Widespread Occurrence of Two Planktonic Ciliate Species (Urotricha, Prostomatida) Originating from High Mountain Lakes
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Sites, Sampling, Cultivation and Identification of the Ciliates
2.2. Molecular Methods and Phylogenetic Analyzes
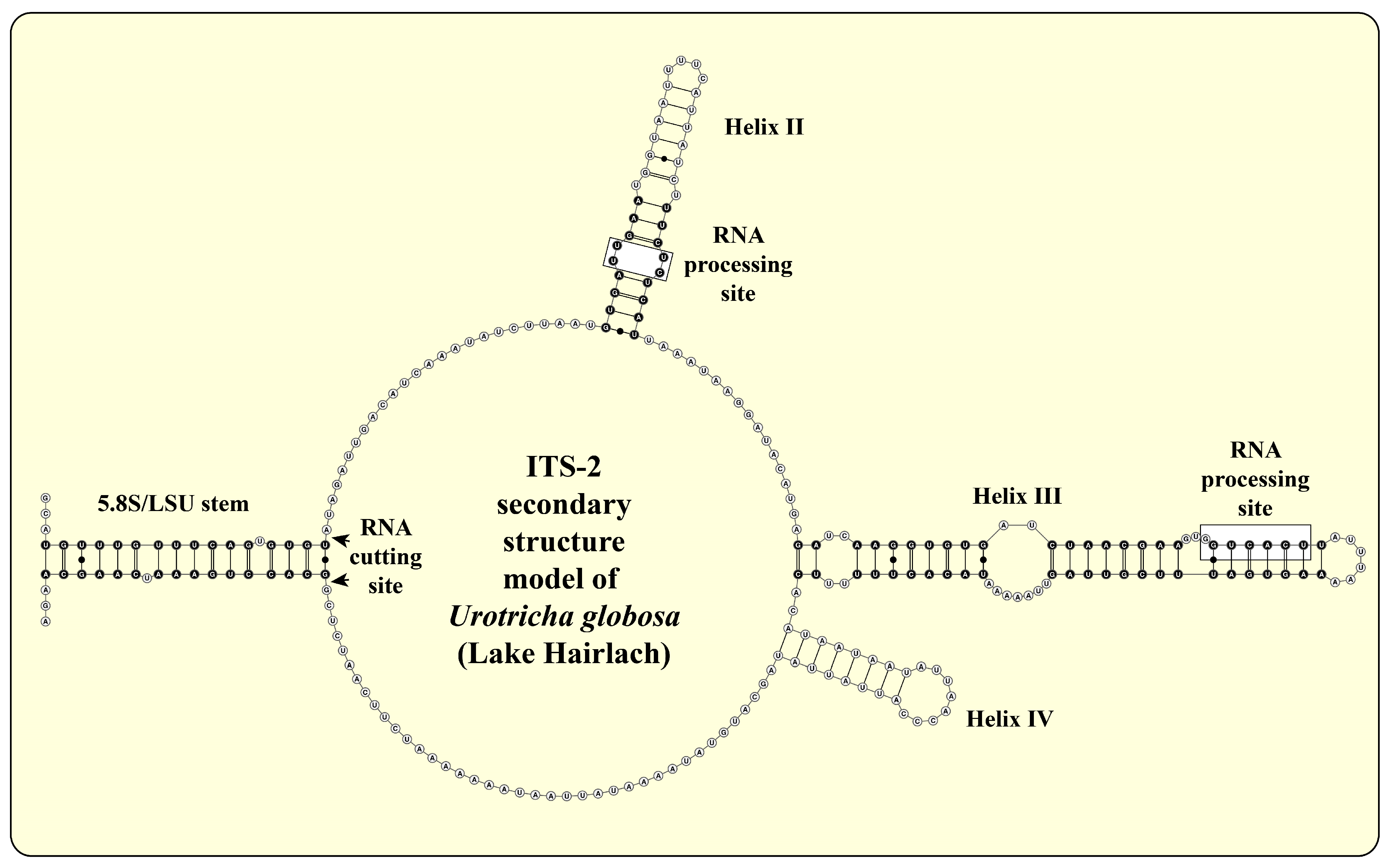
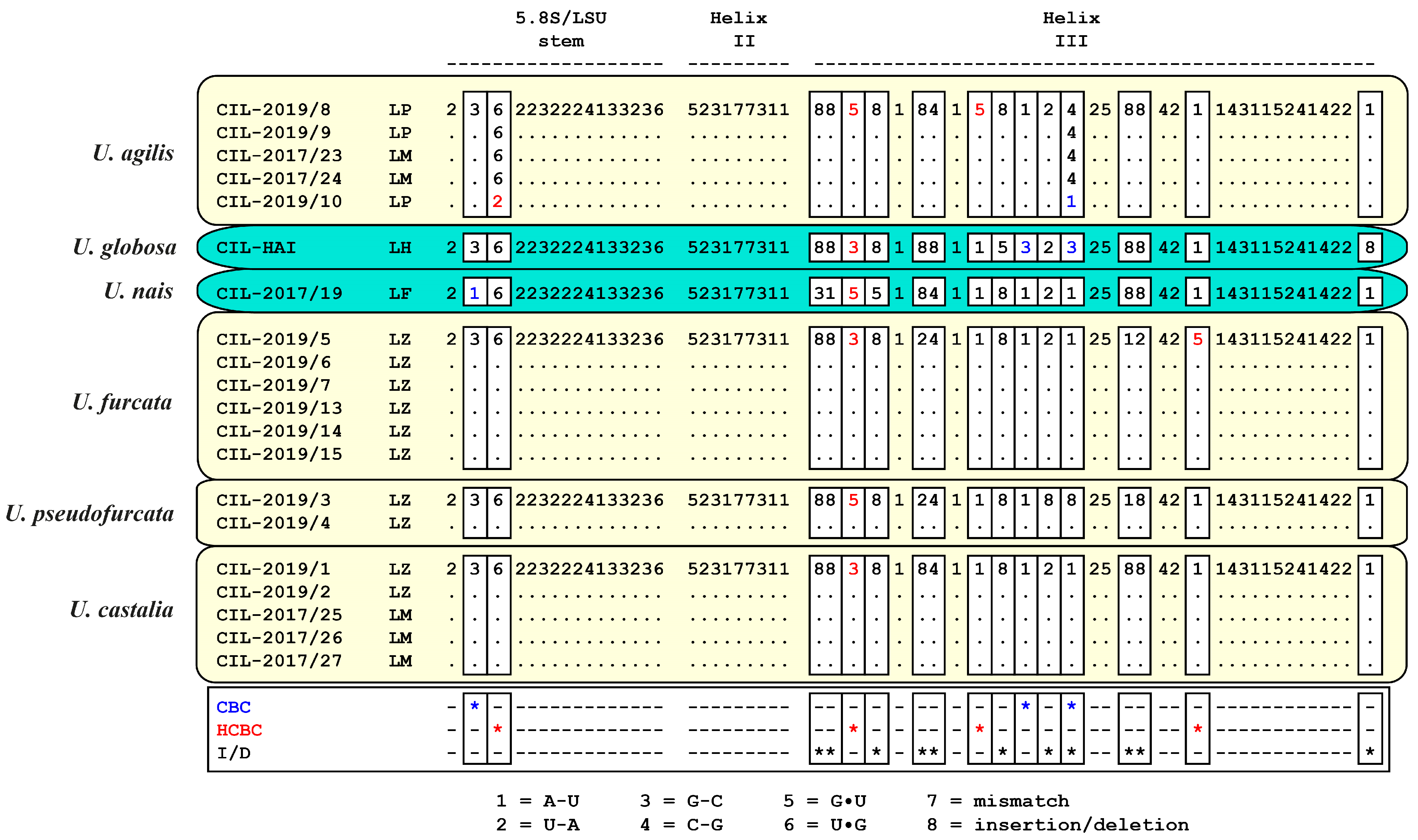
2.3. ITS-2 Secondary Structures and ITS-2/CBC Approach
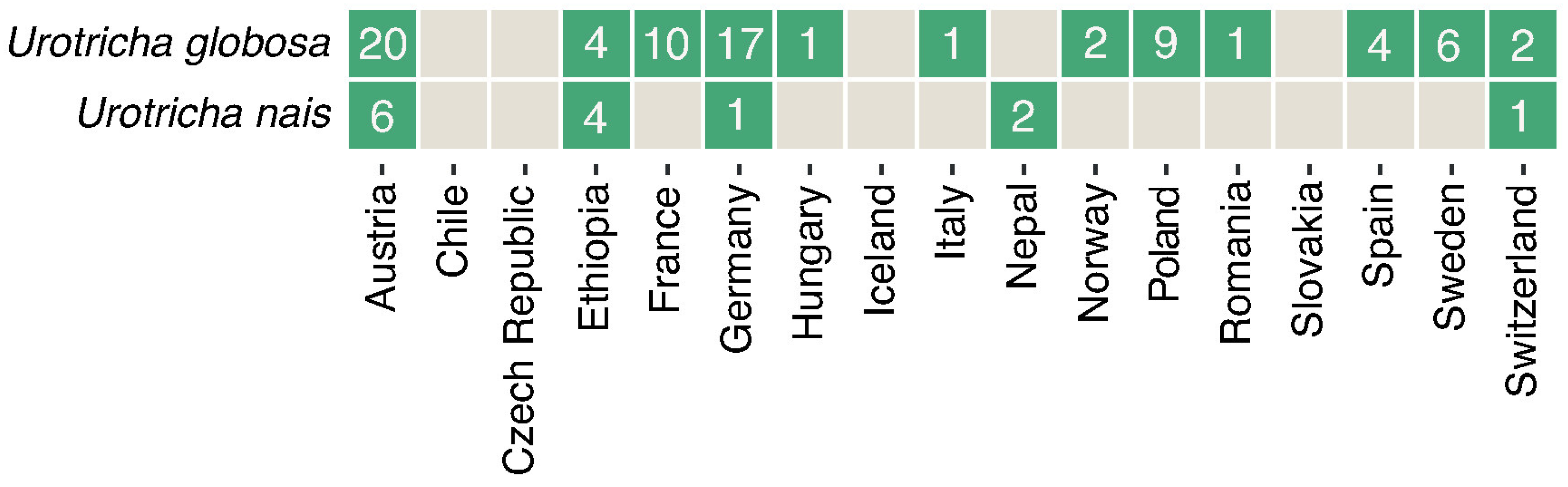
2.4. Screening for Ciliate Biogeography and Correlation to Abiotic Lake Water Parameters
3. Results
3.1. Identification of the Urotrichs
3.2. Molecular Phylogeny
3.3. Biogeography and Correlations to Abiotic Environmental Datasets
4. Discussion
Comparison of the Two Urotricha Species with Congeners
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Frantal, D.; Agatha, S.; Beisser, D.; Boenigk, J.; Darienko, T.; Dirren-Pitsch, G.; Filker, S.; Gruber, M.; Kammerlander, B.; Nachbaur, L.; et al. Molecular data reveal a cryptic diversity in the genus Urotricha (Alveolata, Ciliophora, Prostomatida), a key player in freshwater lakes, with remarks on morphology, food preferences, and distribution. Front. Microbiol. 2022, 12, 787290. [Google Scholar] [CrossRef] [PubMed]
- Müller, H.; Schöne, A.; Pinto-Coelho, R.M.; Schweizer, A.; Weisse, T. Seasonal succession of ciliates in Lake Constance. Microb. Ecol. 1991, 21, 119–138. [Google Scholar] [CrossRef] [PubMed]
- Qu, Z.; Forster, D.; Bruni, E.P.; Frantal, D.; Kammerlander, B.; Nachbaur, L.; Pitsch, G.; Posch, T.; Pröschold, T.; Teubner, K.; et al. Aquatic food webs in deep temperate lakes: Key species establish through their autecological versatility. Mol. Ecol. 2021, 30, 1053–1071. [Google Scholar] [CrossRef]
