM. Extracting Quantitative Information from Images Taken in the Wild: A Case Study of Two Vicariants of the Ophrys aveyronensis Species Complex. Diversity. 2022; 14(5):400. https://doi.org/10.3390/d14050400
Chicago/Turabian StyleGibert, Anais, Florian Louty, Roselyne Buscail, Michel Baguette, Bertrand Schatz, and Joris A. M. Bertrand. 2022. "Extracting Quantitative Information from Images Taken in the Wild: A Case Study of Two Vicariants of the Ophrys aveyronensis Species Complex" Diversity 14, no. 5: 400. https://doi.org/10.3390/d14050400
APA StyleGibert, A., Louty, F., Buscail, R., Baguette, M., Schatz, B., & Bertrand, J. A. M. (2022). Extracting Quantitative Information from Images Taken in the Wild: A Case Study of Two Vicariants of the Ophrys aveyronensis Species Complex. Diversity, 14(5), 400. https://doi.org/10.3390/d14050400








