The Nurse Plant Acacia spirorbis Enriches Ectomycorrhizal Community Composition of a Target Species: Tristaniopsis calobuxus
Abstract
:1. Introduction
2. Materials and Methods
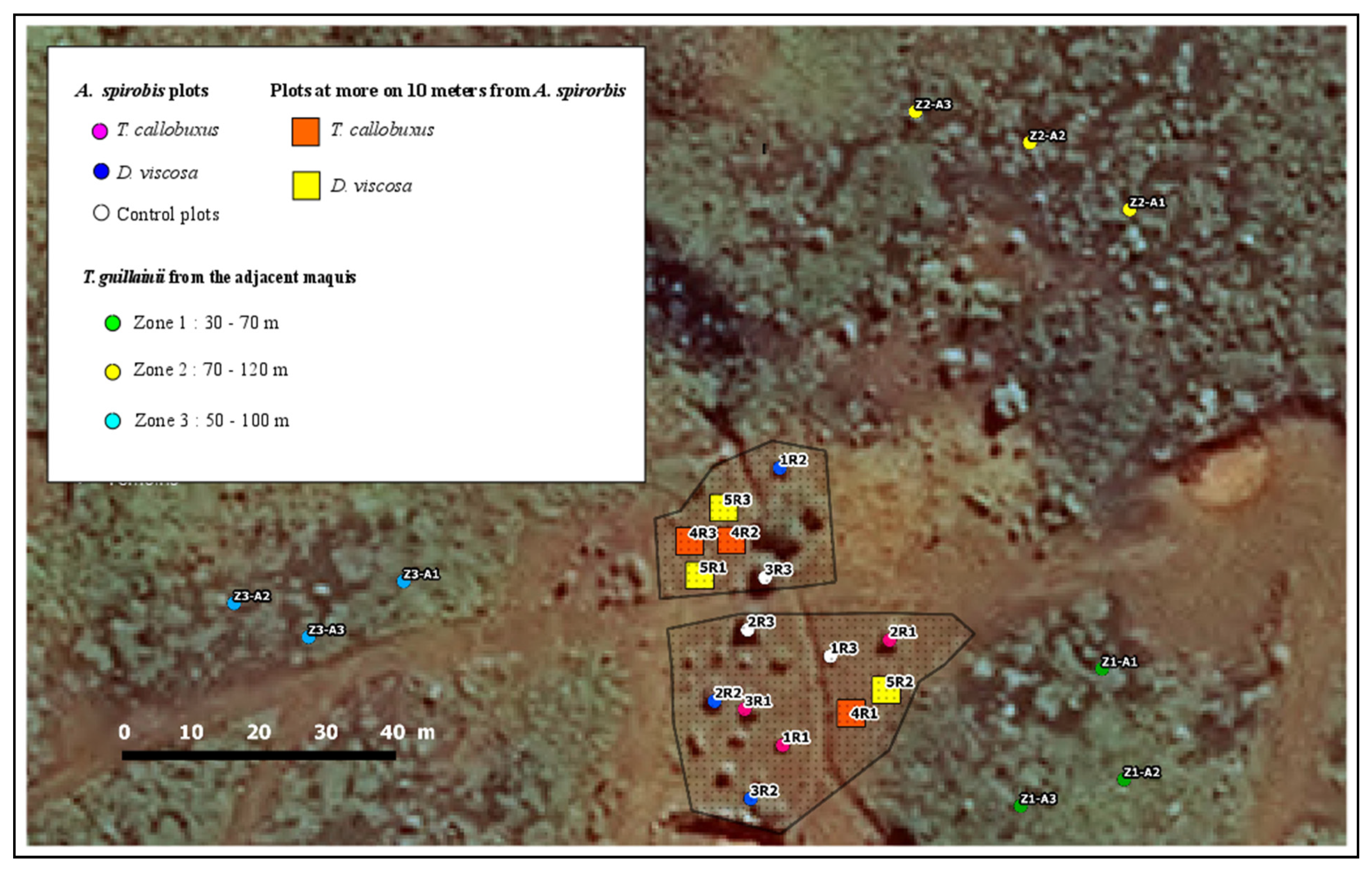
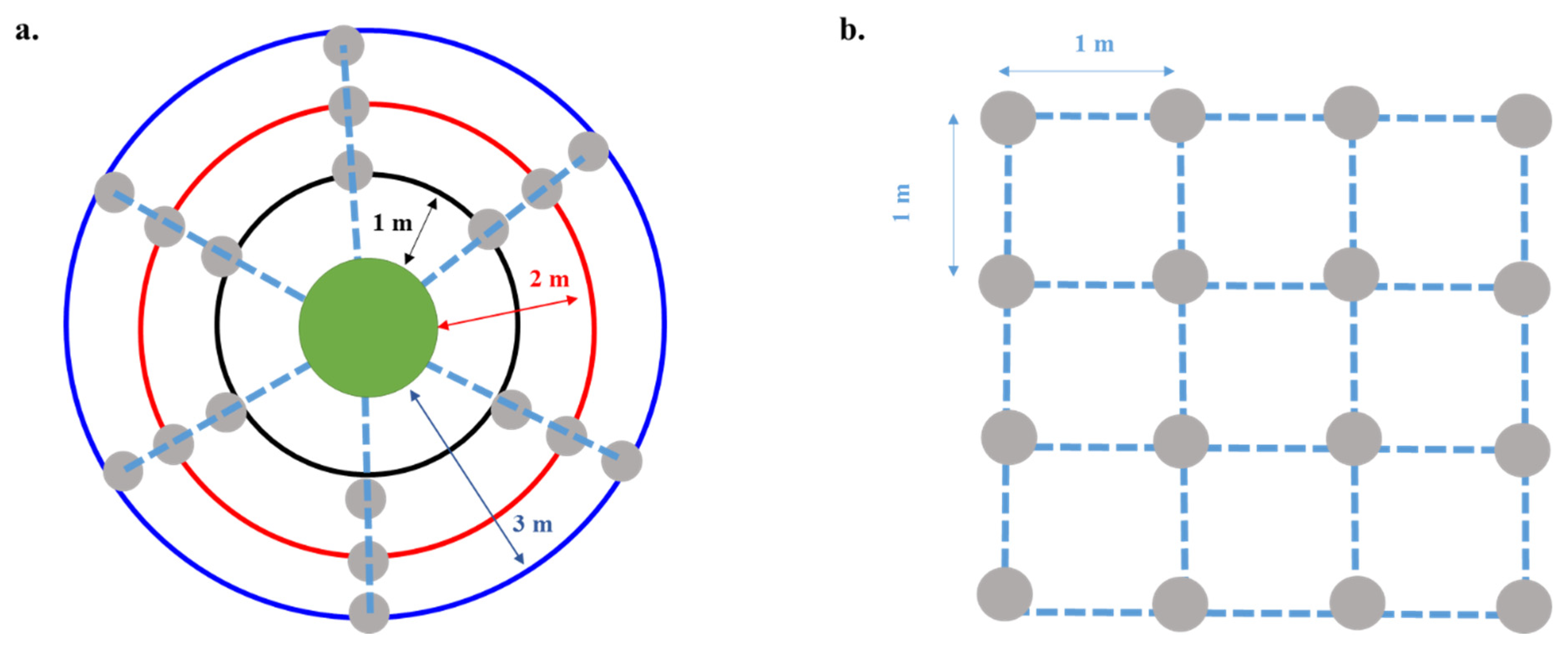
2.1. Study Site and Experimental Design
2.2. Nursery Growth
2.3. Sampling Ectomycorrhizal Root Tips and Molecular Analyses
2.4. Data Analysis
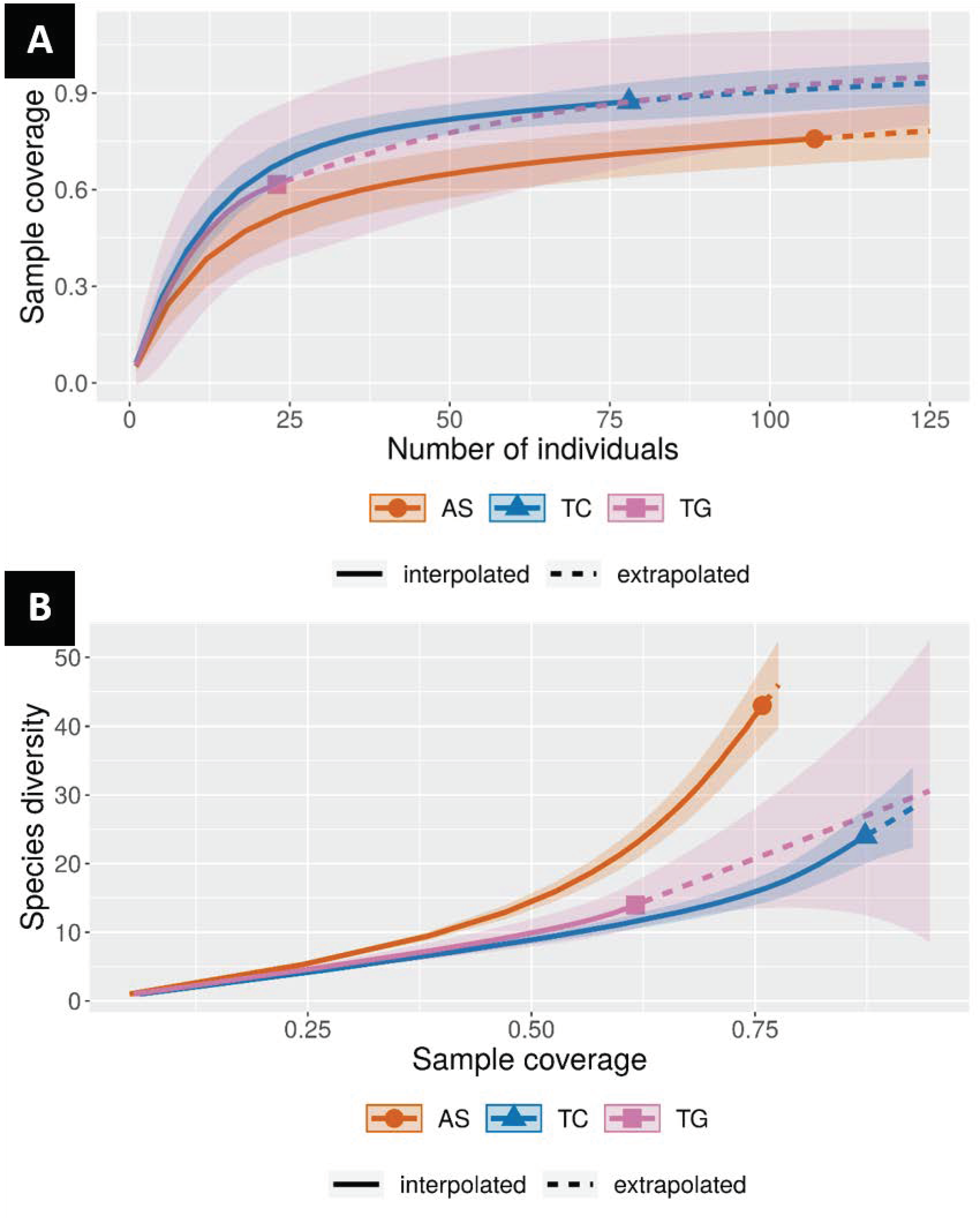
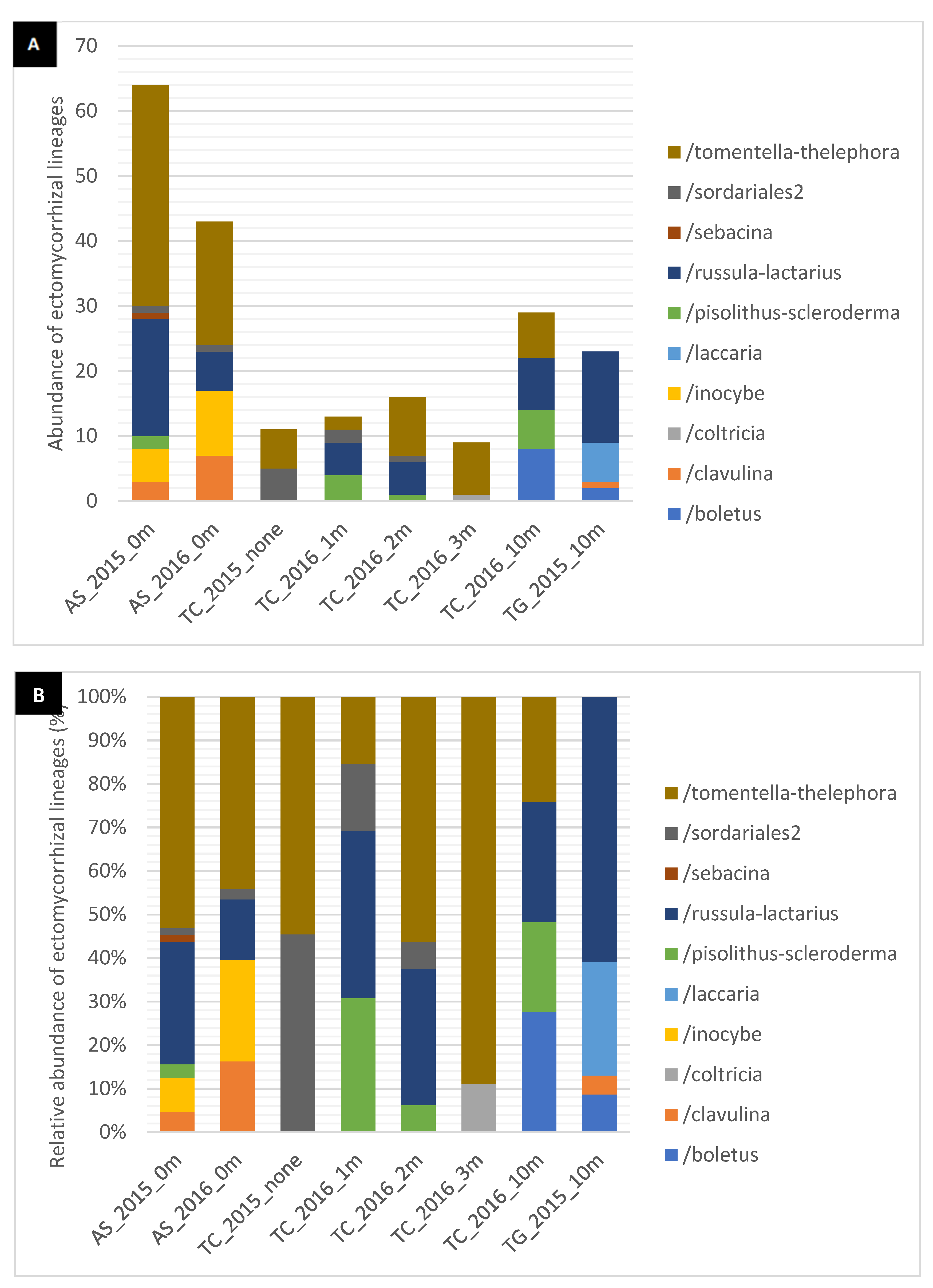
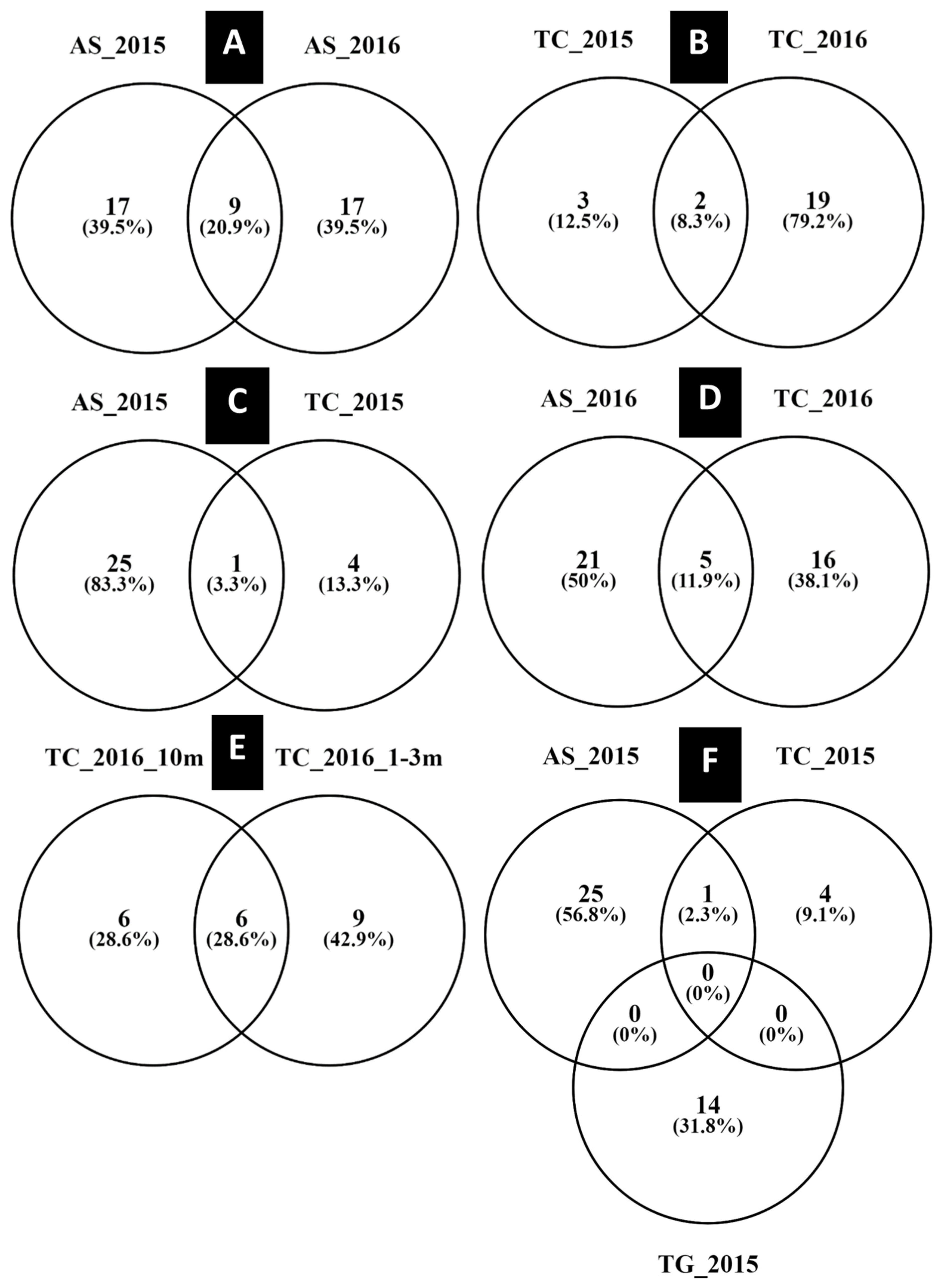
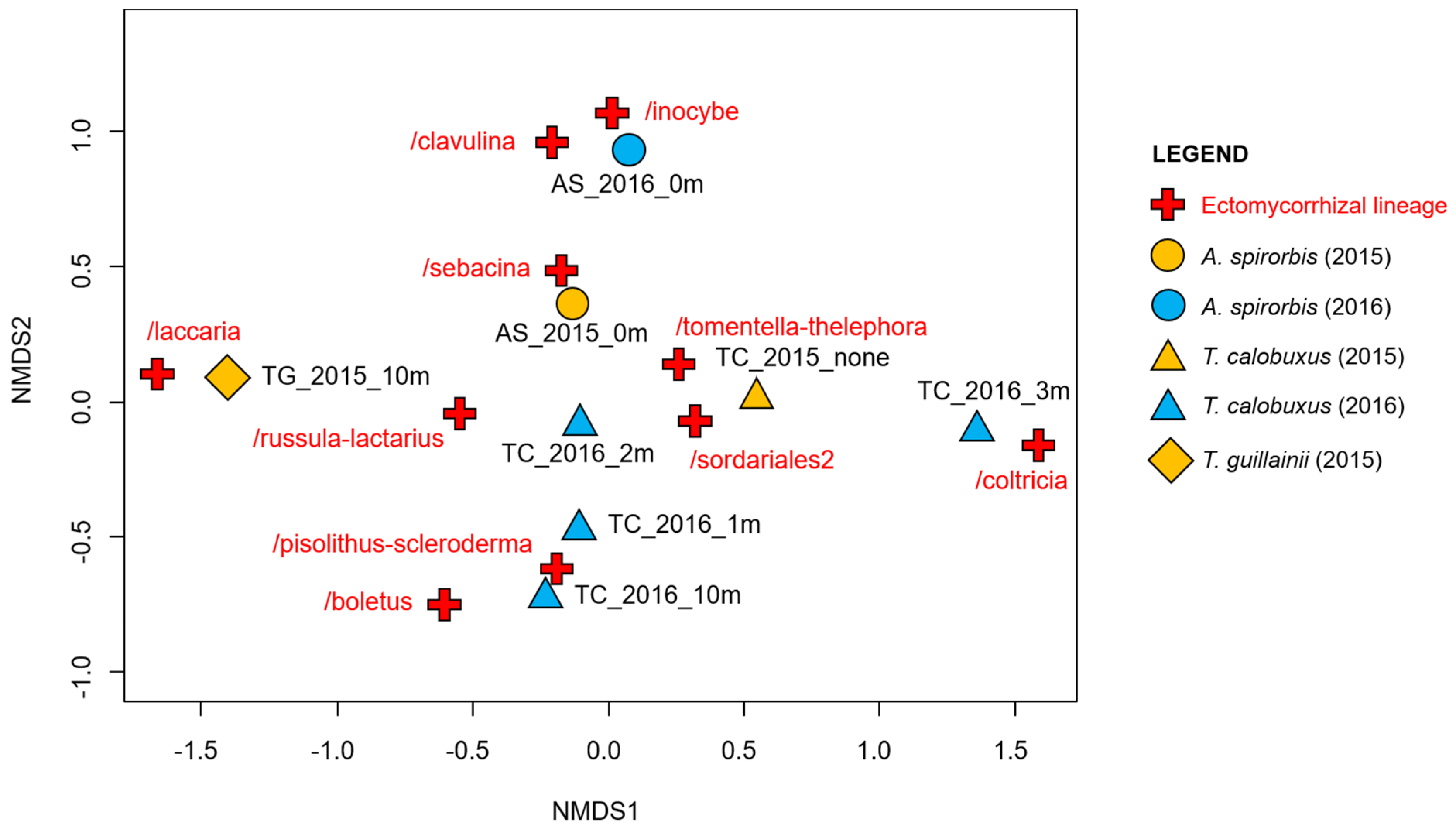
3. Results
Diversity and Comparison of ECM Fungal Communities Associated with the Different ECM Tree Species
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alexander, I.; Selosse, M.-A. Mycorrhizas in Tropical Forests: A neglected research imperative. New Phytol. 2009, 182, 14–16. [Google Scholar] [CrossRef] [PubMed]
- L’Huillier, L.; Jaffré, T.; Wulff, A. Mines et Environnement En Nouvelle-Calédonie: Les Milieux sur Substrats Ultramafiques et Leur Restauration; IAC: Païta, New Caledonia, 2010; ISBN 978-2-9523950-8-3. [Google Scholar]
- Pillon, Y.; Jaffré, T.; Birnbaum, P.; Bruy, D.; Cluzel, D.; Ducousso, M.; Fogliani, B.; Ibanez, T.; Jourdan, H.; Lagarde, L.; et al. Infertile landscapes on an old oceanic island: The biodiversity hotspot of New Caledonia. Biol. J. Linn. Soc. 2021, 133, 317–341. [Google Scholar] [CrossRef]
