Full-Length Transcriptome Comparison Provides Novel Insights into the Molecular Basis of Adaptation to Different Ecological Niches of the Deep-Sea Hydrothermal Vent in Alvinocaridid Shrimps
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and RNA Extraction
2.2. PacBio Library Construction and Sequencing
2.3. Illumina Library Construction, Sequencing, and Gene Expression
2.4. Coding Sequence Detection
2.5. Transcript Function Annotation
2.6. Calculation of GC Content and Codon Usage
2.7. Gene Family Expansion Analysis
2.8. Positive Selection Analysis
3. Results
3.1. The Full-Length Trascripts of S. leurokolos and A. longirostris Using PacBio Sequencing
3.2. Efficient Gene Annotation
3.3. Coding Sequence Prediction and Comparison of GC Content and Codon Usage
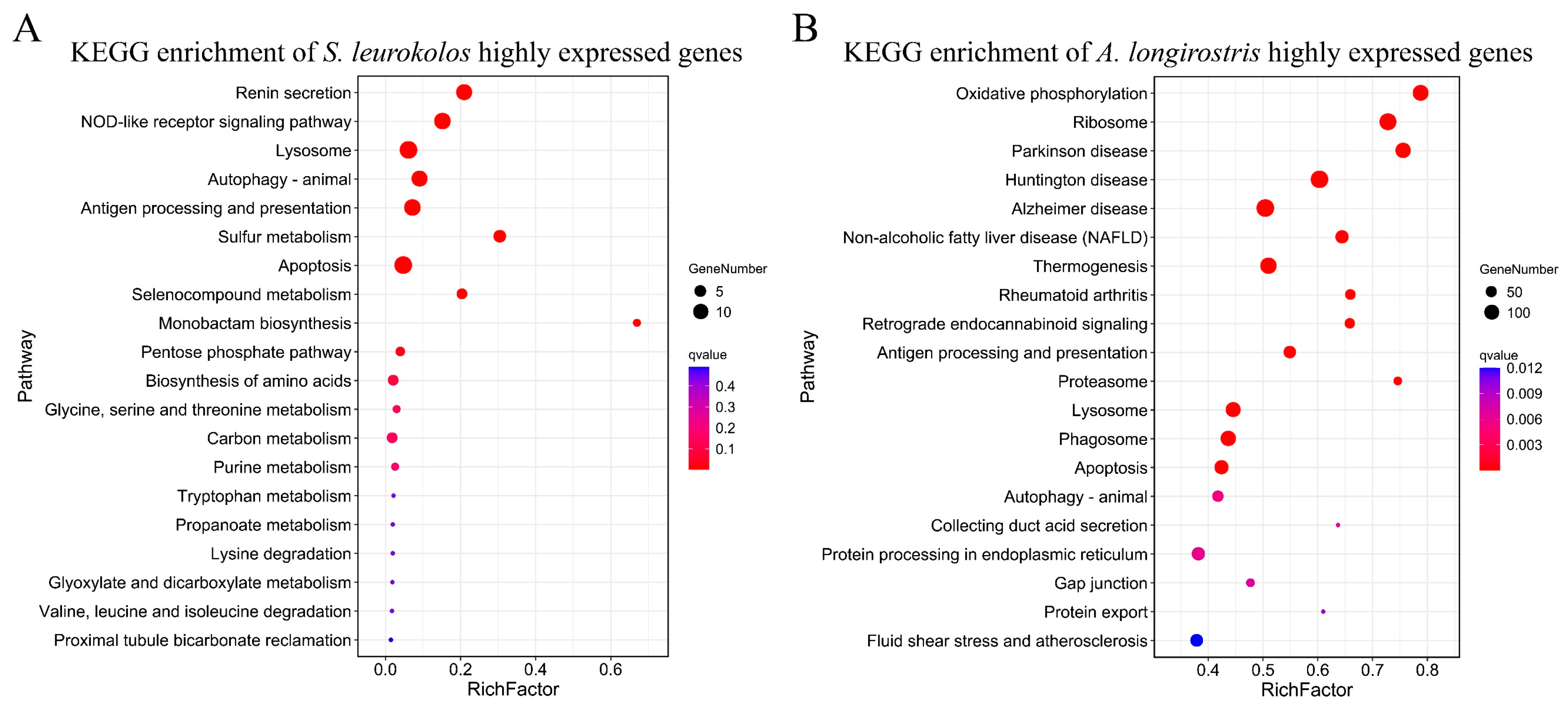
3.4. Highly Expressed Genes and Significantly Expanded Gene Family
3.5. Positive Selection Analysis
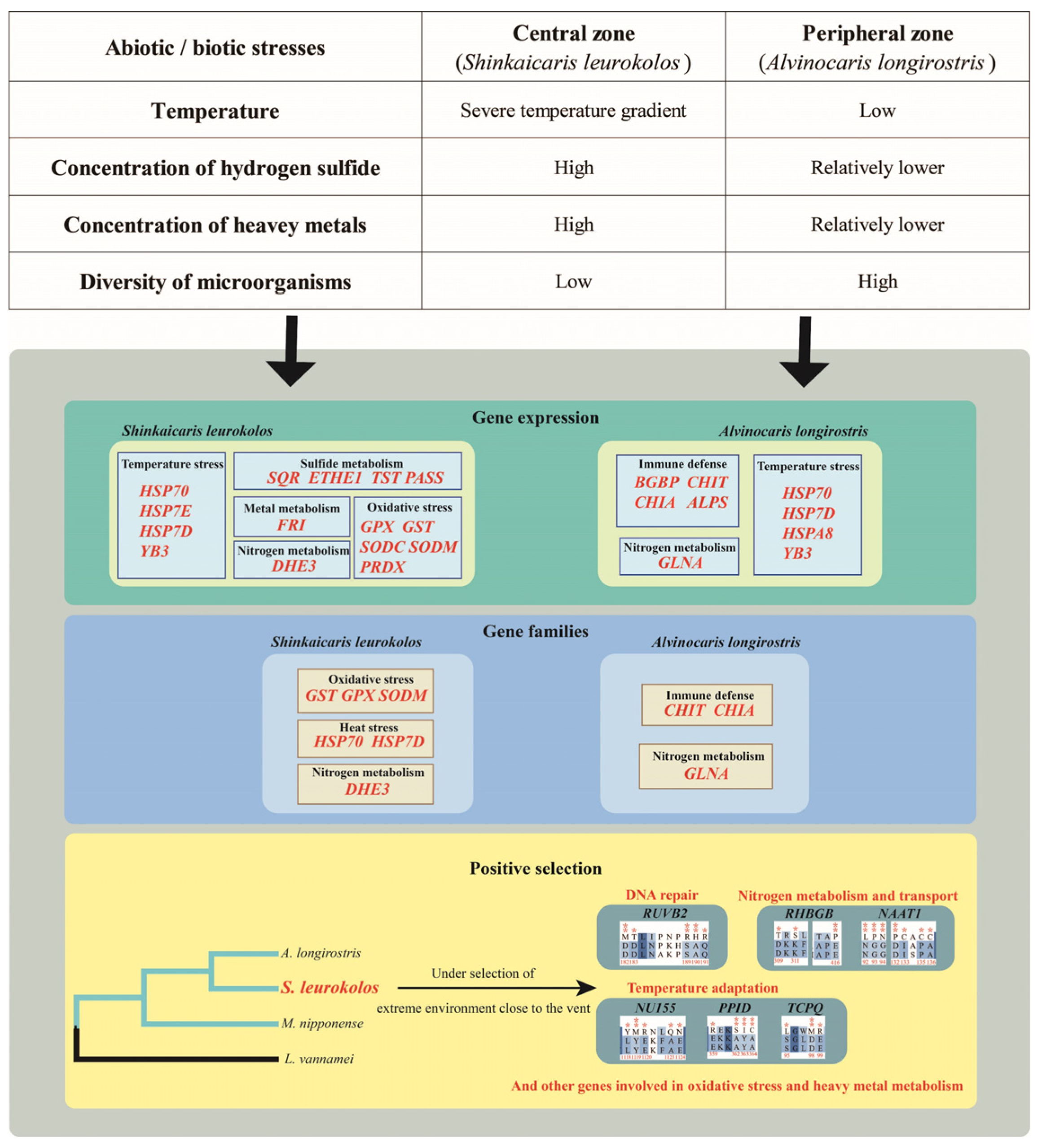
4. Discussion
4.1. Temperature Adaptation
4.2. Substance Metabolism and Detoxification
4.3. Oxidative Stress and DNA Repair
4.4. Innate Immune Response
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Smith, F.; Brown, A.; Mestre, N.C.; Reed, A.J.; Thatje, S. Thermal adaptations in deep-sea hydrothermal vent and shallow-water shrimp. Deep Sea Res. Part II Top. Stud. Oceanogr. 2013, 92, 234–239. [Google Scholar] [CrossRef]
- Little, C.T.S.; Vrijenhoek, R.C. Are hydrothermal vent animals living fossils? Trends Ecol. Evol. 2003, 18, 582–588. [Google Scholar] [CrossRef]
- Hourdez, S.; Weber, R.E. Molecular and functional adaptations in deep-sea hemoglobins. J. Inorg. Biochem. 2005, 99, 130–141. [Google Scholar] [CrossRef] [PubMed]
- Chevaldonné, P.; Desbruyéres, D.; Childress, J.J. … and some even hotter. Nature 1992, 359, 593–594. [Google Scholar] [CrossRef]
- Delaney, J.R.; Robigou, V.; McDuff, R.E.; Tivey, M.K. Geology of a vigorous hydrothermal system on the Endeavour Segment, Juan de Fuca Ridge. J. Geophys. Res. Solid Earth 1992, 97, 19663–19682. [Google Scholar] [CrossRef]
- Cary, S.C.; Shank, T.; Stein, J. Worms bask in extreme temperatures. Nature 1998, 391, 545–546. [Google Scholar] [CrossRef]
- Li, L.; Du, Z.; Zhang, X.; Xi, S.; Wang, B.; Luan, Z.; Lian, C.; Yan, J. In situ Raman spectral characteristics of carbon dioxide in a deep-sea simulator of extreme environments reaching 300 °C and 30 MPa. Appl. Spectrosc. 2018, 72, 48–59. [Google Scholar] [CrossRef]
- Johnson, K.; Childress, J.J.; Hessler, R.R.; Sakamoto-Arnold, C.M.; Beehler, C.L. Chemical and biological interactions in the Rose Garden hydrothermal vent field, Galapagos spreading center. Deep Sea Res. Part A Oceanogr. Res. Pap. 1988, 35, 1723–1744. [Google Scholar] [CrossRef]
- McMullin, E.R.; Bergquist, D.C.; Fisher, C.R. Metazoans in extreme environments: Adaptations of hydrothermal vent and hydrocarbon seep fauna. Gravit. Space Biol. Bull. Publ. Am. Soc. Gravit. Space Biol. 2000, 13, 13–23. [Google Scholar]
- Dover, C.L.V.; Szuts, E.Z.; Chamberlain, S.C.; Cann, J.R. A novel eye in ‘eyeless’ shrimp from hydrothermal vents of the Mid-Atlantic Ridge. Nature 1989, 337, 458–460. [Google Scholar] [CrossRef]
- Ravaux, J.; Gaill, F.; Le Bris, N.; Sarradin, P.-M.; Jollivet, D.; Shillito, B. Heat-shock response and temperature resistance in the deep-sea vent shrimp Rimicaris exoculata. J. Exp. Biol. 2003, 206, 2345–2354. [Google Scholar] [CrossRef] [PubMed]
- Hardivillier, Y.; Leignel, V.; Denis, F.; Uguen, G.; Cosson, R.; Laulier, M. Do organisms living around hydrothermal vent sites contain specific metallothioneins? The case of the genus Bathymodiolus (Bivalvia, Mytilidae). Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2004, 139, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Hourdez, S.; Lallier, F.H. Adaptations to hypoxia in hydrothermal-vent and cold-seep invertebrates. In Life in Extreme Environments; Amils, R., Ellis-Evans, C., Hinghofer-Szalkay, H., Eds.; Springer: Dordrecht, The Netherlands, 2007; pp. 297–313. [Google Scholar]
