Abstract
Lumbrineridae is a family of marine annelids with simple external morphology but complex and diverse jaw apparatuses consisting of paired maxillae and mandibles. Here we present the first phylogeny of lumbrinerids based on combination of nuclear (18S rDNA) and mitochondrial (COI, 16S rDNA) markers utilizing Bayesian inference and Maximum Likelihood approaches. Despite limited taxon sampling, our results support monophyly of the genera Abyssoninoe, Augeneria, Gallardoneris, Lumbrineriopsis, and Ninoe and indicate polyphyly of the genera Lumbrineris (the type genus of the family) and Scoletoma. None of the morphological characters traditionally used in lumbrinerid systematics, such as the presence of connecting plates, four pairs of maxillae, bidentate simple hooded hooks, colorless maxillae IV, and multidentate maxillae IV were found to be exclusive homologies for a well-supported clade and have probably evolved several times independently within Lumbrineridae.
1. Introduction
Lumbrineridae Schmarda, 1861 [1] is a family of marine bristle worms from the larger and well-defined group of jaw bearing annelids–Eunicida. The jaws are composed of dorsal maxillae and ventral mandibles located in the ventral muscular pharynx and are used for feeding. Lumbrinerids exhibit rather simple external morphology with poorly developed parapodia and without prominent prostomial and body appendages (Figure 1). Therefore, the details of maxillary apparatus have proven to be useful as diagnostic characters at genus and species level [2,3]. The maxillae of lumbrinerids are considered to be of symmetrognath type [4] with short paired maxillary carriers, forceps-like maxillae I, multidentate massive maxillae II, and two to four pairs of smaller distal maxillary plates (maxillae III–VI). Accessory plates such as attachment lamellae and connecting plates can be found in some genera but are still poorly described for most species [5,6] (Figure 2).
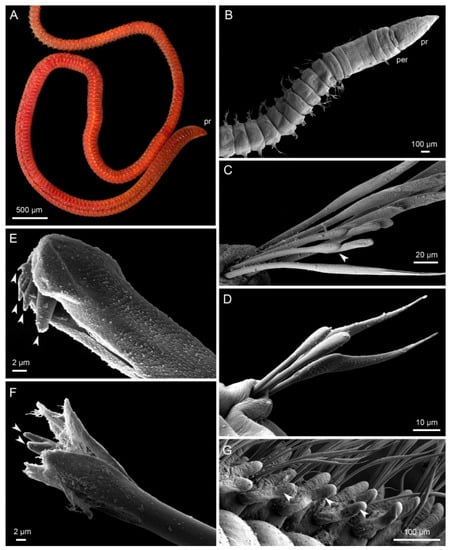
Figure 1.
Morphology of Lumbrineridae, live specimen photograph (A), scanning electron microscopy micrographs (B–G). (A) Scoletoma fragilis, lateral view of anterior part of the body, pr—prostomium ©, Vedenin, A.; (B) Lumbrinerides sp., ventral view of anterior fragment, pr—prostomium, per—peristomium; (C) Lumbrineris sp., limbate chaetae and compound hooks from the 4th parapodium, arrowhead indicates the articulation in the compound hook; (D) Lumbrinerides sp., limbate chaetae and simple hooks from the 4th parapodium; (E) Lumbrineris sp., distal part of the multidentate hooded hook from the 30th parapodium, arrowheads indicate numerous denticles, hood is slightly open; (F) Lumbrineriopsis sp., distal part of the bidentate hook, arrowheads indicate two equal denticles, hood is partly removed; (G), Ninoe armoricana, lateral parapodia with branchiae (arrowheads) from the middle of the body.
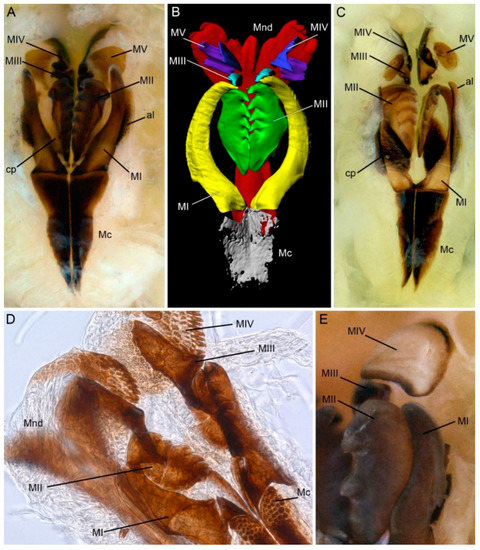
Figure 2.
Morphology of lumbrinerid jaws: mandibles and maxillae, light photographs of dissected jaws (A,C–E), 3D reconstruction of jaws based on micro-CT scans. (A) Lumbrineris sp. showing complete set of maxillary structures; (B) Lumbrineris sp., maxillary carriers partly reconstructed, connecting plates and attachment lamellae omitted; (C) Hilbigneris sp. maxillary apparatus showing extremely sclerotized connecting plates; (D) Lumbrineriopsis sp. distal part of the maxillary apparatus showing multidentate maxillae IV; (E) Augeneria albidentata, part of the maxillary apparatus showing right massive maxilla II and maxilla IV with non-pigmented white central area. al—attachment lamella; cp—connecting plate; Mc—maxillary carriers; Mnd—mandibles; MI–MV—maxillae I–V.