- Amir, H.; Ducousso, M. Les bactéries et les champignons du sol sur roches ultramafiques. In Mines et Environnement en Nouvelle Calédonie: Les Milieux sur Substrats Ultramafiques et Leur Restauration; L’Huillier, L., Jaffré, T., Wulff, A., Eds.; Etudes Synthèses; IAC: Païta, New Caledonia, 2010; pp. 129–145. [Google Scholar]
- Carriconde, F.; Gardes, M.; Bellanger, J.-M.; Letellier, K.; Gigante, S.; Gourmelon, V.; Ibanez, T.; McCoy, S.; Goxe, J.; Read, J.; et al. Host effects in high ectomycorrhizal diversity tropical rainforests on ultramafic soils in New Caledonia. Fungal Ecol. 2019, 39, 201–212. [Google Scholar] [CrossRef]
- Jaffré, T.; Bouchet, P.; Veillon, J.M. Threatened plants of New Caledonia: Is the system of protected areas adequate? Biodivers. Conserv. 1998, 7, 109–135. [Google Scholar] [CrossRef]
- Jaffré, T.; Munzinger, J.; Lowry, P.P. Threats to the conifer species found on New Caledonia’s ultramafic massifs and proposals for urgently needed measures to improve their protection. Biodivers. Conserv. 2010, 19, 1485–1502. [Google Scholar] [CrossRef]
- Losfeld, G.; L’Huillier, L.; Fogliani, B.; Jaffré, T.; Grison, C. Mining in New Caledonia: Environmental stakes and restoration opportunities. Environ. Sci Pollut Res 2015, 22, 5592–5607. [Google Scholar] [CrossRef]
- Ibanez, T.; Birnbaum, P.; Gâteblé, G.; Hequet, V.; Isnard, S.; Munzinger, J.; Pillon, Y.; Pouteau, R.; Vandrot, H.; Jaffré, T. Twenty years after Jaffré et al. (1998), is the system of protected areas now adequate in New Caledonia? Biodivers. Conserv. 2019, 28, 245–254. [Google Scholar] [CrossRef]