- Zeng, Z.; Ma, Y.; Wang, X.; Chen, C.-T.A.; Yin, X.; Zhang, S.; Zhang, J.; Jiang, W. Elemental compositions of crab and snail shells from the Kueishantao hydrothermal field in the southwestern Okinawa Trough. J. Mar. Syst. 2018, 180, 90–101. [Google Scholar] [CrossRef]
- Lan, Y.; Sun, J.; Xu, T.; Chen, C.; Tian, R.; Qiu, J.-W.; Qian, P.-Y. De novo transcriptome assembly and positive selection analysis of an individual deep-sea fish. BMC Genom. 2018, 19, 394. [Google Scholar] [CrossRef]
- Zhang, Y.; Sun, J.; Chen, C.; Watanabe, H.K.; Feng, D.; Zhang, Y.; Chiu, J.M.Y.; Qian, P.-Y.; Qiu, J.-W. Adaptation and evolution of deep-sea scale worms (Annelida: Polynoidae): Insights from transcriptome comparison with a shallow-water species. Sci. Rep. 2017, 7, 46205. [Google Scholar] [CrossRef]
- Zheng, P.; Wang, M.; Li, C.; Sun, X.; Wang, X.; Sun, Y.; Sun, S. Insights into deep-sea adaptations and host–symbiont interactions: A comparative transcriptome study on Bathymodiolus mussels and their coastal relatives. Mol. Ecol. 2017, 26, 5133–5148. [Google Scholar] [CrossRef]
- Hui, M.; Song, C.; Liu, Y.; Li, C.; Cui, Z. Exploring the molecular basis of adaptive evolution in hydrothermal vent crab Austinograea alayseae by transcriptome analysis. PLoS ONE 2017, 12, e0178417. [Google Scholar] [CrossRef]
- Martin, J.W.; Haney, T.A. Decapod crustaceans from hydrothermal vents and cold seeps: A review through 2005. Zool. J. Linn. Soc. 2005, 145, 445–522. [Google Scholar] [CrossRef]
- Yahagi, T.; Watanabe, H.; Kojima, S.; Beedessee, G.; Komai, T. First record and a new species of Alvinocaris Williams & Chace, 1982 (Crustacea: Decapoda: Caridea: Alvinocarididae) from the Indian Ocean. Zootaxa 2014, 3893, 101–113. [Google Scholar] [CrossRef][Green Version]
- Yahagi, T.; Watanabe, H.; Ishibashi, J.-I.; Kojima, S. Genetic population structure of four hydrothermal vent shrimp species (Alvinocarididae) in the Okinawa Trough, Northwest Pacific. Mar. Ecol. Prog. Ser. 2015, 529, 159–169. [Google Scholar] [CrossRef]
- Miyazaki, J.; Kawagucci, S.; Makabe, A.; Takahashi, A.; Kitada, K.; Torimoto, J.; Matsui, Y.; Tasumi, E.; Shibuya, T.; Nakamura, K.; et al. Deepest and hottest hydrothermal activity in the Okinawa Trough: The Yokosuka site at Yaeyama Knoll. R. Soc. Open Sci. 2017, 4, 171570. [Google Scholar] [CrossRef] [PubMed]
- Tokuda, G.; Yamada, A.; Nakano, K.; Arita, N.; Yamasaki, H. Occurrence and recent long-distance dispersal of deep-sea hydrothermal vent shrimps. Biol. Lett. 2005, 2, 257–260. [Google Scholar] [CrossRef] [PubMed]
- Komai, T.; Segonzac, M. Taxonomic Review of the Hydrothermal Vent Shrimp Genera Rimicaris Williams & Rona and Chorocaris Martin & Hessler (Crustacea: Decapoda: Caridea: Alvinocarididae). J. Shellfish Res. 2008, 27, 21–41. [Google Scholar] [CrossRef]
- Komai, T.; Segonzac, M. A revision of the genus Alvinocaris Williams and Chace (Crustacea: Decapoda: Caridea: Alvinocarididae), with descriptions of a new genus and a new species of Alvinocaris. Ann. Mag. Nat. Hist. 2005, 39, 1111–1175. [Google Scholar] [CrossRef]
- Komai, T.; Chan, T.-Y. A new genus and two new species of alvinocaridid shrimps (Crustacea: Decapoda: Caridea) from a hydrothermal vent field off northeastern Taiwan. Zootaxa 2010, 2372, 15–32. [Google Scholar] [CrossRef]
- Watanabe, H.; Yahagi, T.; Nagai, Y.; Seo, M.; Kojima, S.; Ishibashi, J.-I.; Yamamoto, H.; Fujikura, K.; Mitarai, S.; Toyofuku, T. Different thermal preferences for brooding and larval dispersal of two neighboring shrimps in deep-sea hydrothermal vent fields. Mar. Ecol. 2016, 37, 1282–1289. [Google Scholar] [CrossRef]