The currently accepted system of Lumbrineridae was proposed by Carrera-Parra [7] who provided diagnoses for 19 genera including six new genera. Recent review of the family by Oug et al. [6] listed updated diagnoses for all known lumbrinerid genera largely based on the revision by Carrera-Parra [7] but also utilizing more recent publications and the original descriptions (Table 1). The current total species count ranges between about 200 [6] and 280 [5] species; however, even the latter number might be highly underestimated.

Table 1.
Accepted genera of Lumbrineridae with the number of species in each genus and number of species used in the phylogenetic analysis in the present study. Species count for each genus follows Oug et al. [3].
Carrera-Parra [7] suggested a phylogenetic reconstruction of Lumbrineridae based on 38 morphological characters using genera as terminal taxa. Thus, the hypotheses of monophyly of individual genera was not tested. All genera were grouped into three sub-clades, except for Lysarete and Arabellonereis lacking bi- or multidentate hooks, a character present in all other lumbrinerids. The first sub-clade was represented by Ninoe, a genus with well-developed palmate parapodial branchiae. The second sub-clade combined genera with connecting plates–maxillary accessory structures placed between MI and MII (Figure 2A,C): Scoletoma, Lumbrineris, Hilbigneris, Kuwaita, Lumbricalus, Sergioneris and Eranno. Nevertheless, the shape, position, and degree of sclerotization of the connecting plates in all seven genera varies greatly and homology of these structures is not clearly understood. The remaining nine genera (Abyssoninoe, Cenogenus, Lumbrinerides, Lumbrineriopsis, Augeneria, Loboneris, Gallardoneris, Helmutneris, and Gesaneris) were grouped into the third sub-clade based on the presence of only four pairs of maxillae, although this character state was not exclusive to the clade and was also reported in Sergioneris.
Lumbrineris is the type and the most species-rich genus of the family. Carrera-Parra [7] recognized 36 valid species based on 57 known names but the number may well reach 50 [6]. Originally, Lumbrineris included only three species [20] but due to very wide definition subsequently encompassed a very large number of species [23,24,25]. Reevaluation of maxillary and fine chaetal structures allowed restricting of the genus diagnosis and led to transferring of part of the species to several different genera [2,7,16]. Nevertheless, many of the early described lumbrinerids lacking detailed information on their jaw morphology are still referred to Lumbrineris [6] making this genus a taxon of questionable monophyly.
Very little molecular data for lumbrinerids is available in public databases (e.g., GenBank, Barcode of Life Database—BOLD) mostly for the genera Lumbrineris, Scoletoma and Ninoe. For nine genera no genetic information is published up to date. Several species were used in reconstructing large-scale phylogenies of Eunicida [26,27] or Annelida [28,29,30] based on several markers or genomic data but no molecular phylogenetic reconstruction of relationships between the genera is available. In the present study, we publish new genetic data for 18 species of lumbrinerids and propose the first molecular phylogeny of Lumbrineridae based on independent evidence such as combination of mitochondrial and nuclear markers. We also discuss the importance of selected morphological characters in systematics of Lumbrineridae.
2. Materials and Methods
2.1. Taxon Sampling
We have sampled 45 specimens from 18 species of lumbrinerids representing 10 of the 19 currently known genera. Sequences for eight more species from the sampled genera were obtained from the GenBank and BOLD (Table 2). Nine genera were not included in the analysis due to unavailability of material suitable for DNA extraction. Three species from other eunicids families and Glycera alba (Glyceridae) were selected as outgroup taxa. Voucher specimens are deposited in the Invertebrate Collection, University Museum of Bergen, University of Bergen, Bergen, Norway (ZMBN), Zoological Museum of Moscow State University, White Sea Biological Station, Moscow, Russia (WSBS), and in the annelid collection of the Shirshov Institute of Oceanology, Russian Academy of Sciences, Moscow, Russia (IORAS). Detailed information of the museum catalogue numbers and geographical localities of each specimen can be found in the Supplementary Material (Table S1).

Table 2.
List of taxa used in the phylogenetic analyses with BOLD process IDs and GenBank accession numbers.
2.2. DNA Extraction, PCR Amplification and DNA Sequencing
Genomic DNA was extracted from 96% ethanol fixed samples using glass milk methods or QuickExtract™. Fragments of mitochondrial COI and 16S genes were amplified using primer pairs: 16SarL (forward) + 16Brh (reverse) [53], polyLCO (forward) + polyHCO (reverse) [50], HCO (forward) + LCO (reverse) [54]. For the nuclear marker 18S we designed specific primers (Table S2). PCR cycling protocol are shown in the Table S2. After visualization on a 1% agarose gel, the PCR products were purified using Ethanol/EDTA/Sodium Acetate Precipitation and bidirectionally Sanger sequenced using an Applied Biosystems automated sequencer. Chromatograms of forward and reverse sequences were aligned and proofread in Geneious R6 [55].
2.3. Sequence Alignment
Alignment was conducted using default parameters in MUSCLE [56] implemented in MEGA7 [57]. Aligned COI sequences were translated into amino-acid sequences using the invertebrate mitochondrial code (NCBI translation code 5) to ensure stop codons or frameshift mutations were not present. The sequences were then back translated into nucleotides for further phylogenetic analyses. For 16S and 18S markers we used Gblocks V.0.91b [58] to eliminate poorly aligned positions in the original alignments. The following parameters were used while masking: minimum number of sequences for a conserved position–49; minimum number of sequences for a flanking position–81; maximum number of contiguous non conserved positions–8; minimum length of a block–10; allowed gap positions–with half.