- Myers, N.; Mittermeier, R.A.; Mittermeier, C.G.; da Fonseca, G.A.B.; Kent, J. Biodiversity hotspots for conservation priorities. Nature 2000, 403, 853–858. [Google Scholar] [CrossRef]
- Isnard, S.; L’Huillier, L.; Rigault, F.; Jaffré, T. How did the ultramafic soils shape the flora of the New Caledonian hotspot? Plant Soil 2016, 403, 53–76. [Google Scholar] [CrossRef]
- Jaffré, T.; Rigault, F.; Sarrailh, J.-M. La végétalisation des anciens sites miniers. Bois For. Trop. 1994, 242, 45–57. [Google Scholar]
- Jaffré, T.; Veillon, J.M.; Cherrier, J.F. Sur la présence de deux Cupressaceae, Neocallitropsis pancheri (Carr.) Laubenf. et Libocedrus austrocaledonica Brongn. & Gris dans le Massif du Paéoua et localités nouvelles de Gymnospermes en Nouvelle-Calédonie. Adansonia 1987, 3, 273–288. [Google Scholar]
- McCoy, S.; Jaffré, T.; Rigault, F.; Ash, J.E. Fire and succession in the ultramafic maquis of New Caledonia. J. Biogeogr. 1999, 26, 579–594. [Google Scholar] [CrossRef]
- Wulff, A.S.; Hollingsworth, P.M.; Ahrends, A.; Jaffré, T.; Veillon, J.-M.; L’Huillier, L.; Fogliani, B. Conservation priorities in a biodiversity hotspot: Analysis of narrow endemic plant species in New Caledonia. PLoS ONE 2013, 8, e73371. [Google Scholar] [CrossRef] [Green Version]
- Houlès, A.; Vincent, B.; David, M.; Ducousso, M.; Galiana, A.; Juillot, F.; Hannibal, L.; Carriconde, F.; Fritsch, E.; Jourand, P. Ectomycorrhizal communities associated with the legume Acacia spirorbis growing on contrasted edaphic constraints in New Caledonia. Microb. Ecol. 2018, 76, 964–975. [Google Scholar] [CrossRef] [PubMed]