- Wille, A.; Sonntag, B.; Sattler, B.; Psenner, R. Abundance, biomass and size structure of the microbial assemblage in the high mountain lake Gossenköllesee (Tyrol, Austria) during the ice-free period. J. Limnol. 1999, 58, 117–126. [Google Scholar] [CrossRef]
- Sonntag, B.; Summerer, M.; Sommaruga, R. Factors involved in the distribution pattern of ciliates in the water column of a transparent alpine lake. J. Plankton Res. 2011, 33, 541–546. [Google Scholar] [CrossRef][Green Version]
- Sonntag, B.; Kammerlander, B.; Summerer, M. Bioaccumulation of ultraviolet sunscreen compounds (mycosporine-like amino acids) by the heterotrophic freshwater ciliate Bursaridium living in alpine lakes. Inland Waters 2017, 7, 55–64. [Google Scholar] [CrossRef]
- Kammerlander, B.; Koinig, K.A.; Rott, E.; Sommaruga, R.; Tartarotti, B.; Trattner, F.; Sonntag, B. Ciliate community structure and interactions within the planktonic food web in two alpine lakes of contrasting transparency. Freshwat. Biol. 2016, 61, 1950–1965. [Google Scholar] [CrossRef]
- Sommer, F.; Sonntag, B.; Rastl, N.; Summerer, M.; Tartarotti, B. Ciliates in man-made mountain reservoirs. Front. Microbiol. 2022; under revision. [Google Scholar]
- Rose, K.C.; Williamson, C.E.; Saros, J.E.; Sommaruga, R.; Fischer, J.M. Differences in UV transparency and thermal structure between alpine and subalpine lakes: Implications for organisms. Photochem. Photobiol. 2009, 8, 1244–1256. [Google Scholar] [CrossRef]
- Kammerlander, B.; Breiner, H.W.; Filker, S.; Sommaruga, R.; Sonntag, B.; Stoeck, T. High diversity of protistan plankton communities in remote high mountain lakes in the European Alps and the Himalayan mountains. FEMS Microbiol. Ecol. 2015, 91, fiv010. [Google Scholar] [CrossRef][Green Version]
- Foissner, W. An update of ‘basic light and scanning electron microscopic methods for taxonomic studies of ciliated protozoa’. Int. J. Syst. Evol. Microbiol. 2014, 64, 271–292. [Google Scholar] [CrossRef]