2.4. Phylogenetic Analyses
2.4.1. Bayesian Inference (BI)
Best-fit partitions and models were inferred with PartitionFinder2 [59] on XSEDE v1.6.10. Models were: GTR + G + I for second codon in COI and for 16S, HKY + G for 18S, SYM + G for first codon in COI, HKY + G + I for third codon in COI. Trees were reconstructed in MrBayes v. 3.2.1 [60] implementing PartitionFinder results which assigned codon position 1, 2, and 3 to different partitions. Two independent and simultaneous runs with flat prior probabilities and four chains were run for 5,000,000 generations. Trees were sampled every 1000th generation. Stationarity of each chain was checked with TRACER v.1.7 [61] and the first 25% discarded as burn-in after visualizing a plot of likelihood score. The remaining trees were summarized into a majority rule consensus tree with posterior probabilities (PP) indicating the support value for each clade.
2.4.2. Maximum Likelihood (ML)
The same dataset was used for phylogeny inference using the maximum likelihood criterion implemented in IQ-Tree on XSEDE v1.6.10 [62]. IQ-TREE was used to select models based on the Akaike information criterion (AIC), model was GTR + F + I + G4 for both partitions COI + 16S and 18S. IQ Tree was run through the CIPRES Science Gateway V3.3. Ultrafast bootstrap was performed in 10,000 iterations.
3. Results
The combined dataset had 1722 aligned positions (659 positions of COI, 552 positions of 16S rDNA, and 511 positions of 18S rDNA). After applying Gblocks, the new 16S rDNA alignment retained 415 positions (75%), 18S rDNA alignment retained 481 positions (95%), Gbloks was not applied to the COI alignment. Both Bayesian and Maximum likelihood approaches yielded trees with very similar topologies and node support (Figure 3 and Figure S1). The following differences between the resulting trees from the two analyses were observed: the polytomy comprising clades A, B and C in the Bayesian tree versus resolved and well supported relationships (A–C) in the ML tree; the polytomy comprising clades (A–E), (F–H), and I in the Bayesian tree versus resolved but very poorly supported relationships between these clades in the ML tree. Regardless of the method used, the following clades were obtained. Lumbrineridae is monophyletic (PP 1.00, BP 98). The following genera are monophyletic: Gallardoneris (clade D, PP 1.00, BP 100), Abyssoninoe (clade G, PP 1.00, BP 100), Ninoe (clade J, PP 1.00, BP 100), Augeneria (clade L, PP 1.00, BP 96), Lumbrineriopsis (clade M, PP 1.00, BP 97). Helmutneris (clade I), Hilbigneris (clade F), Lumbrinerides (clade K), and a single specimen that could not be attributed to any of the described genera, Lumbrineridae gen. sp. (clade C), were represented by a single species, thus their monophyly was not tested. The type genus of the family, Lumbrineris (clades A and H), and the genus Scoletoma (clades B and E) appeared polyphyletic. Lumbrineris mixochaeta formed a well-supported (PP 1.00, BP 100) clade with Scoletoma fragilis and Lumbrineridae gen. sp. while Lumbrineris latreilli, L. inftalta, and Lumbrineris sp. formed own clade (PP 0.89, BP 73) sister to Abyssoninoe/Hilbigneris clade (PP 0.85, BP 81). Nevertheless, these sister relationships were poorly supported (PP 0.60, BP 52). Scoletoma tetraura and S. zonata (clade E) were sister to the clade comprising Gallardoneris, L. mixochaeta, S. fragilis and Lumbrineridae gen. sp. although these relationships were also poorly supported (PP 0.81, BP 81). Three highly supported sister relationships were recovered in both analyses: Ninoe and Lumbrinerides (PP 0.99, BP 87), Augeneria and Lumbrineriopsis (PP 0.96, BP 74), and Gallardoneris and (L. mixochaeta, S. fragilis and Lumbrineridae gen. sp.) clade (PP 1.00, BP 98).
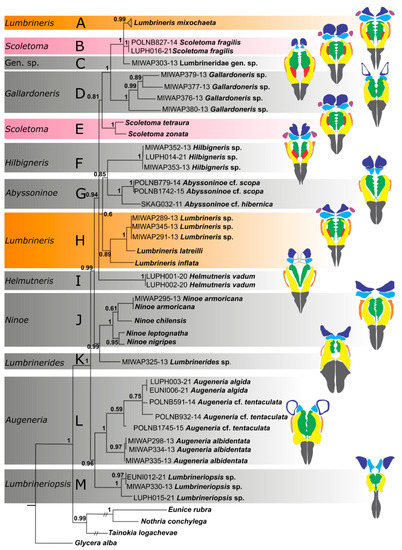
Figure 3.
Consensus tree from the Bayesian analysis of the combined COI, 16S rDNA and 18S rDNA dataset; numbers on nodes indicate Bayesian posterior probabilities; capital letters (A–M) correspond with the clades discussed in the text. Colored bars indicate polyphyletic genera. Sequences of fourteen specimens of Lumbrineris mixochaeta were collapsed into a single triangle for better presentation. Color codes for maxillary elements: grey—maxillary carriers, yellow—maxillae I, green—maxillae II, light blue—maxillae III, navy—maxillae IV, purple—maxillae V, red—connecting plates, orange—attachment lamellae of maxillae I.