- Peay, K.G.; Schubert, M.G.; Nguyen, N.H.; Bruns, T.D. Measuring ectomycorrhizal fungal dispersal: Macroecological patterns driven by microscopic propagules. Mol. Ecol. 2012, 21, 4122–4136. [Google Scholar] [CrossRef] [PubMed]
- Vaario, L.-M.V.; Tervonen, A.T.; Haukioja, K.H.; Haukioja, M.H.; Pennanen, T.P.; Timonen, S.T. The effect of nursery substrate and fertilization on the growth and ectomycorrhizal status of containerized and outplanted seedlings of Picea abies. Can. J. For. Res. 2008, 39, 64–75. [Google Scholar] [CrossRef]
- Kazantseva, O.; Bingham, M.; Simard, S.W.; Berch, S.M. Effects of growth medium, nutrients, water, and aeration on mycorrhization and biomass allocation of greenhouse-grown interior Douglas-fir seedlings. Mycorrhiza 2009, 20, 51–66. [Google Scholar] [CrossRef]
- El Karkouri, K.; Martin, F.; Emmanuel Douzery, J.P.; Mousain, D. Diversity of ectomycorrhizal fungi naturally established on containerised Pinus seedlings in nursery conditions. Microbiol. Res. 2005, 160, 47–52. [Google Scholar] [CrossRef]
- Trocha, L.K.; Rudawska, M.; Leski, T.; Dabert, M. Genetic diversity of naturally established ectomycorrhizal fungi on Norway Spruce seedlings under nursery conditions. Microb. Ecol. 2006, 52, 418–425. [Google Scholar] [CrossRef]
- Fernández, N.V.; Marchelli, P.; Fontenla, S.B. Ectomycorrhizas naturally established in Nothofagus nervosa seedlings under different cultivation practices in a forest nursery. Microb. Ecol. 2013, 66, 581–592. [Google Scholar] [CrossRef]