- Watanabe, H.; Kojima, S. Vent Fauna in the Okinawa Trough. In Subseafloor Biosphere Linked to Hydrothermal Systems: TAIGA Concept; Ishibashi, J.-I., Okino, K., Sunamura, M., Eds.; Springer: Tokyo, Japan, 2015; pp. 449–459. [Google Scholar]
- Hui, M.; Cheng, J.; Sha, Z. Adaptation to the deep-sea hydrothermal vents and cold seeps: Insights from the transcriptomes of Alvinocaris longirostris in both environments. Deep Sea Res. Part I Oceanogr. Res. Pap. 2018, 135, 23–33. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Jin, S.; Bian, C.; Jiang, S.; Han, K.; Xiong, Y.; Zhang, W.; Shi, C.; Qiao, H.; Gao, Z.; Li, R.; et al. A chromosome-level genome assembly of the oriental river prawn, Macrobrachium nipponense. GigaScience 2021, 10, giaa160. [Google Scholar] [CrossRef]
- De Bie, T.; Cristianini, N.; Demuth, J.P.; Hahn, M.W. CAFE: A computational tool for the study of gene family evolution. Bioinformatics 2006, 22, 1269–1271. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; dos Reis, M. Statistical Properties of the Branch-Site Test of Positive Selection. Mol. Biol. Evol. 2011, 28, 1217–1228. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wong, W.S.W.; Nielsen, R. Bayes Empirical Bayes Inference of amino acid sites under positive selection. Mol. Biol. Evol. 2005, 22, 1107–1118. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Yang, Z.; Nielsen, R. Codon-Substitution Models for Detecting Molecular Adaptation at Individual Sites Along Specific Lineages. Mol. Biol. Evol. 2002, 19, 908–917. [Google Scholar] [CrossRef]
- Zhu, F.-C.; Sun, J.; Yan, G.-Y.; Huang, J.-M.; Chen, C.; He, L.-S. Insights into the strategy of micro-environmental adaptation: Transcriptomic analysis of two alvinocaridid shrimps at a hydrothermal vent. PLoS ONE 2020, 15, e0227587. [Google Scholar] [CrossRef]
- Ayala-del-Río Héctor, L.; Chain Patrick, S.; Grzymski Joseph, J.; Ponder Monica, A.; Ivanova, N.; Bergholz Peter, W.; Di Bartolo, G.; Hauser, L.; Land, M.; Bakermans, C.; et al. The Genome Sequence of Psychrobacter arcticus 273-4, a Psychroactive Siberian Permafrost Bacterium, Reveals Mechanisms for Adaptation to Low-Temperature Growth. Appl. Environ. Microbiol. 2010, 76, 2304–2312. [Google Scholar] [CrossRef]
- Zhao, J.-S.; Deng, Y.; Manno, D.; Hawari, J. Shewanella spp. genomic evolution for a cold marine lifestyle and in-situ explosive biodegradation. PLoS ONE 2010, 5, e9109. [Google Scholar] [CrossRef]
- Saunders, N.F.; Thomas, T.; Curmi, P.M.; Mattick, J.S.; Kuczek, E.; Slade, R.; Davis, J.; Franzmann, P.D.; Boone, D.; Rusterholtz, K.; et al. Mechanisms of Thermal Adaptation Revealed From the Genomes of the Antarctic Archaea Methanogenium frigidum and Methanococcoides burtonii. Genome Res. 2003, 13, 1580–1588. [Google Scholar] [CrossRef]
- Yang, L.-L.; Tang, S.-K.; Huang, Y.; Zhi, X.-Y. Low Temperature Adaptation Is Not the Opposite Process of High Temperature Adaptation in Terms of Changes in Amino Acid Composition. Genome Biol. Evol. 2015, 7, 3426–3433. [Google Scholar] [CrossRef]
- Van Dover, C.L.; German, C.R.; Speer, K.G.; Parson, L.M.; Vrijenhoek, R.C. Evolution and Biogeography of Deep-Sea Vent and Seep Invertebrates. Science 2002, 295, 1253–1257. [Google Scholar] [CrossRef] [PubMed]
- Cottin, D.; Ravaux, J.; Léger, N.; Halary, S.; Toullec, J.-Y.; Sarradin, P.-M.; Gaill, F.; Shillito, B. Thermal biology of the deep-sea vent annelid Paralvinella grasslei:in vivostudies. J. Exp. Biol. 2008, 211, 2196–2204. [Google Scholar] [CrossRef] [PubMed]
- Davies, T.J.; Meiri, S.; Barraclough, T.G.; Gittleman, J.L. Species co-existence and character divergence across carnivores. Ecol. Lett. 2007, 10, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Somero, G.N. Linking biogeography to physiology: Evolutionary and acclimatory adjustments of thermal limits. Front. Zool. 2005, 2, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Collier, R.J.; Gebremedhin, K.G. Thermal Biology of Domestic Animals. Annu. Rev. Anim. Biosci. 2015, 3, 513–532. [Google Scholar] [CrossRef]