4. Discussion
Although our dataset was rather limited, covering only 12 out of 19 described genera and 26 species (~10% of total species count), certain clades and relationships were well supported. Most genera were found monophyletic and several of them were highly supported. Ninoe clade (J) comprised four species, all sharing the presence of the parapodial palmate branchiae in the anterior region of the body (Figure 1G). Augeneria clade (L) included three species, all sharing massive maxillae II with few rounded teeth and large maxillae IV with white central areas (Figure 2E). Specimens identified as Augeneria cf. tentaculata did not form a sub-clade withing the genus dues to large amount of missing sequence data (Table 2). Gallardoneris clade (D) included four specimens with four pairs of maxillae, maxillae IV with unpigmented central areas, and maxillae II with few rounded teeth and missing attachment lamellae. These specimens were provisionally identified as Gallardoneris iberica Martins, Carrera-Parra, Quintino & Rodrigues, 2012 [63] collected from the western African waters. Despite very similar morphology, all four specimens showed very divergent sequences indicating a possible case of cryptic speciation in the genus which should be investigated further based on larger sampling size. Lumbrineriopsis clade (M) included two species, both showing similar maxillary morphology with prolonged maxillary carriers and multidentate maxillae IV (Figure 2D). Abyssoninoe clade (G) comprised two species sharing the presence of anterior prolonged hooded hooks and fused maxillae IV and V–the two main diagnostic characters of the genus. Notably, a single specimen that was not identified to the genus level (Lumbrineridae gen. sp.) also exhibited joined maxillae IV and V and elongated anterior hooks. However, this specimen could not be assigned to Abyssoninoe due to the presence of very strongly sclerotized connecting plates and short maxillae II (significantly shorter than maxillae I). The latter two characters are not reported for any of Abyssoninoe species, including the type species, Abyssoninoe abyssorum (McIntosh, 1885) [8]. Furthermore, Oug et al. [6] noted that the fusion of maxillae IV and V occurs also in other genera and thus should be reconsidered as the main diagnostic character in Abyssoninoe. Lumbrineridae gen. sp. could potentially represent a new genus of lumbrinerids, however we suggest that the new genera are erected after complete revision of the existing system of the family.
Two most species-rich genera within Lumbrineridae, Lumbrineris and Scoletoma, were recovered as polyphyletic taxa forming two unrelated clades each. These two genera have very similar maxillary morphology (five pairs of maxillae, maxillae I and II almost equal in length, connecting plates present) and were split based on the presence of compound multidentate hooks (Figure 1C) in the former and only simple multidentate hooks in the later (Frame 1992). Lumbrineris latreilli, the type species of the genus, formed a clade with two other Lumbrineris species and this clade presumably can be considered the part of Lumbrineris sensu stricto. Nevertheless, L. latreilli is a species with problematic status. It has been originally described from the Atlantic Ocean off France [20] but later reported widely in the world [3]. Sequences of L. latreilli used in our analyses were downloaded from the GenBank and although at least one of them was obtained from a specimen collected from the South European Atlantic coast, the identification of this species requires further clarification. Scoletoma fragilis (O.F. Müller, 1776), the type species of the genus, formed a well-supported clade with L. mixochaeta and Lumbrineridae gen. sp., however we were not able to identify morphological characters common for these three taxa. Lack of own material and reliable published sequence data on several markers obtained from the same species prevented from including more species of Lumbrineris and Scoletoma into the analyses. Although our data indicate that both genera are polyphyletic and require revision, more complete dataset is needed for formal decision on the systematics of both genera.
Augeneria, Gallardoneris and Helmutneris, together with Gesaneris and Loboneris (both absent in the present analysis), were also seen as a group of closely related genera sharing maxillae III with unpigmented white central area [7] (Figure 2E). General architecture of maxillary apparatuses in these five genera is rather dissimilar. Loboneris has very thin and prolonged maxillary carriers and wing-shaped colorless maxillae IV. Gesaneris demonstrates maxillae II, significantly shorter than maxillae I, which is not characteristic for other genera in this group. Helmutneris has maxillae I and II almost equal in length but maxillae II bear numerous (6–7) pointed teeth, while in Augeneria and Gallardoneris, the maxillae II have only three rounded and widely distributed teeth. Maxillae of the latter two genera are most similar with each other, however in Augeneria, the basal parts of maxillae I form a clear locking system characteristic to all lumbrinerids [4] while in Gallardoneris, this system appears to be absent (pers. observation). Our results do not support close relationships between Augeneria, Gallardoneris and Helmutneris suggesting an independent origin of colorless maxillae IV at least in these three genera.
Lumbrinerides and Lumbrineriopsis were traditionally seen as closely related genera [7] due to their minute size (less than 1 mm in width), elongated prostomium (Figure 1B) and bidentate simple hooded hooks (Figure 1F)–a condition reported for these two genera only. Nonetheless, these genera did not form sister relationships on the present tree suggesting their independent miniaturization and acquisition of bidentate hooks. Maxillary apparatuses of Lumbrinerides and Lumbrineriopsis are also very dissimilar with short carriers, massive maxillae I and very short maxillae II in the former and elongated carriers and multidentate maxillae III in the latter. Notably, Lumbrineriopsis was sister to Augeneria while Lumbrinerides–sister to Ninoe, both relationships were highly supported on the molecular-based tree although we could not identify morphological characters supporting these two clades.