- Henry, C.; Raivoarisoa, J.-F.; Razafimamonjy, A.; Ramanankierana, H.; Andrianaivomahefa, P.; Ducousso, M.; Selosse, M.-A. Transfer to forest nurseries significantly affects mycorrhizal community composition of Asteropeia mcphersonii wildings. Mycorrhiza 2017, 27, 321–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selosse, M.-A.; Jacquot, D.; Bouchard, D.; Martin, F.; Tacon, F.L. Temporal persistence and spatial distribution of an American inoculant strain of the ectomycorrhizal basidiomycete Laccaria bicolor in a French forest plantation. Mol. Ecol. 1998, 7, 561–573. [Google Scholar] [CrossRef] [Green Version]
- Karkouri, K.E.; Martin, F.; Mousain, D. Dominance of the mycorrhizal fungus Rhizopogon rubescens in a plantation of Pinus pinea seedlings inoculated with Suillus collinitus. Ann. For. Sci. 2002, 59, 197–204. [Google Scholar] [CrossRef] [Green Version]
- Walker, L.R.; del Moral, R. Primary Succession and Ecosystem Rehabilitation, 1st ed.; Cambridge University Press: Cambridge, UK, 2003; ISBN 978-0-521-80076-1. [Google Scholar]
- Nara, K.; Hogetsu, T. Ectomycorrhizal fungi on established shrubs facilitate subsequent seedling establishment of successional plant species. Ecology 2004, 85, 1700–1707. [Google Scholar] [CrossRef]