- Wojda, I. Temperature stress and insect immunity. J. Therm. Biol. 2017, 68, 96–103. [Google Scholar] [CrossRef]
- Madeira, D.; Narciso, L.; Cabral, H.N.; Diniz, M.S.; Vinagre, C. Thermal tolerance of the crab Pachygrapsus marmoratus: Intraspecific differences at a physiological (CTMax) and molecular level (Hsp70). Cell Stress Chaperones 2012, 17, 707–716. [Google Scholar] [CrossRef]
- Yuan, K.; Yuan, F.-H.; He, H.-H.; Bi, H.-T.; Weng, S.-P.; He, J.-G.; Chen, Y.-H. Heat shock 70 kDa protein cognate 5 involved in WSSV toleration of Litopenaeus vannamei. Dev. Comp. Immunol. 2017, 72, 9–20. [Google Scholar] [CrossRef]
- Liu, Z.-M.; Zhu, X.-L.; Lu, J.; Cai, W.-J.; Ye, Y.-P.; Lv, Y.-P. Effect of high temperature stress on heat shock protein expression and antioxidant enzyme activity of two morphs of the mud crab Scylla paramamosain. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2018, 223, 10–17. [Google Scholar] [CrossRef]
- Sornchuer, P.; Junprung, W.; Yingsunthonwattana, W.; Tassanakajon, A. Heat shock factor 1 regulates heat shock proteins and immune-related genes in Penaeus monodon under thermal stress. Dev. Comp. Immunol. 2018, 88, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Cottin, D.; Shillito, B.; Chertemps, T.; Thatje, S.; Léger, N.; Ravaux, J. Comparison of heat-shock responses between the hydrothermal vent shrimp Rimicaris exoculata and the related coastal shrimp Palaemonetes varians. J. Exp. Mar. Biol. Ecol. 2010, 393, 9–16. [Google Scholar] [CrossRef]
- Kwon, K.; Lee, N.; Yu, K.O. Lethal (2) Essential for Life l(2)efl Gene in the Two-spotted Cricket, Gryllus bimaculatus (Orthoptera: Gryllidae). J. Life Sci. 2021, 31, 671–676. [Google Scholar] [CrossRef]
- Rollenhagen, C.; Hodge, C.A.; Cole, C.N. The Nuclear Pore Complex and the DEAD Box Protein Rat8p/Dbp5p Have Nonessential Features Which Appear To Facilitate mRNA Export following Heat Shock. Mol. Cell. Biol. 2004, 24, 4869–4879. [Google Scholar] [CrossRef] [PubMed]
- Qian, B.; Xue, L. Liver transcriptome sequencing and de novo annotation of the large yellow croaker ( Larimichthy crocea ) under heat and cold stress. Mar. Genom. 2016, 25, 95–102. [Google Scholar] [CrossRef]
- Johnson, K.S.; Beehler, C.L.; Sakamoto-Arnold, C.M.; Childress, J.J. In Situ Measurements of Chemical Distributions in a Deep-Sea Hydrothermal Vent Field. Science 1986, 231, 1139–1141. [Google Scholar] [CrossRef]
- Luther, G.W.; Rozan, T.F.; Taillefert, M.; Nuzzio, D.B.; Di Meo, C.; Shank, T.M.; Lutz, R.A.; Cary, S.C. Chemical speciation drives hydrothermal vent ecology. Nature 2001, 410, 813–816. [Google Scholar] [CrossRef]
- Levesque, C.; Juniper, S.K.; Limén, H. Spatial organization of food webs along habitat gradients at deep-sea hydrothermal vents on Axial Volcano, Northeast Pacific. Deep Sea Res. Part I Oceanogr. Res. Pap. 2006, 53, 726–739. [Google Scholar] [CrossRef]
- Tunnicliffe, V. The biology of hydrothermal vents: Ecology and evolution. Oceanog. Mar. Biol. 1991, 29, 319–407. [Google Scholar]
- Compère, P.; Martinez, A.-S.; Charmantier-Daures, M.; Toullec, J.-Y.; Goffinet, G.; Gaill, F. Does sulphide detoxication occur in the gills of the hydrothermal vent shrimp, Rimicaris exoculata? Comptes Rendus. Biol. 2002, 325, 591–596. [Google Scholar] [CrossRef]
- Powell, M.A.; Somero, G.N. Adaptations to sulfide by hydrothermal vent animals: Sites and mechanisms of detoxification and metabolism. Biol. Bull. 1986, 171, 274–290. [Google Scholar] [CrossRef]
- Xin, Y.; Liu, H.; Cui, F.; Liu, H.; Xun, L. Recombinant Escherichia coli with sulfide:quinone oxidoreductase and persulfide dioxygenase rapidly oxidises sulfide to sulfite and thiosulfate via a new pathway. Environ. Microbiol. 2016, 18, 5123–5136. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Xin, Y.; Xun, L. Distribution, Diversity, and Activities of Sulfur Dioxygenases in Heterotrophic Bacteria. Appl. Environ. Microbiol. 2014, 80, 1799–1806. [Google Scholar] [CrossRef] [PubMed]