None of the morphological characters traditionally used in lumbrinerid systematics could be assigned to a clade higher than the genus level. The morphological characters used by Carrera-Parra [7] to define large groups of genera, such as the presence of connecting plates and only four pairs of maxillae did not support any of the clades recovered in our analyses. On the contrary, the genera showing these character states were scattered across the whole tree (clades A + B + C, E, F, and H with connecting plates and clades D, G, I, J + K, and L + M with four pairs of maxillae) suggesting their homoplasy (Figure 3).
5. Conclusions
Insufficient taxon sampling and moderate molecular data allowed only partial revision of Lumbrineridae based on molecular phylogeny results. Nevertheless, our data corroborate monophyletic status of the genera Abyssoninoe, Augeneria, Gallardoneris, Lumbrineriopsis, and Ninoe and indicate polyphyly of the genera Lumbrineris and Scoletoma. Being the type genus of the family, Lumbrineris requires further study with inclusion of higher number of species and subsequent definition of a monophyletic group inholding the type species, L. latreilli, as the carrier of the genus name.
Strongly supported relationships between several clades suggest that the characters, such as the presence of bidentate simple hooded hooks, non-pigmented whitish central areas in maxillae IV, multidentate maxillae IV, the presence of connecting plates and four or five pairs of maxillae have evolved several times independently within Lumbrineridae and do not represent exclusive homologies defining groups of closely related genera.
Supplementary Materials
The following are available online at www.mdpi.com/article/10.3390/d14020083/s1, Table S1: List of specimens used in this study with GenBank Accession numbers, BOLD process ID, and data on their sampling and storage, Table S2: Primer sequences and PCR parameters used for amplification of COI, 18S rDNA and 16S rDNA, Figure S1: Consensus tree from the Maximum likelihood analysis of the combined COI, 16S rDNA and 18S rDNA dataset; numbers on nodes indicate bootstrap support; capital letters correspond with the clades (A–M) discussed in the text. Colored bars indicate polyphyletic genera. Sequences of fourteen specimens of Lumbrineris mixochaeta were collapsed into a single triangle for better presentation.
Author Contributions
Conceptualization, N.B. and P.B.; methodology, P.B. and N.B.; formal analysis, P.B.; data curation, P.B. and N.B.; writing—original draft preparation, P.B. and N.B.; writing—review and editing, P.B. and N.B.; visualization, P.B. and N.B.; supervision, N.B.; project administration, P.B. and N.B.; funding acquisition, P.B. and N.B. All authors have read and agreed to the published version of the manuscript.
Funding
The reported study was funded by RFBR according to the research projects No. 20-34-90053 and No. 19-04-00501.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Statement: Sequences obtained in this study are uploaded to GenBank and BOLD (Table S2).
Acknowledgments
We would like to thank the staff of the University Museum of Bergen, University of Bergen (UM, UiB), Jon A. Kongsrud, Endre Willassen, Tom Alvestad and Katrine Kongshavn for providing access and assisting in sorting, identification and curating of the diverse collections of lumbrinerids used in the present study. We are also grateful to Irine Ekimova and Dmitry Schepetov (Moscow State University), Louise Lindblom and Kenneth Meland (UiB) for help in molecular laboratory work and with sequencing; Irene Heggstad for help with SEM (UiB); Dina Gilyazetdinova and Dmitry Korost (Petroleum Geology department of Faculty of Geology, Moscow State University) for assistance with micro-computed tomography; Nikolay Neretin (Moscow State University) for providing access to materials from the White Sea Biological Station collection; and Evind Oug (Norwegian Institute of Water Research) for identification of some specimens. We are very grateful to the NORAD-funded EAF-Nansen programme, and the Guinea Current Large Marine Ecosystem (GCLME) and Canary Current Large Marine Ecosystem (CCLME) partners for depositing unsorted and unidentified samples in the University Museum of Bergen. We particularly thank Jens-Otto Krakstad (Institute of Marine Research, Norway) for their coordinating role in the cooperation between EAF-Nansen and UM, UiB. Sorting, identification and barcoding of west African specimens was supported by the JRS Biodiversity