- Skibbe, O. An improved quantitative protargol stain for ciliates and other planktonic protists. Arch. Hydrobiol. 1994, 130, 339–347. [Google Scholar] [CrossRef]
- Foissner, W.; Berger, H.; Kohmann, F. Taxonomische und Ökologische Revision der Ciliaten des Saprobiensystems. Band III: Hymenostomata, Prostomatida, Nassulida. Informationsber. Bayer; Landesamt für Wasserwirtschaft: Munich, Germany, 1994; Volume 1/94, pp. 1–548. [Google Scholar]
- Foissner, W.; Berger, H.; Schaumburg, J. Identification and Ecology of Limnetic Plankton Ciliates. Informationsber. Bayer; Landesamt für Wasserwirtschaft: Munich, Germany, 1999; Volume 3/99, pp. 1–793. [Google Scholar]
- Swofford, D.L. PAUP* Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4.0b10; Sinauer Associates: Sunderland, MA, USA, 2002. [Google Scholar]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Jow, H.; Hudelot, C.; Rattray, M.; Higgs, P. Bayesian phylogenetics using an RNA substitution model applied to early mammalian evolution. Mol. Biol. Evol. 2002, 19, 1591–1601. [Google Scholar] [CrossRef]
- Higgs, P.; Jameson, D.; Jow, H.; Rattray, M. The evolution of tRNA-Leu genes in animal mitochondrial genomes. J. Mol. Evol. 2003, 57, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Hudelot, C.; Gowri-Shankar, V.; Jow, H.; Rattray, M.; Higgs, P. RNA-based phylogenetic methods: Application to mammalian mitochondrial RNA sequences. Mol. Phylogen. Evol. 2003, 28, 241–252. [Google Scholar] [CrossRef]
- Gibson, A.; Gowri-Shankar, V.; Higgs, P.; Rattray, M. A comprehensive analysis of mammalian mitochondrial genome base composition and improved phylogenetic methods. Mol. Biol. Evol. 2005, 22, 251–264. [Google Scholar] [CrossRef]