- Hildebrandt, T.M.; Grieshaber, M.K. Three enzymatic activities catalyze the oxidation of sulfide to thiosulfate in mammalian and invertebrate mitochondria. FEBS J. 2008, 275, 3352–3361. [Google Scholar] [CrossRef] [PubMed]
- Doeller, J.E.; Grieshaber, M.K.; Kraus, D.W. Chemolithoheterotrophy in a metazoan tissue: Thiosulfate production matches ATP demand in ciliated mussel gills. J. Exp. Biol. 2001, 204, 3755–3764. [Google Scholar] [CrossRef]
- Cosson, R.P.; Thiébaut, E.; Company, R.; Castrec-Rouelle, M.; Colaço, A.; Martins, I.; Sarradin, P.-M.; Bebianno, M.J. Spatial variation of metal bioaccumulation in the hydrothermal vent mussel Bathymodiolus azoricus. Mar. Environ. Res. 2008, 65, 405–415. [Google Scholar] [CrossRef]
- Kádár, E.; Costa, V.; Santos, R.S. Distribution of micro-essential (Fe, Cu, Zn) and toxic (Hg) metals in tissues of two nutritionally distinct hydrothermal shrimps. Sci. Total Environ. 2006, 358, 143–150. [Google Scholar] [CrossRef]
- Liu, X.-L.; Ye, S.; Li, H.-W.; Lu, B.; Yu, Y.-Q.; Fan, Y.-P.; Yang, W.-J.; Yang, J.-S. An H-ferritin from the hydrothermal vent shrimp Rimicaris exoculata and its potential role in iron metabolism. BioMetals 2019, 32, 251–264. [Google Scholar] [CrossRef]
- Mao, H.; Wang, D.-H.; Yang, W.-X. The involvement of metallothionein in the development of aquatic invertebrate. Aquat. Toxicol. 2012, 110, 208–213. [Google Scholar] [CrossRef]
- Alvarez-Carreño, C.; Alva, V.; Becerra, A.; Lazcano, A. Structure, function and evolution of the hemerythrin-like domain superfamily. Protein Sci. 2018, 27, 848–860. [Google Scholar] [CrossRef]
- Lee, R.W.; Robinson, J.J.; Cavanaugh, C.M. Pathways of inorganic nitrogen assimilation in chemoautotrophic bacteria-marine invertebrate symbioses: Expression of host and symbiont glutamine synthetase. J. Exp. Biol. 1999, 202, 289–300. [Google Scholar] [CrossRef]
- Reitzer, L.J. Ammonia assimilation and the biosynthesis of glutamine, glutamate, aspartate, asparagine, L-alanine and D-alanine. In Escherichia coli and Salmonella typhimurium:Cellular and Molecular Biology; American Society for Microbiology: Washington, DC, USA, 1987; pp. 302–320. [Google Scholar]
- Fu, K.-Y.; Guo, W.-C.; Ahmat, T.; Li, G.-Q. Knockdown of a nutrient amino acid transporter gene LdNAT1 reduces free neutral amino acid contents and impairs Leptinotarsa decemlineata pupation. Sci. Rep. 2015, 5, 18124. [Google Scholar] [CrossRef] [PubMed]
- Mirandela, G.D.; Tamburrino, G.; Hoskisson, P.A.; Zachariae, U.; Javelle, A. The lipid environment determines the activity of the Escherichia coli ammonium transporter AmtB. FASEB J. 2019, 33, 1989–1999. [Google Scholar] [CrossRef] [PubMed]
- Soupene, E.; Inwood, W.; Kustu, S. Lack of the Rhesus protein Rh1 impairs growth of the green alga Chlamydomonas reinhardtii at high CO2. Proc. Natl. Acad. Sci. USA 2004, 101, 7787–7792. [Google Scholar] [CrossRef] [PubMed]
- Lushchak, V.I. Environmentally induced oxidative stress in aquatic animals. Aquat. Toxicol. 2011, 101, 13–30. [Google Scholar] [CrossRef]
- Sharma, P.; Jha, A.B.; Dubey, R.S.; Pessarakli, M. Reactive Oxygen Species, Oxidative Damage, and Antioxidative Defense Mechanism in Plants under Stressful Conditions. J. Bot. 2012, 2012, 217037. [Google Scholar] [CrossRef]
- Duan, Y.; Liu, P.; Li, J.; Li, J.; Chen, P. Expression profiles of selenium dependent glutathione peroxidase and glutathione S-transferase from Exopalaemon carinicauda in response to Vibrio anguillarum and WSSV challenge. Fish Shellfish Immunol. 2013, 35, 661–670. [Google Scholar] [CrossRef]