Foundation. Sorting, identification and generation of COI sequences for a number of specimens from the Nordic Seas were performed through the projects supported by the Norwegian Taxonomy Initiative (Polychaetes in Skagerrak, project no. 70184216; Polychaetes in the Norwegian Sea, project no. 70184227) in collaboration with the Norwegian Barcode of Life Network (NorBOL) funded by the Research Council of Norway (project no. 226134/F50) and the Norwegian Biodiversity Information Centre (project no. 70184209). Examination of lumbrinerid specimens in the Invertebrate collection, University Museum of Bergen by P.B. was supported via the Cooperation Programme between Norway and Russia “Multidisciplinary EDUcation and reSearch in mArine biology in Norway and Russia” (MEDUSA)–funded by Direktoratet for høyere utdanning og kompetanse (HK-dir).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Schmarda, L.K. Neue wirbellose thiere beobachtet und gesammelt auf einer reise um die erde 1853 bis 1857. In Turbellarien, Rotatorien und Anneliden; Verlag von Wilhelm Engelmann: Leipzig, Germany, 1861. [Google Scholar]
- Orensanz, J.M. The eunicemorph polychaete annelids from antarctic and subantarctic seas: With addenda to the Eunicemorpha of Argentina, Chile, New Zealand, Australia, and the Southern Indian Ocean. In Biology of the Antarctic Seas XXI; American Geophysical Union (AGU): New Orleans, LA, USA, 1990; pp. 1–183. ISBN 978-1-118-66779-8. [Google Scholar]
- Carrera-Parra, L.F. Revision of Lumbrineris de Blainville, 1828 (Polychaeta: Lumbrineridae) Sars. Zootaxa 2006, 1336, 1–64. [Google Scholar] [CrossRef]
- Paxton, H. Phylogeny of Eunicida (Annelida) Based on Morphology of Jaws. Zoosymposia 2009, 2, 241–264. [Google Scholar] [CrossRef] [Green Version]
- Zanol, J.; Carrera-Parra, L.F.; Steiner, T.M.; Amaral, A.C.Z.; Wiklund, H.; Ravara, A.; Budaeva, N. The Current State of Eunicida (Annelida) Systematics and Biodiversity. Diversity 2021, 13, 74. [Google Scholar] [CrossRef]
- Oug, E.; Budaeva, N.; Borisova, P. Lumbrineridae Schmarda, 1861. In Handbook of Zoology Annelida Volume 4: Pleistoannelida, Errantia II; Purschke, G., Böggemann, M., Westheide, W., Eds.; De Gruyter: Berlin, Germany, 2022; pp. 1–35. [Google Scholar]
- Carrera-Parra, L.F. Phylogenetic Analysis of Lumbrineridae Schmarda, 1861 (Annelida: Polychaeta). Zootaxa 2006, 1332, 1–36. [Google Scholar] [CrossRef]
- McIntosh, W.C. Report on the Annelida Polychaeta Collected by HMS ”Challenger” during the Years 1873-76. Reports on the Scientific Results of the Voyage of HMS ”Challenger”. Zoology 1885, 12, 1–554. [Google Scholar]
- Hartmann-Schröder, G. Die polychaeten der tropischen nordwestküste Australiens (zwischen derby im norden und port hedland im süden). teil 2. In Zur Kenntnis Des Eulitorals Der Australischen Küsten Unter Besonder Berücksichtigung Der Polychaeten Und Ostracoden. (Teil 2 Und Teil 3); Hartmann-Schröder, G., Hartmann, G., Eds.; Mitteilungen aus dem Hamburgischen Zoologischen Museum und Institut: Hamburg, Germany, 1979; Volume 76, pp. 77–218. [Google Scholar]
- Monro, C.C.A. Polychaete Worms. Discov. Rep. Camb. 1930, 2, 1–222. [Google Scholar]
- Chamberlin, R.V. The Annelida Polychaeta; Museum of Comparative Zoölogy at Harvard College: Cambridge, MA, USA, 1919; Volume 48. [Google Scholar]
- Kinberg, J.G.H. Annulata Nova, Eunicea; Fversi gt af Kongl Vetenskaps-Akademiens Forhandlingar: Stockholm, Sweden, 1865; Volume 21, pp. 559–574. [Google Scholar]
- Gallardo, V.A. Polychaeta from the Bay of Nha Trang, South Viet Nam. Naga Rep. 1968, 4, 35–279. [Google Scholar]
- Fage, L. Sur l’association d’un Annélide Polychète, Lumbriconereis flabellicola n. sp. et d’un Madrépore Flabellum pavoninum distinctum e. et h. XII Congrs Int. De Zool. Lisb. 1936, 2, 941–945. [Google Scholar]
- Mohammad, M.-B.M. New Species and Records of Polychaete Annelids from Kuwait, Arabian Gulf. Zool. J. Linn. Soc. 1973, 52, 23–44. [Google Scholar] [CrossRef]
- Frame, A.B. The Lumbrinerids (Annelida: Polychaeta) Collected in Two Northwestern Atlantic Surveys with Descriptions of a New Genus and Two New Species. Proc. Biol. Soc. Wash. 1992, 105, 185–218. [Google Scholar]
- Grube, A.E. Untersuchungen Über Die Familie Eunicea. Jahresber. Schlesichen Ges. Fr Vaterlndische Kult. 1878, 55, 79–104. [Google Scholar]
- Orensanz, J.M. Los Anélidos Poliquetos de La Provincia Biogeográfica Argentina. IV. Lumbrineridae. [The Polychaetous Annelids of the Argentine Biogeographic Province. IV. Lumbrineridae]. Physis 1973, 32, 343–393. [Google Scholar]