- Richard, F.; Millot, S.; Gardes, M.; Selosse, M.-A. Diversity and specificity of ectomycorrhizal fungi retrieved from an old-growth mediterranean forest dominated by Quercus iIlex. New Phytol. 2005, 166, 1011–1023. [Google Scholar] [CrossRef] [PubMed]
- Ryberg, M.; Larsson, E.; Molau, U. Ectomycorrhizal diversity on Dryas octopetala and Salix reticulata in an alpine cliff ecosystem. Arct. Antarct. Alp. Res. 2009, 41, 506–514. [Google Scholar] [CrossRef]
- Diédhiou, A.G.; Selosse, M.-A.; Galiana, A.; Diabaté, M.; Dreyfus, B.; Bâ, A.M.; Faria, S.M.D.; Béna, G. Multi-host ectomycorrhizal fungi are predominant in a Guinean tropical rainforest and shared between canopy trees and seedlings. Environ. Microbiol. 2010, 12, 2219–2232. [Google Scholar] [CrossRef]
- Smith, M.E.; Henkel, T.W.; Aime, M.C.; Fremier, A.K.; Vilgalys, R. Ectomycorrhizal fungal diversity and community structure on three co-occurring leguminous canopy tree species in a neotropical rainforest. New Phytol. 2011, 192, 699–712. [Google Scholar] [CrossRef]
- Smith, M.E.; Henkel, T.W.; Uehling, J.K.; Fremier, A.K.; Clarke, H.D.; Vilgalys, R. The ectomycorrhizal fungal community in a neotropical forest dominated by the endemic dipterocarp Pakaraimaea dipterocarpacea. PLoS ONE 2013, 8, e55160. [Google Scholar] [CrossRef]
- Perrier, N.; Amir, H.; Colin, F. Occurence of mycorrhizal symbioses in the metal-rich lateritic soils of the Koniambo Massif, New Caledonia. Mycorrhiza 2006, 16, 449–458. [Google Scholar] [CrossRef]