- Gomezanduro, G.; Barillasmury, C.; Peregrino-Uriarte, A.B.; Gupta, L.; Gollasgalvan, T.; Hernandezlopez, J.; Yepizplascencia, G. The cytosolic manganese superoxide dismutase from the shrimp Litopenaeus vannamei: Molecular cloning and expression. Dev. Comp. Immunol. 2006, 30, 893–900. [Google Scholar] [CrossRef]
- Aertsen, A.; Van Houdt, R.; Vanoirbeek, K.; Michiels, C.W. An SOS Response Induced by High Pressure in Escherichia coli. J. Bacteriol. 2004, 186, 6133–6141. [Google Scholar] [CrossRef]
- Rothschild, L.J.; Mancinelli, R.L. Life in extreme environments. Nature 2001, 409, 1092–1101. [Google Scholar] [CrossRef]
- Hartwig, A. Carcinogenicity of metal compounds: Possible role of DNA repair inhibition. Toxicol. Lett. 1998, 102, 235–239. [Google Scholar] [CrossRef]
- D’Orazi, G.; Cecchinelli, B.; Bruno, T.; Manni, I.; Higashimoto, Y.; Saito, S.; Gostissa, M.; Coen, S.; Marchetti, A.; Del Sal, G.; et al. Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat. Cell Biol. 2001, 4, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Kuwano, Y.; Nishida, K.; Akaike, Y.; Kurokawa, K.; Nishikawa, T.; Masuda, K.; Rokutan, K. Homeodomain-Interacting Protein Kinase-2: A Critical Regulator of the DNA Damage Response and the Epigenome. Int. J. Mol. Sci. 2016, 17, 1638. [Google Scholar] [CrossRef] [PubMed]
- Reuven, N.; Adler, J.; Porat, Z.; Polonio-Vallon, T.; Hofmann, T.G.; Shaul, Y. The Tyrosine Kinase c-Abl Promotes Homeodomain-interacting Protein Kinase 2 (HIPK2) Accumulation and Activation in Response to DNA Damage. J. Biol. Chem. 2015, 290, 16478–16488. [Google Scholar] [CrossRef]
- Wieland, I.; Arden, K.C.; Michels, D.; Klein-Hitpass, L.; Böhm, M.; Viars, C.S.; Weidle, U.H. Isolation of DICE1: A gene frequently affected by LOH and downregulated in lung carcinomas. Oncogene 1999, 18, 4530–4537. [Google Scholar] [CrossRef] [PubMed]
- Wieland, I.; Röpke, A.; Stumm, M.; Sell, C.; Weidle, U.; Wieacker, P. Molecular Characterization of the DICE1 (DDX26) Tumor Suppressor Gene in Lung Carcinoma Cells. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2001, 12, 491–500. [Google Scholar] [CrossRef]
- Hannan, K.M.; Soo, P.; Wong, M.S.; Lee, J.K.; Hein, N.; Evers, M.; Wysoke, K.D.; Williams, T.D.; Montellese, C.; Smith, L.K.; et al. Nuclear stabilisation of p53 requires a functional nucleolar surveillance pathway. bioRxiv 2021. [Google Scholar] [CrossRef]
- Jha, S.; Dutta, A. RVB1/RVB2: Running Rings around Molecular Biology. Mol. Cell 2009, 34, 521–533. [Google Scholar] [CrossRef]
- Nano, N.; Houry, W.A. Chaperone-like activity of the AAA+ proteins Rvb1 and Rvb2 in the assembly of various complexes. Philos. Trans. R. Soc. B Biol. Sci. 2013, 368, 20110399. [Google Scholar] [CrossRef]
- Yi, F.; Wang, Z.; Liu, J.; Zhang, Y.; Wang, Z.; Xu, H.; Li, X.; Bai, N.; Cao, L.; Song, X. Structural Maintenance of Chromosomes protein 1: Role in Genome Stability and Tumorigenesis. Int. J. Biol. Sci. 2017, 13, 1092–1099. [Google Scholar] [CrossRef]
- Langsfeld, E.S.; Bodily, J.; Laimins, L.A. The Deacetylase Sirtuin 1 Regulates Human Papillomavirus Replication by Modulating Histone Acetylation and Recruitment of DNA Damage Factors NBS1 and Rad51 to Viral Genomes. PLoS Pathog. 2015, 11, e1005181. [Google Scholar] [CrossRef]
- Terauchi, K.; Kobayashi, H.; Yatabe, K.; Yui, N.; Fujiya, H.; Niki, H.; Musha, H.; Yudoh, K. The NAD-Dependent Deacetylase Sirtuin-1 Regulates the Expression of Osteogenic Transcriptional Activator Runt-Related Transcription Factor 2 (Runx2) and Production of Matrix Metalloproteinase (MMP)-13 in Chondrocytes in Osteoarthritis. Int. J. Mol. Sci. 2016, 17, 1019. [Google Scholar] [CrossRef] [PubMed]
- Van Dover, C.L.; Lutz, R.A. Experimental ecology at deep-sea hydrothermal vents: A perspective. J. Exp. Mar. Biol. Ecol. 2004, 300, 273–307. [Google Scholar] [CrossRef]
- Tokuda, G.; Yamada, A.; Nakano, K.; Arita, N.O.; Yamasaki, H. Colonization of Sulfurovum sp. on the gill surfaces of Alvinocaris longirostris, a deep-sea hydrothermal vent shrimp. Mar. Ecol. 2008, 29, 106–114. [Google Scholar] [CrossRef]