- Ehlers, E. Die bodensässigen anneliden aus den sammlungen der Deutschen tiefsee-expedition. In Wissenschaftliche Ergebnisse der Deutschen Tiefsee-Expedition auf dem Dampfer ”Valdivia” 1898–1899; Chun, C., Ed.; Verlag von Gustav Fischer: Stuttgart, Germany, 1908; pp. 1–168. [Google Scholar]
- De Blainville, H. Mollusques, vers et zoophytes. In Dictionnaire des Sciences Naturelles; Leuvrant, F.G., Ed.; Dictionnaire des Sciences Naturelles: Strasbourg, France, 1828; Volume 57, p. 628. [Google Scholar]
- Audouin, J.V.; Milne-Edwards, H. Recherches Pour Servir L’histoire Naturelle Du Littoral de La France, Ou Recueil de Mémoires Sur L’anatomie, La Physiologie, La Classification et Les Moeurs Des Animaux de Nos Côtes; Ouvrage Accompagné Des Planches Faites de Après Nature. 2. Annélides; Crochard Libraire: Paris, France, 1834. [Google Scholar]
- Müller, O.F. Zoologiae Danicae Prodromus: Seu Animalium Daniae et Norvegiae Indigenarum Characteres, Nomina, et Synonyma Imprimis Popularium; Typis Havniæ [Copenhagen], Hallageriis: Canberra, Austrilia, 1776. [Google Scholar]
- Hartman, O. Polychaetous Annelids, Est. 5, Eunicea. Allan Hancock Pacif. Exped 1944, 10, 1–238. [Google Scholar]
- Fauchald, K. Polychaetous Annelids of the Families Eunicidae, Lumbrineridae, Iphitimidae, Arabellidae, Lysaretidae and Dorvilleidae from Western Mexico; Allan Hancock Monographs in Marine Biology: Los Angeles, CA, USA, 1970; pp. 1–112. [Google Scholar]
- Fauchald, K. The Polychaete Worms. Definitions and Keys to the Orders, Families and Genera. Nat. Hist. Mus. Los Angeles Cty. Sci. Ser. 1977, 28, 1–198. [Google Scholar]
- Struck, T.H.; Purschke, G.; Halanych, K.M. Phylogeny of Eunicida (Annelida) and Exploring Data Congruence Using a Partition Addition Bootstrap Alteration (PABA) Approach. Syst. Biol. 2006, 55, 1–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tilic, E.; Bartolomaeus, T.; Rouse, G.W. Chaetal Type Diversity Increases during Evolution of Eunicida (Annelida). Org. Divers. Evol. 2016, 16, 105–119. [Google Scholar] [CrossRef]
- Struck, T.H. Direction of Evolution within Annelida and the Definition of Pleistoannelida. J. Zool. Syst. Evol. Res. 2011, 49, 340–345. [Google Scholar] [CrossRef]
- Struck, T.H.; Golombek, A.; Weigert, A.; Franke, F.A.; Westheide, W.; Purschke, G.; Bleidorn, C.; Halanych, K.M. The Evolution of Annelids Reveals Two Adaptive Routes to the Interstitial Realm. Curr. Biol. 2015, 25, 1993–1999. [Google Scholar] [CrossRef] [Green Version]
- Tilic, E.; Stiller, J.; Campos, E.; Pleijel, F.; Rouse, G.W. Phylogenomics Resolves Ambiguous Relationships within Aciculata (Errantia, Annelida). Mol. Phylogenetics Evol. 2022, 166, 107339. [Google Scholar] [CrossRef]
- Oug, E. A New Small Species of Lumbrineris from Northern Norway and Arctic Waters, with Comments on L. minuta (Théel, 1879) and L. vanhoeefeni (Michaelsen, 1898) (Polychaeta: Lumbrineridae). Ophelia 1998, 49, 147–162. [Google Scholar] [CrossRef]
- Zhou, H.; Zhang, Z.; Chen, H.; Sun, R.; Wang, H.; Guo, L.; Pan, H. Integrating a DNA Barcoding Project with an Ecological Survey: A Case Study on Temperate Intertidal Polychaete Communities in Qingdao, China. Chin. J. Oceanol. Limnol. 2010, 28, 899–910. [Google Scholar] [CrossRef]
- McIntosh, W.C. Notes from the Gatty Marine Laboratory, St. Andrews. No. 25. 1. On the Eunicidae Dredged by HMS ‘Porcupine’ in 1869 and 1870. 2. On Canadian Eunicidae Dredged by Dr. Whiteaves, of the Canadian Geological Survey, in 1871–1873. 3. On Norwegian Eunicidae Collected by Canon Norman DCL, FRS. Ann. Mag. Nat. Hist. 1903, 12, 128–165. [Google Scholar]
- Paul, C.; Halanych, K.M.; Tiedemann, R.; Bleidorn, C. Molecules Reject an Opheliid Affinity for Travisia (Annelida). Syst. Biodivers. 2010, 8, 507–512. [Google Scholar] [CrossRef]
- Johnson, H.P. The Polychaeta of the Puget Sound Region. Proc. Boston Soc. Nat. Hist. 1901, 29, 381–437. [Google Scholar]
- Fauchald, K. Deep-Water Errant Polychaetes from Hardangerfjorden, Western Norway. Sarsia 1974, 57, 1–32. [Google Scholar] [CrossRef] [Green Version]
- Moore, J.P. The Polychaetous Annelids Dredged by the U.S.S. “Albatross” off the Coast of Southern California in 1904. III. Euphrosynidae to Goniadidae. Proc. Acad. Nat. Sci. Phila. 1911, 63, 234–318. [Google Scholar]
- Struck, T.H.; Purschke, G. The Sister Group Relationship of Aeolosomatidae and Potamodrilidae (Annelida: “Polychaeta”)—A Molecular Phylogenetic Approach Based on 18S RDNA and Cytochrome Oxidase I. Zool. Anz. A J. Comp. Zool. 2005, 243, 281–293. [Google Scholar] [CrossRef]