- Telford, M.J.; Wise, M.J.; Gowri-Shankar, V. Consideration of RNA secondary structure significantly improves likelihood-based estimates of phylogeny: Examples from the bilateria. Mol. Biol. Evol. 2005, 22, 1129–1136. [Google Scholar] [CrossRef] [PubMed]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acid Res. 2003, 31, 3406–3615. [Google Scholar] [CrossRef]
- Byun, Y.; Han, K. PseudoViewer. Web application and web service for visualizing RNA pseudoknots and secondary structures. Nucleic Acids Res. 2006, 34, W416–W422. [Google Scholar] [CrossRef]
- Do, C.B.; Woods, D.A.; Batzoglou, S. CONTRAfold: RNA secondary structure prediction without physics-based models. Bioinformatics 2006, 22, e90–e98. [Google Scholar] [CrossRef]
- Coleman, A.W. ITS2 is a double-edged tool for eukaryote evolutionary comparisons. Trends Genet. 2003, 19, 370–375. [Google Scholar] [CrossRef]
- Coleman, A.W. Pan-eukaryote ITS2 homologies revealed by RNA secondary structure. Nucleic Acids Res. 2007, 35, 3322–3329. [Google Scholar] [CrossRef] [PubMed]
- Cote, C.A.; Greer, C.L.; Peculis, B.A. Dynamic conformational model for the role of ITS2 in pre-rRNA processing in yeast. RNA 2002, 8, 786–797. [Google Scholar] [PubMed]
- Coleman, A.W. Paramecium aurelia revisited. J. Eukaryot. Microbiol. 2005, 52, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Darienko, T.; Gustavs, L.; Eggert, A.; Wolf, W.; Pröschold, T. Evaluating the species boundaries of green microalgae (Coccomyxa, Trebouxiophyceae, Chlorophyta) using integrative taxonomy and DNA barcoding with further implications for the species identification in environmental samples. PLoS ONE 2015, 10, e0127838. [Google Scholar] [CrossRef]
- Boenigk, J.; Wodniok, S.; Bock, C.; Beisser, D.; Hempel, C.; Grossmann, L.; Lange, A.; Jensen, M. Geographic distance and mountain range structure freshwater protist communities on a European scale. Metabarcoding Metagenom. 2018, 2, e21519. [Google Scholar] [CrossRef]