- Jourand, P.; Hannibal, L.; Majorel, C.; Mengant, S.; Ducousso, M.; Lebrun, M. Ectomycorrhizal Pisolithus albus inoculation of Acacias spirorbis and Eucalyptus globulus grown in ultramafic topsoil enhances plant growth and mineral nutrition while limits metal uptake. J. Plant Physiol. 2014, 171, 164–172. [Google Scholar] [CrossRef]
- Carriconde, F.; Gardes, M.; Jargeat, P.; Heilmann-Clausen, J.; Mouhamadou, B.; Gryta, H. Population evidence of cryptic species and geographical structure in the cosmopolitan ectomycorrhizal fungus, Tricholoma scalpturatum. Microb. Ecol. 2008, 56, 513–524. [Google Scholar] [CrossRef] [PubMed]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Consortium, F.B. Nuclear ribosomal Internal Transcribed Spacer (ITS) region as a universal DNA barcode marker for fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for Basidiomycetes—Application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols; Elsevier: Amsterdam, The Netherlands, 1990; pp. 315–322. ISBN 978-0-12-372180-8. [Google Scholar]
- Li, W.; Godzik, A. Cd-Hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tedersoo, L.; May, T.W.; Smith, M.E. Ectomycorrhizal lifestyle in fungi: Global diversity, distribution, and evolution of phylogenetic lineages. Mycorrhiza 2010, 20, 217–263. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. 2020. Available online: https://cran.r-project.org/package=vegan (accessed on 23 December 2021).
- Hervé, M. RVAideMemoire: Testing and Plotting Procedures for Biostatistics (0.9-79) [Computer Software]. Available online: https://cran.r-project (accessed on 23 December 2021).