- Borisova, P.; Budaeva, N. Helmutneris vadum, a New Species of Lumbrineridae (Annelida) from Lizard Island, Great Barrier Reef, Australia. Zootaxa 2020, 4877, 413–421. [Google Scholar] [CrossRef]
- Glémarec, M. Ninoe armoricana n. sp., Polychète Lumbrineridae de La «Grande Vasière»(Golfe de Gascogne); Vie Et Milieu: Banyuls-sur-Mer, France, 1968; pp. 315–322. [Google Scholar]
- Aylagas, E.; Borja, Á.; Irigoien, X.; Rodríguez-Ezpeleta, N. Benchmarking DNA Metabarcoding for Biodiversity-Based Monitoring and Assessment. Front. Mar. Sci. 2016, 3, 96. [Google Scholar] [CrossRef] [Green Version]
- Maturana, C.S.; Moreno, R.A.; Labra, F.A.; González-Wevar, C.A.; Rozbaczylo, N.; Carrasco, F.D.; Poulin, E. DNA Barcoding of Marine Polychaetes Species of Southern Patagonian Fjords. Rev. Biol. Mar. Oceanogr. 2011, 46, 35–42. [Google Scholar] [CrossRef] [Green Version]
- Ehlers, E. Magellanischen Anneliden Gesammelt Während Der Schwedischen Expedition Nach Den Magellansländern. Nachr. Königliche Ges. Wiss. Göttingen Math. Phys. Kl. 1900, 1900, 206–223. [Google Scholar]
- Verrill, A.E. XVIII. Report upon the Invertebrate Animals of Vineyard Sound and the Adjacent Waters, with an Account of the Physical Characters of the Region; Report on the Condition of the Sea Fisheries of the South Coast of New England; Government Printing Office: Washington, DC, USA, 1873. [Google Scholar]
- Lobo, J.; Costa, P.M.; Teixeira, M.A.L.; Ferreira, M.S.G.; Costa, M.H.; Costa, F.O. Enhanced Primers for Amplification of DNA Barcodes from a Broad Range of Marine Metazoans. BMC Ecol. 2013, 13, 34. [Google Scholar] [CrossRef] [Green Version]
- Rousset, V.; Pleijel, F.; Rouse, G.W.; Erséus, C.; Siddall, M.E. A Molecular Phylogeny of Annelids. Cladistics 2007, 23, 41–63. [Google Scholar] [CrossRef]
- Aylagas, E.; Borja, A.; Rodríguez-Ezpeleta, N. Environmental Status Assessment Using DNA Metabarcoding: Towards a Genetics Based Marine Biotic Index (GAMBI). PLoS ONE 2014, 9, e90529. [Google Scholar] [CrossRef] [Green Version]
- Zanol, J.; Halanych, K.M.; Struck, T.H.; Fauchald, K. Phylogeny of the Bristle Worm Family Eunicidae (Eunicida, Annelida) and the Phylogenetic Utility of Noncongruent 16S, COI and 18S in Combined Analyses. Mol. Phylogenet. Evol. 2010, 55, 660–676. [Google Scholar] [CrossRef]
- Sars, M. Beskrivelser og Iagttagelser Over Nogle Mærkelige Eller Nye i Havet Ved Den Bergenske Kyst Levende Dyr af Polypernes, Acalephernes, Radiaternes, Annelidernes, og Molluskernes Classer: Med en Kort Oversigt Over de Hidtil af Forfatteren Sammesteds Fundne Arter og Deres Forekommen; Thorstein Hallagers Forlag hos Chr. Dahl, R.S.: Bergen, Norway, 1835. [Google Scholar]
- Carr, C.M.; Hardy, S.M.; Brown, T.M.; Macdonald, T.A.; Hebert, P.D.N. A Tri-Oceanic Perspective: DNA Barcoding Reveals Geographic Structure and Cryptic Diversity in Canadian Polychaetes. PLoS ONE 2011, 6, e22232. [Google Scholar] [CrossRef] [Green Version]
- Budaeva, N.; Schepetov, D.; Zanol, J.; Neretina, T.; Willassen, E. When Molecules Support Morphology: Phylogenetic Reconstruction of the Family Onuphidae (Eunicida, Annelida) Based on 16S RDNA and 18S RDNA. Mol. Phylogenet. Evol. 2016, 94, 791–801. [Google Scholar] [CrossRef]
- Ravara, A.; Cunha, M.R. An Unusual New Species of Oenonidae (Polychaeta, Eunicida) from the North Atlantic Ocean. Mar. Biodivers. 2017, 48, 1037–1043. [Google Scholar] [CrossRef]
- Palumbi, S. Simple Fool’s Guide to PCR; Department of Zoology and Kewalo Marine Laboratory, University of Hawaii: Honolulu, HI, USA, 1991. [Google Scholar]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA Primers for Amplification of Mitochondrial Cytochrome c Oxidase Subunit I from Diverse Metazoan Invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castresana, J. Selection of Conserved Blocks from Multiple Alignments for Their Use in Phylogenetic Analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New Methods for Selecting Partitioned Models of Evolution for Molecular and Morphological Phylogenetic Analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef] [Green Version]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [Green Version]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Martins, R.; Carrera-Parra, L.F.; Quintino, V.; Rodrigues, A.M. Lumbrineridae (Polychaeta) from the Portuguese Continental Shelf (NE Atlantic) with the Description of Four New Species. Zootaxa 2012, 3416, 1–21. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).