- Oikonomou, A.; Filker, S.; Breiner, H.-W.; Stoeck, T. Protistan diversity in a permanently stratified meromictic lake (Lake Alatsee, SW Germany). Environ. Microbiol. 2015, 17, 2144–2157. [Google Scholar] [CrossRef]
- Filker, S.; Sommaruga, R.; Vila, I.; Stoeck, T. Microbial eukaryote plankton communities of high-mountain lakes from three different continents exhibit strong biogeographic patterns. Mol. Ecol. 2016, 25, 2286–2301. [Google Scholar] [CrossRef]
- Weisse, T.; Karstens, N.; Meyer, V.C.L.; Janke, L.; Lettner, S.; Teichgräber, K. Niche separation in common prostome freshwater ciliates: The effect of food and temperature. Aquat. Microb. Ecol. 2001, 26, 167–179. [Google Scholar] [CrossRef]
- Muñoz, A.; Téllez, C.; Fernández-Galiano, D. Morphology and infraciliature in Urotricha nais sp. n. and Urotricha castalia sp. n. (Ciliophora, Prorodontida). Acta Protozool. 1987, 26, 197–204. [Google Scholar]
- Song, W.; Wilbert, N. Taxonomische Untersuchungen an Aufwuchsciliaten (Protozoa, Ciliophora) im Poppelsdorfer Weiher, Bonn. Lauterbornia 1989, 3, 2–221. [Google Scholar]
- Krainer, K.-H. Taxonomische Untersuchungen an neuen und wenig bekannten planktischen Ciliaten (Protozoa: Ciliophora) aus Baggerseen in Österreich. Lauterbornia 1995, 21, 39–68. [Google Scholar]
- Foissner, W. Ecological and systematic studies on the neuston of alpine pools, with special regard to ciliated protozoa. Int. Revue Ges. Hydrobiol. 1979, 64, 99–140. [Google Scholar] [CrossRef]
- Sonntag, B.; Posch, T.; Klammer, S.; Teubner, K.; Psenner, R. Phagotrophic ciliates and flagellates in an oligotrophic, deep, alpine lake: Contrasting variability with seasons and depths. Aquat. Microb. Ecol. 2006, 43, 193–207. [Google Scholar] [CrossRef]