- Petersen, J.M.; Ramette, A.; Lott, C.; Cambon-Bonavita, M.-A.; Zbinden, M.; Dubilier, N. Dual symbiosis of the vent shrimp Rimicaris exoculata with filamentous gamma- and epsilonproteobacteria at four Mid-Atlantic Ridge hydrothermal vent fields. Environ. Microbiol. 2010, 12, 2204–2218. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.-l.; Zeng, Z.-g.; Chen, S.; Sun, L. First comparative analysis of the community structures and carbon metabolic pathways of the bacteria associated with Alvinocaris longirostris in a hydrothermal vent of okinawa trough. PLoS ONE 2016, 11, e0154359. [Google Scholar] [CrossRef]
- Sievert, S.; Brinkhoff, T.; Muyzer, G.; Ziebis, W.; Kuever, J. Spatial Heterogeneity of Bacterial Populations along an Environmental Gradient at a Shallow Submarine Hydrothermal Vent near Milos Island (Greece). Appl. Environ. Microbiol. 1999, 65, 3834–3842. [Google Scholar] [CrossRef]
- Pagé, A.; Tivey, M.K.; Stakes, D.S.; Reysenbach, A.-L. Temporal and spatial archaeal colonization of hydrothermal vent deposits. Environ. Microbiol. 2008, 10, 874–884. [Google Scholar] [CrossRef]
- Goncalves, P.; Vernal, J.; Rosa, R.D.; Yepiz-Plascencia, G.; de Souza, C.; Barracco, M.A.; Perazzolo, L.M. Evidence for a novel biological role for the multifunctional β-1,3-glucan binding protein in shrimp. Mol. Immunol. 2012, 51, 363–367. [Google Scholar] [CrossRef]
- Kawabata, S.; Beisel, H.G.; Huber, R.; Bode, W.; Gokudan, S.; Muta, T.; Tsuda, R.; Koori, K.; Kawahara, T.; Seki, N.; et al. Role of tachylectins in host defense of the Japanese horseshoe crab Tachypleus Tridentatus. In Phylogenetic Perspectives on the Vertebrate Immune System; Beck, G., Sugumaran, M., Cooper, E.L., Eds.; Springer: Boston, MA, USA, 2001; pp. 195–202. [Google Scholar]
- Liu, X.-L.; Ye, S.; Cheng, C.-Y.; Li, H.-W.; Lu, B.; Yang, W.-J.; Yang, J.-S. Identification and characterization of a symbiotic agglutination-related C-type lectin from the hydrothermal vent shrimp Rimicaris exoculata. Fish Shellfish Immunol. 2019, 92, 1–10. [Google Scholar] [CrossRef]
- Gu, H.-j.; Sun, Q.-l.; Jiang, S.; Zhang, J.; Sun, L. First characterization of an anti-lipopolysaccharide factor (ALF) from hydrothermal vent shrimp: Insights into the immune function of deep-sea crustacean ALF. Dev. Comp. Immunol. 2018, 84, 382–395. [Google Scholar] [CrossRef]
- Emani, C.; Garcia, J.M.; Lopata-Finch, E.; Pozo, M.J.; Uribe, P.; Kim, D.-J.; Sunilkumar, G.; Cook, D.R.; Kenerley, C.M.; Rathore, K.S. Enhanced fungal resistance in transgenic cotton expressing an endochitinase gene from Trichoderma virens. Plant Biotechnol. J. 2003, 1, 321–336. [Google Scholar] [CrossRef] [PubMed]
- Boot, R.G.; Blommaart, E.F.C.; Swart, E.; der Vlugt, K.G.-V.; Bijl, N.; Moe, C.; Place, A.; Aerts, J. Identification of a Novel Acidic Mammalian Chitinase Distinct from Chitotriosidase. J. Biol. Chem. 2001, 276, 6770–6778. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Zheng, T.; Homer, R.J.; Kim, Y.-K.; Chen, N.Y.; Cohn, L.; Hamid, Q.; Elias, J.A. Acidic Mammalian Chitinase in Asthmatic Th2 Inflammation and IL-13 Pathway Activation. Science 2004, 304, 1678–1682. [Google Scholar] [CrossRef] [PubMed]
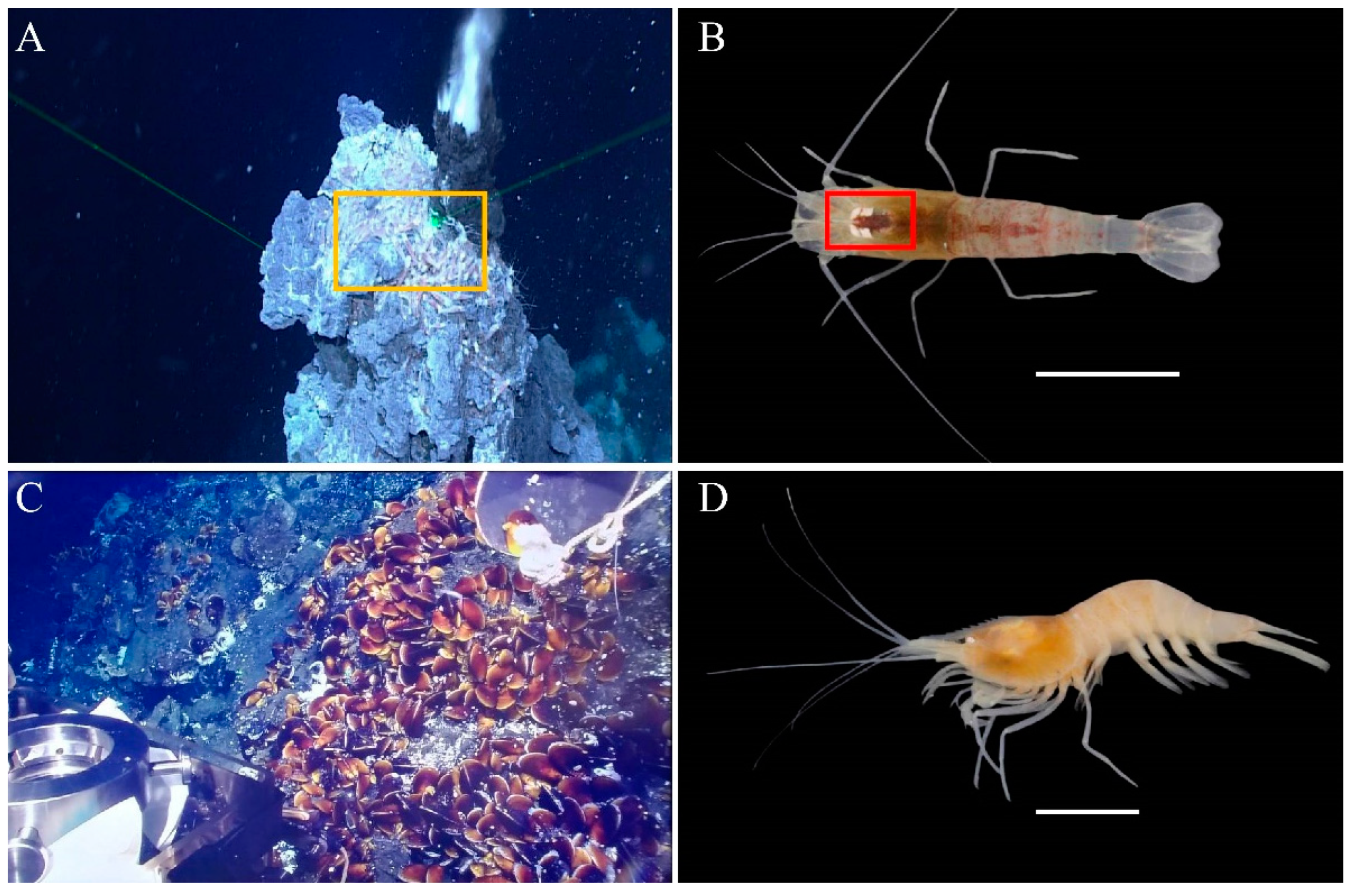
| Parameters | S. leurokolos | A. longirostris |
|---|---|---|
| Total bases | 91.61 G | 80.72 G |
| Number of polymerases reads | 57,789,107 | 48,178,061 |
| Polymerases reads N50 value | 2180 | 2228 |
| Reads of Insert (ROI) | 1,059,004 | 929,230 |
| Number of full-length non-chimeric reads | 612,807 | 389,605 |
| Number of consensus isoforms | 23,682 | 18,939 |
| Number of polished high-quality isoforms | 23,414 | 18,794 |
| Number of unigenes | 16,037 | 13,666 |
| Unigenes N50 length | 2880 bp | 3046 bp |
| Annotation Database | S. leurokolos | A. longirostris |