- Chao, A.; Chiu, C.-H.; Jost, L. Statistical challenges of evaluating diversity patterns across environmental gradients in mega-diverse communities. J. Veg. Sci. 2016, 27, 437–438. [Google Scholar] [CrossRef]
- Hsieh, T.C.; Ma, K.H.; Chao, A. INEXT: An R package for rarefaction and extrapolation of species diversity (Hill numbers). Methods Ecol. Evol. 2016, 7, 1451–1456. [Google Scholar] [CrossRef]
- Zack, J.; Willig, M. Fungal biodiversity and patterns. In Biodiversity of Fungi: Inventory and Monitoring Methods; Elsevier: Amsterdam, The Netherlands, 2004; pp. 59–75. ISBN 978-0-08-047026-9. [Google Scholar]
- Oliveros, J.C. VENNY. An Interactive Tool for Comparing Lists with Venn Diagrams. 2007. Available online: http://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 23 December 2021).
- Smith, E.P.; van Belle, G. Nonparametric estimation of species richness. Biometrics 1984, 40, 119–129. [Google Scholar] [CrossRef] [Green Version]
- Waseem, M.; Ducousso, M.; Prin, Y.; Domergue, O.; Hannibal, L.; Majorel, C.; Jourand, P.; Galiana, A. ectomycorrhizal fungal diversity associated with endemic Tristaniopsis spp. (Myrtaceae) in ultramafic and volcano-sedimentary soils in New Caledonia. Mycorrhiza 2017, 27, 407–413. [Google Scholar] [CrossRef]
- Helbert; Turjaman, M.; Nara, K. Ectomycorrhizal fungal communities of secondary tropical forests dominated by Tristaniopsis in Bangka Island, Indonesia. PLoS ONE 2019, 14, e0221998. [Google Scholar] [CrossRef] [PubMed]
- Epp Schmidt, D.J.; Pouyat, R.; Szlavecz, K.; Setälä, H.; Kotze, D.J.; Yesilonis, I.; Cilliers, S.; Hornung, E.; Dombos, M.; Yarwood, S.A. Urbanization erodes ectomycorrhizal fungal diversity and may cause microbial communities to converge. Nat. Ecol. Evol. 2017, 1, 0123. [Google Scholar] [CrossRef] [PubMed]
- Rudawska, M.; Leski, T. Ectomycorrhizal Fungal Assemblages of nursery-grown Scots pine are influenced by age of the seedlings. Forests 2021, 12, 134. [Google Scholar] [CrossRef]
- Hilszczanska, D.; Sierota, Z. Persistence of ectomycorrhizas by Telephora terrestris on outplanted Scots pine seedlings. Acta Mycol. 2006, 41, 313–318. [Google Scholar] [CrossRef] [Green Version]
| Diversity Indexes | |||||
|---|---|---|---|---|---|
| Treatment | Richness 0D | Chao1 | ACE | Shannon 1D | Simpson 2D |
| AS_2015_0m | 26 | 50 ± 16 | 54 ± 3.5 | 2.8 | 0.90 |
| AS_2016_0m | 26 | 57 ± 20 | 67 ± 4.3 | 3.0 | 0.94 |
| TG_2015_10m | 14 | 26 ± 11 | 27 ± 2.4 | 2.5 | 0.90 |
| TC_2015_none | 5 | 5.5 ± 1.3 | 6.4 ± 1.1 | 1.5 | 0.74 |
| TC_2016_1m | 9 | 30 ± 17 | 27 ± 1.8 | 2.1 | 0.85 |
| TC_2016_2m | 6 | 6.5 ± 1.2 | 7.2 ± 1.0 | 1.6 | 0.78 |
| TC_2016_3m | 6 | 9.0 ± 4.1 | 12 ± 1.49 | 1.7 | 0.79 |
| TC_2016_10m | 13 | 13 ± 1.6 | 15 ± 1.7 | 2.3 | 0.88 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Houlès, A.; Gotty, K.; Joussemet, F.; Vincent, B.; Hannibal, L.; Patrois, M.; Jourand, P.; Ducousso, M. The Nurse Plant Acacia spirorbis Enriches Ectomycorrhizal Community Composition of a Target Species: Tristaniopsis calobuxus. Diversity 2022, 14, 107. https://doi.org/10.3390/d14020107
Houlès A, Gotty K, Joussemet F, Vincent B, Hannibal L, Patrois M, Jourand P, Ducousso M. The Nurse Plant Acacia spirorbis Enriches Ectomycorrhizal Community Composition of a Target Species: Tristaniopsis calobuxus. Diversity. 2022; 14(2):107. https://doi.org/10.3390/d14020107
Chicago/Turabian StyleHoulès, Anne, Karine Gotty, François Joussemet, Bryan Vincent, Laure Hannibal, Magali Patrois, Philippe Jourand, and Marc Ducousso. 2022. "The Nurse Plant Acacia spirorbis Enriches Ectomycorrhizal Community Composition of a Target Species: Tristaniopsis calobuxus" Diversity 14, no. 2: 107. https://doi.org/10.3390/d14020107
APA StyleHoulès, A., Gotty, K., Joussemet, F., Vincent, B., Hannibal, L., Patrois, M., Jourand, P., & Ducousso, M. (2022). The Nurse Plant Acacia spirorbis Enriches Ectomycorrhizal Community Composition of a Target Species: Tristaniopsis calobuxus. Diversity, 14(2), 107. https://doi.org/10.3390/d14020107