- Sichrowsky, U.; Schabetsberger, R.; Sonntag, B.; Stoyneva, M.; Maloney, A.E.; Nelson, D.B.; Richey, J.N.; Sachs, J.P. Limnological characterization of volcanic crater lakes on Uvea Island (Wallis and Futuna, South Pacific). Pac. Sci. 2014, 68, 333–343. [Google Scholar] [CrossRef]
- Sommaruga, R.; Psenner, R. Nanociliates of the order Prostomatida: Their relevance in the microbial food web of a mesotrophic lake. Aquat. Sci. 1993, 55, 179–187. [Google Scholar] [CrossRef]
- Sonntag, B.; Foissner, W. Urotricha psenneri n. sp. and Amphileptus piger (Vuxanovici, 1962) n. comb., two planktonic ciliates (Protozoa, Ciliophora) from an oligotrophic lake in Austria. J. Eukaryot. Microbiol. 2004, 51, 670–677. [Google Scholar] [CrossRef]
- Foissner, W. Urotricha spetai nov. spec., a new plankton ciliate (Ciliophora, Prostomatea) from a fishpond in the Seidlwinkel Valley, Rauris, Austrian Central Alps. Verh. Zool.-Bot.-Ges. Österreich 2012, 148/149, 173–184. [Google Scholar]
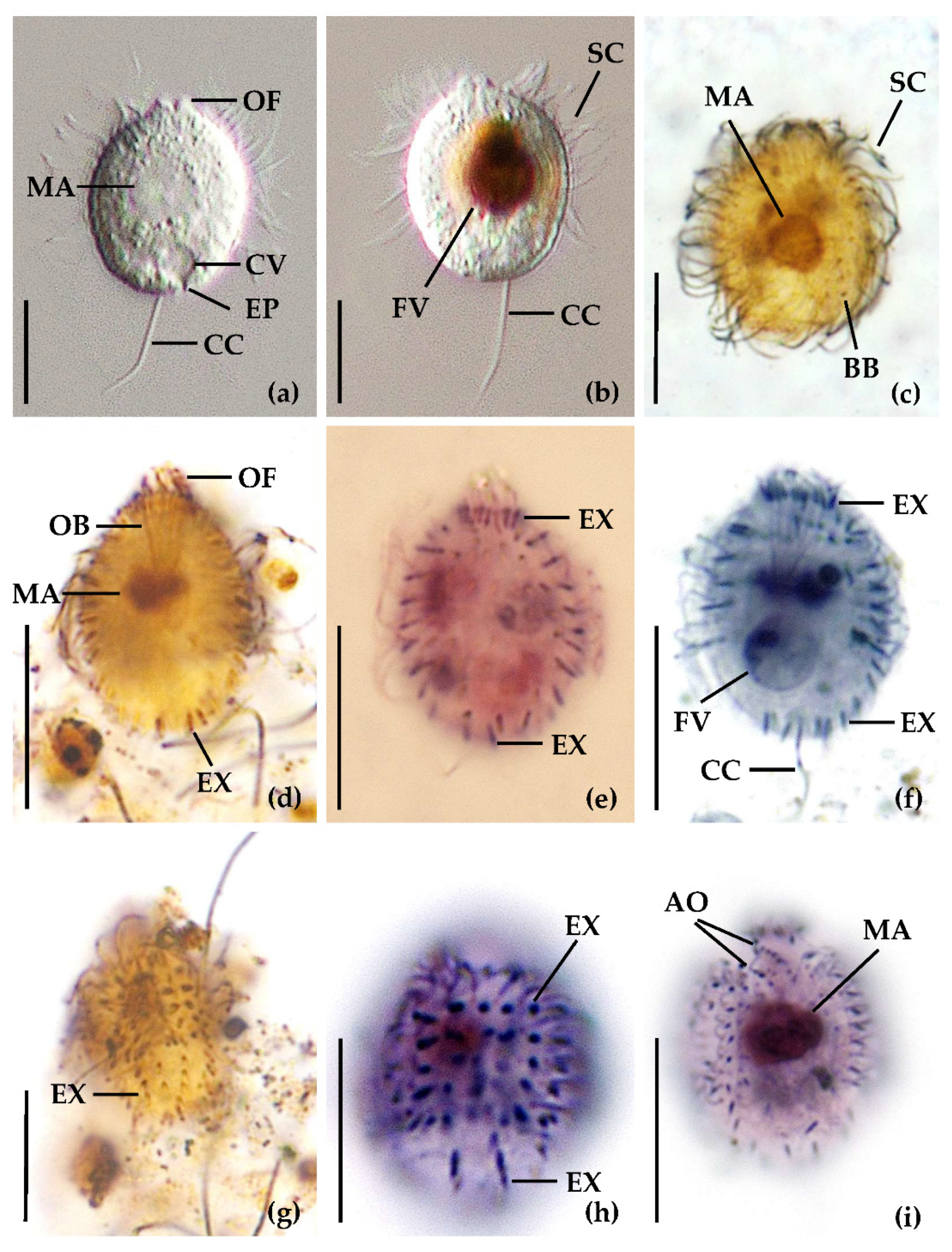
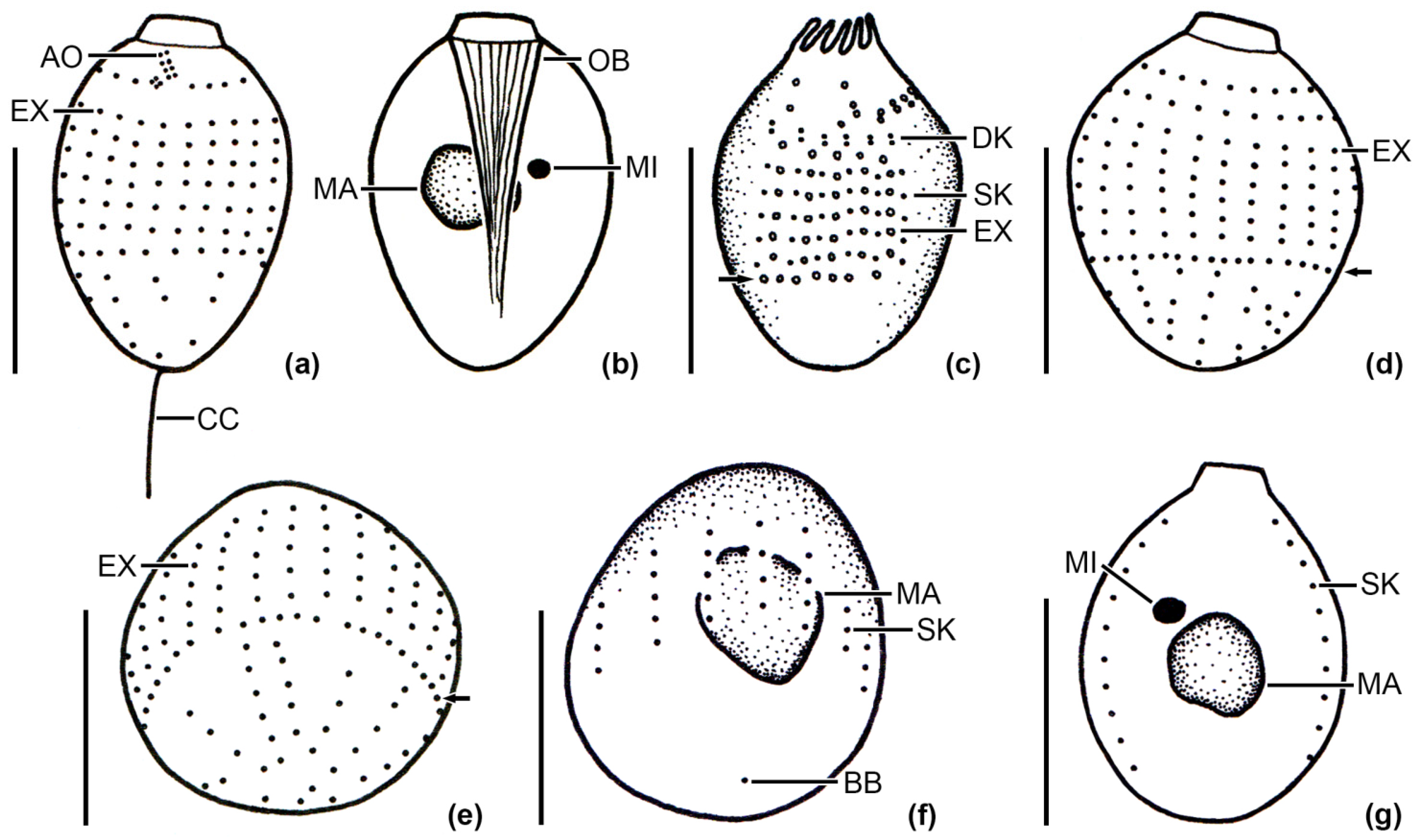
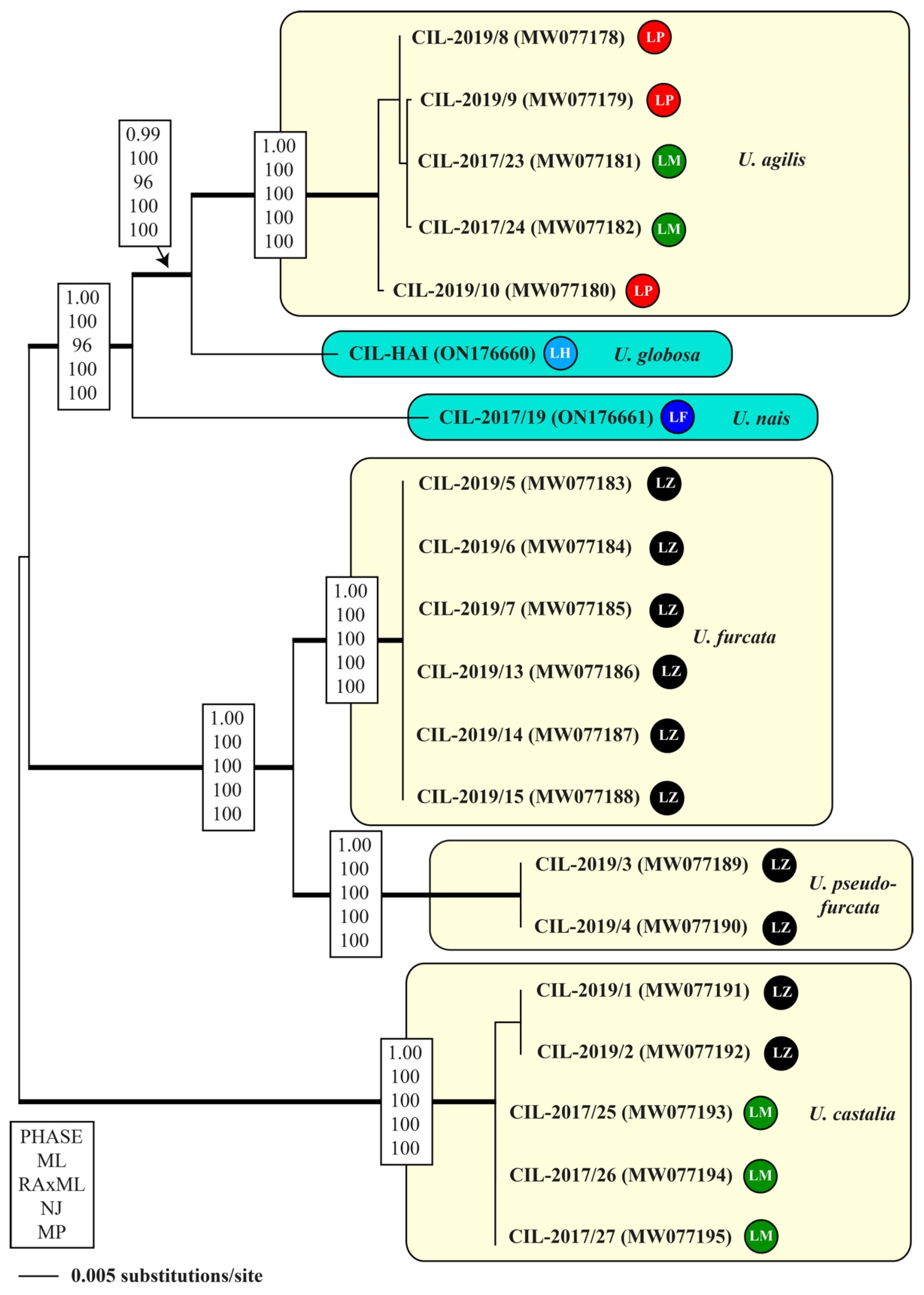
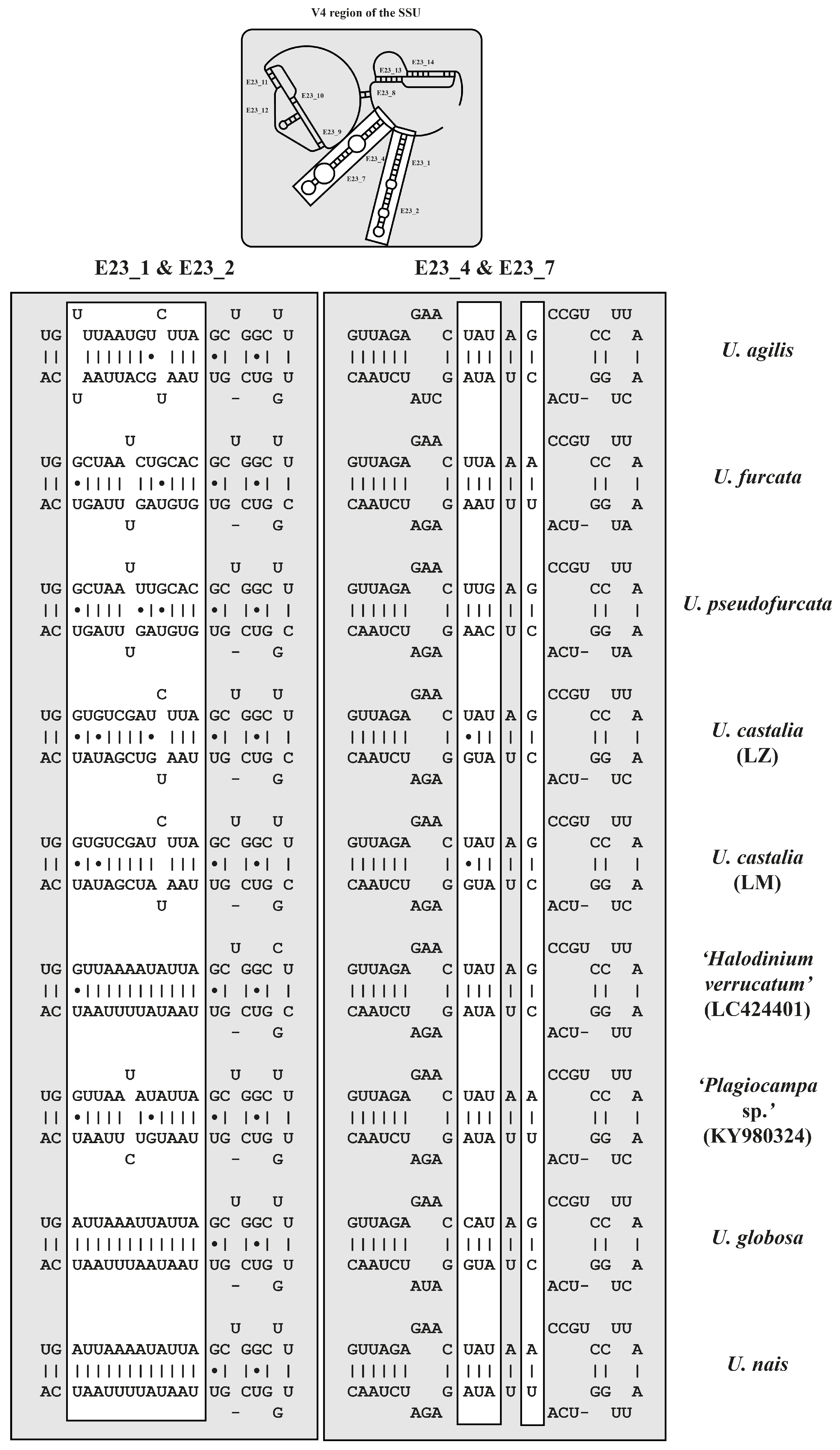

| V4 (218 bp) | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
| 1 | U. agilis | - | 10 | 12 | 26 | 24 | 18 | 19 | 12 | 10 |
| 2 | U. globosa | 0.04587 | - | 8 | 26 | 23 | 19 | 18 | 9 | 12 |
| 3 | U. nais | 0.05505 | 0.03670 | - | 21 | 23 | 19 | 18 | 5 | 6 |
| 4 | U. furcata | 0.11927 | 0.11927 | 0.09633 | - | 5 | 33 | 32 | 22 | 22 |
| 5 | U. pseudofurcata | 0.11009 | 0.10550 | 0.10550 | 0.02294 | - | 31 | 30 | 20 | 24 |
| 6 | U. castalia (LZ) | 0.08257 | 0.08716 | 0.08716 | 0.15138 | 0.14220 | - | 1 | 16 | 16 |
| 7 | U. castalia (LM) | 0.08716 | 0.08257 | 0.08257 | 0.14679 | 0.13761 | 0.00459 | - | 15 | 17 |