|---|---|---|
| NR_Annotation | 11,302 (70.47%) | 10,001 (73.18%) |
| Swissprot_Annotation | 9596 (59.84%) | 8263 (60.46%) |
| KOG_Annotation | 8465 (52.78%) | 7084 (51.84%) |
| KEGG_Annotation | 7918 (49.37%) | 6717 (49.15%) |
| GO_Annotation | 7007 (43.69%) | 5720 (41.86%) |
| NT_Annotation | 4809 (29.99%) | 3561 (26.06%) |
| All_Annotated | 11,718 (73.07%) | 10,243 (74.95%) |
| Parameters | AA | S. leurokolos | A. longirostris | M. nipponense |
|---|---|---|---|---|
| Non-polar AA | Leu # | 8.12 | 8.28 | 9.55 |
| Ala | 6.87 | 6.63 | 6.93 | |
| Val | 6.43 | 6.53 | 6.46 | |
| Ile | 4.94 | 4.99 | 4.26 | |
| Phe # | 3.44 | 3.54 | 1.49 | |
| Met | 2.42 | 2.35 | 2.04 | |
| Trp | 1.03 | 1.05 | 1.52 | |
| Polar-uncharged AA | Ser | 7.94 | 8.35 | 8.81 |
| Gly | 6.89 | 6.91 | 6.23 | |
| Thr | 6.04 | 6.14 | 5.53 | |
| Pro | 5.81 | 5.66 | 6.28 | |
| Asn | 4.17 | 4.33 | 3.40 | |
| Gln | 4.13 | 4.20 | 4.15 | |
| Tyr | 2.87 | 3.00 | 2.41 | |
| Cys | 1.81 | 2.01 | 1.70 | |
| Negative-charged AA | Glu | 7.31 | 6.82 | 6.34 |
| Asp | 5.79 | 5.62 | 5.21 | |
| Positive-charged AA | Lys # | 6.18 | 5.74 | 8.03 |
| Arg # | 5.14 | 5.11 | 7.00 | |
| His | 2.49 | 2.55 | 2.46 |
| Gene Name | Gene Description | Function | Adjusted p Value |
|---|---|---|---|
| DNA repair | |||
| HIPK2 | Homeodomain-interacting protein kinase 2 | Activate DNA damage response | 1.94 × 10−3 |
| INT6 | Integrator complex subunit 6 | DNA damage repair | 3.73 × 10−8 |
| HEAT3 | HEAT repeat-containing protein 3 | Responding DNA damage | 3.67 × 10−8 |
| RUVB2 | RuvB-like 2 | DNA damage repair | 2.18 × 10−6 |
| SMC1A | Structural maintenance of chromosomes protein 1A | DNA damage repair | 4.74 × 10−10 |
| SIR1 | NAD-dependent protein deacetylase sirtuin-1 | DNA damage repair | 0.00 |
| Oxidative stress | |||
| CCD51 | Mitochondrial potassium channel | Reduce ROS production | 0.01 |
| RT05 | 28S ribosomal protein S5 | Reduce ROS production | 7.80 × 10−5 |
| DHC24 | Delta (24)-sterol reductase | ROS scavenging | 1.45 × 10−9 |
| IMP2L | Mitochondrial inner membrane protease subunit 2 | Regulate ROS production | 0.03738529 |
| VA0E | V-type proton ATPase subunit e | Prevent V-ATPase oxidative damage | 0.006203782 |
| Temperature stress | |||
| PPID | Peptidyl-prolyl cis-trans isomerase D | Heat shock response | 1.13 × 10−8 |
| EXL1 | Chloride intracellular channel exl-1 | Heat shock response | 0.01 |
| L2EFL | Protein lethal (2) essential for life | Heat shock response | 1.85 × 10−7 |
| NU155 | Nuclear pore complex protein Nup155 | Cold shock response | 9.69 × 10−8 |
| TCPQ | T-complex protein 1 subunit theta | Adaptation to low temperature | 3.72 × 10−7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, A.; Sha, Z.; Hui, M. Full-Length Transcriptome Comparison Provides Novel Insights into the Molecular Basis of Adaptation to Different Ecological Niches of the Deep-Sea Hydrothermal Vent in Alvinocaridid Shrimps. Diversity 2022, 14, 371. https://doi.org/10.3390/d14050371
Wang A, Sha Z, Hui M. Full-Length Transcriptome Comparison Provides Novel Insights into the Molecular Basis of Adaptation to Different Ecological Niches of the Deep-Sea Hydrothermal Vent in Alvinocaridid Shrimps. Diversity. 2022; 14(5):371. https://doi.org/10.3390/d14050371
Chicago/Turabian StyleWang, Aiyang, Zhongli Sha, and Min Hui. 2022. "Full-Length Transcriptome Comparison Provides Novel Insights into the Molecular Basis of Adaptation to Different Ecological Niches of the Deep-Sea Hydrothermal Vent in Alvinocaridid Shrimps" Diversity 14, no. 5: 371. https://doi.org/10.3390/d14050371
APA StyleWang, A., Sha, Z., & Hui, M. (2022). Full-Length Transcriptome Comparison Provides Novel Insights into the Molecular Basis of Adaptation to Different Ecological Niches of the Deep-Sea Hydrothermal Vent in Alvinocaridid Shrimps. Diversity, 14(5), 371. https://doi.org/10.3390/d14050371