| 8 | Halodinium | 0.05505 | 0.04128 | 0.02294 | 0.10092 | 0.09174 | 0.07339 | 0.06881 | - | 9 |
| 9 | Plagiocampa | 0.04587 | 0.05505 | 0.02752 | 0.10092 | 0.11009 | 0.07339 | 0.07798 | 0.04128 | - |
| V9 (98 bp) | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
| 1 | U. agilis | - | 0 | 5 | 10 | 10 | 11 | 12 | 6 | 7 |
| 2 | U. globosa | 0.00000 | - | 5 | 10 | 10 | 11 | 12 | 6 | 7 |
| 3 | U. nais | 0.05102 | 0.05102 | - | 11 | 11 | 14 | 13 | 4 | 5 |
| 4 | U. furcata | 0.10204 | 0.10204 | 0.11224 | - | 0 | 11 | 10 | 12 | 11 |
| 5 | U. pseudofurcata | 0.10204 | 0.10204 | 0.11224 | 0.00000 | - | 11 | 10 | 12 | 11 |
| 6 | U. castalia (LZ) | 0.11224 | 0.11224 | 0.14286 | 0.11224 | 0.11224 | - | 1 | 13 | 12 |
| 7 | U. castalia (LM) | 0.12245 | 0.12245 | 0.13265 | 0.10204 | 0.10204 | 0.01020 | - | 12 | 11 |
| 8 | Halodinium | 0.06122 | 0.06122 | 0.04082 | 0.12245 | 0.12245 | 0.13265 | 0.12245 | - | 3 |
| 9 | Plagiocampa | 0.07143 | 0.07143 | 0.05102 | 0.11224 | 0.11224 | 0.12245 | 0.11224 | 0.03061 | - |
| Characteristic | U. nais Lake Faselfad | U. globosa Lake Hairlach | U. agilis Complex | U. nais | U. globosa | U. ristoi | U. ovata | U. farcta |
|---|---|---|---|---|---|---|---|---|
| Size range (length × width), µm, in vivo | 17–35 × 15–29 | n.d. | 13–23 × 9–16 | n.d. | 18–24 × 15–25 | 15–25 × 10–25 | 25–40 × 20–38 | 15–30 × 15–25 |
| Size range (length × width), µm | 10–18 × 8–14 | 11–17 × 9–15 | 9–16 × 6–14 | 16–34 × 12–32 | * | 12–18 × 10–20 | * | * |
| Somatic kineties, number | 15–19 | 16–24 | 14–24 | 18–21 | 17–25 | 25–28 | 19–27 | 20–30 |
| Cilia in a somatic kinety, number | 8–11 | 5–8 | 5–11 | 5–11 | 13–15 | 9–14 | ca. 19 | 10–16 |
| Circumoral dikinetids, number | n.d. | 8–13 | 8–9 | 9–10 | 11–13 | 5–6 | ca. 10 | 10–14 |
| Adoral organelles, number | 2 * | 2 * | 3 | 2 | 3 | 2 | 3 | 2–3 |
| Adoral organelle 1, dikinetid number | 3–4 | 4 | 3–5 | 4 | n.d. | 4 | n.d. | 3–4 |
| Adoral organelle 2, dikinetid number | 2 | 2 | 2–3 | 2 | n.d. | 4 | n.d. | 3–4 |
| Adoral organelle 3, dikinetid number | - | - | 1–2 | - | n.d. | - | n.d. | 2 |
| Extrusomes present | yes | yes | yes | yes | yes | ‘not seen’ | yes | yes |
| Extrusomes, length, µm | 1.0–2.7 | 1.0–1.5 | 1.0–1.5 | n.d. | short | - | 1.5–2.0 | 1.5–2.0 |
| Reference | This study | This study | [1] | [34] | [35] | [36] | [37] | [13] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sonntag, B.; Frantal, D.; Kammerlander, B.; Darienko, T.; Filker, S.; Stoeck, T.; Gruber, M.; Pröschold, T. Widespread Occurrence of Two Planktonic Ciliate Species (Urotricha, Prostomatida) Originating from High Mountain Lakes. Diversity 2022, 14, 362. https://doi.org/10.3390/d14050362
Sonntag B, Frantal D, Kammerlander B, Darienko T, Filker S, Stoeck T, Gruber M, Pröschold T. Widespread Occurrence of Two Planktonic Ciliate Species (Urotricha, Prostomatida) Originating from High Mountain Lakes. Diversity. 2022; 14(5):362. https://doi.org/10.3390/d14050362
Chicago/Turabian StyleSonntag, Bettina, Daniela Frantal, Barbara Kammerlander, Tatyana Darienko, Sabine Filker, Thorsten Stoeck, Michael Gruber, and Thomas Pröschold. 2022. "Widespread Occurrence of Two Planktonic Ciliate Species (Urotricha, Prostomatida) Originating from High Mountain Lakes" Diversity 14, no. 5: 362. https://doi.org/10.3390/d14050362
APA StyleSonntag, B., Frantal, D., Kammerlander, B., Darienko, T., Filker, S., Stoeck, T., Gruber, M., & Pröschold, T. (2022). Widespread Occurrence of Two Planktonic Ciliate Species (Urotricha, Prostomatida) Originating from High Mountain Lakes. Diversity, 14(5), 362. https://doi.org/10.3390/d14050362







