Recent Changes in Genetic Diversity, Structure, and Gene Flow in a Passerine Experiencing a Rapid Population Decline, the Dupont’s Lark (Chersophilus duponti)
Abstract
1. Introduction
2. Materials and Methods
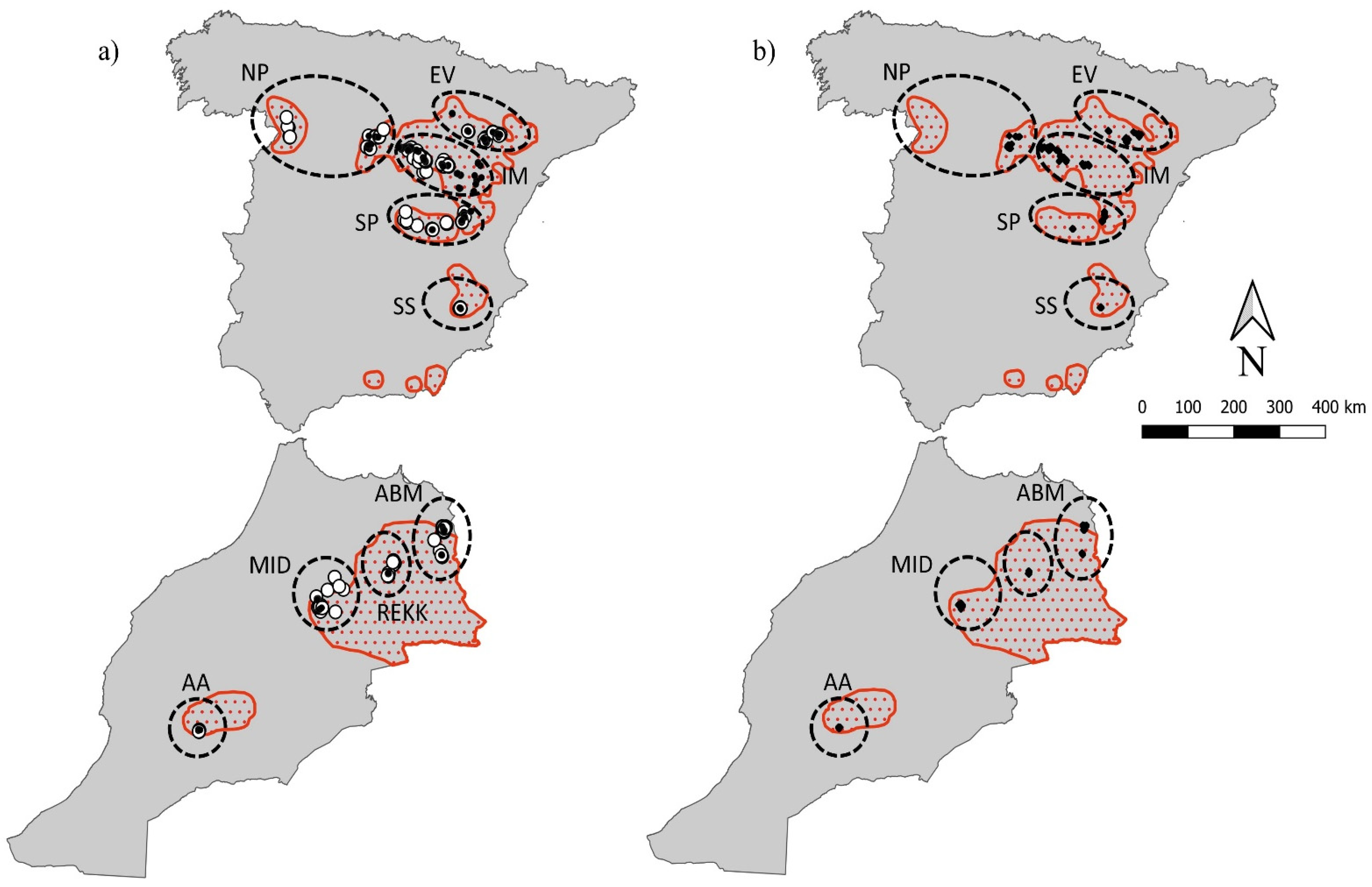
2.1. Study Area and Sampling
2.2. Molecular Analysis
2.3. Genetic Diversity
2.4. Genetic Structure of Populations
2.5. Gene Flow Estimation
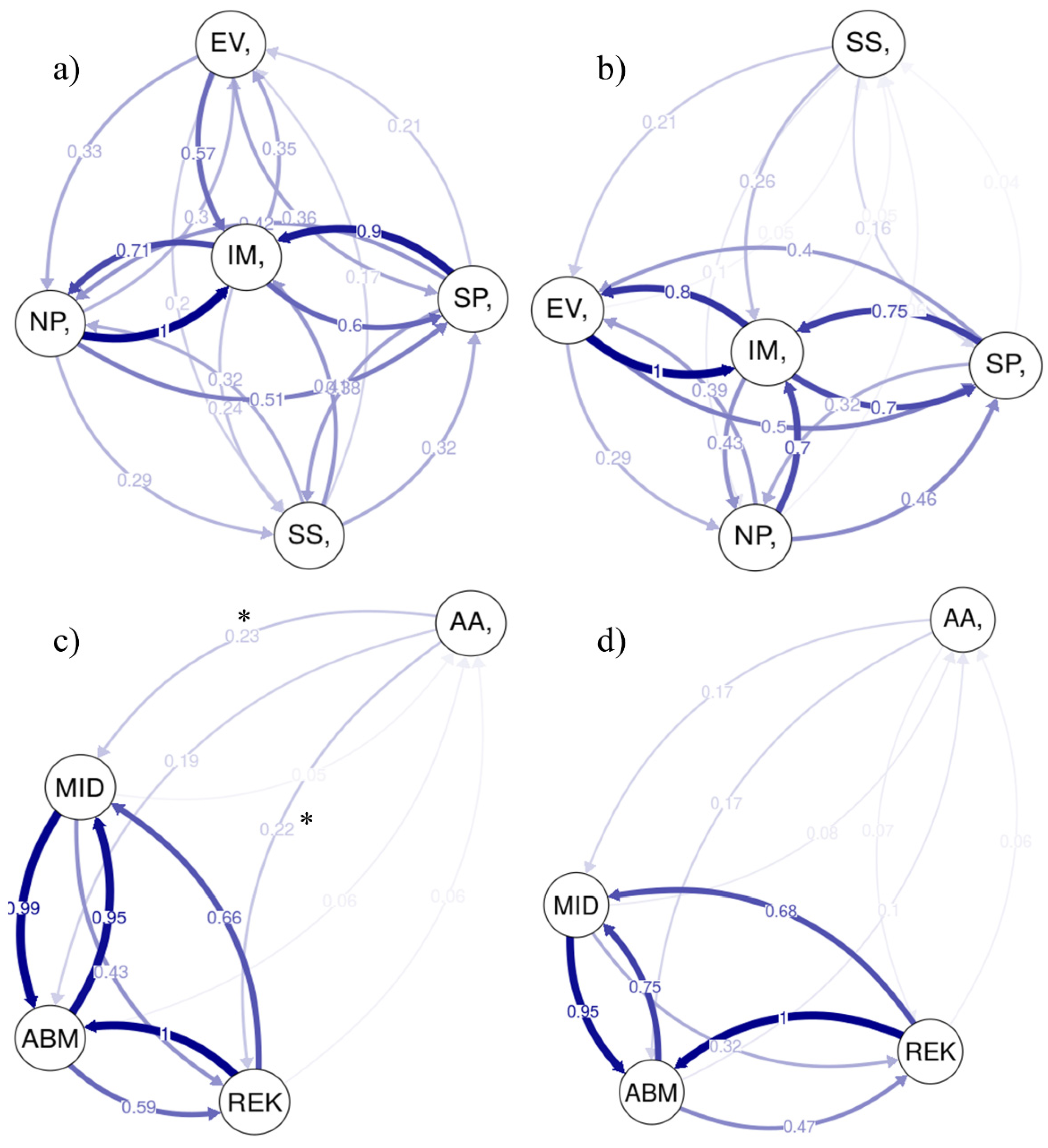
3. Results
3.1. Genetic Diversity and Effective Population Size Estimates
3.2. Temporal Variation of Genetic Diversity
3.3. Genetic Structure of Populations
3.4. Direction of Gene Flow
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Heath, D.D.; Busch, C.; Kelly, J.; Atagi, D.Y. Temporal Change in Genetic Structure and Effective Population Size in Steelhead Trout (Oncorhynchus Mykiss). Mol. Ecol. 2002, 11, 197–214. [Google Scholar] [CrossRef] [PubMed]
- Collier, N.; Gardner, M.; Adams, M.; McMahon, C.R.; Benkendorff, K.; Mackay, D.A. Contemporary Habitat Loss Reduces Genetic Diversity in an Ecologically Specialized Butterfly. J. Biogeogr. 2010, 37, 1277–1287. [Google Scholar] [CrossRef]
- Kleinhans, C.; Willows-Munro, S. Low Genetic Diversity and Shallow Population Structure in the Endangered Vulture, Gyps Coprotheres. Sci. Rep. 2019, 9, 5536. [Google Scholar] [CrossRef] [PubMed]
- Frankham, R.; Ballou, S.E.J.D.; Briscoe, D.A.; Ballou, J.D. Introduction to Conservation Genetics; Cambridge University Press: Cambridge, UK, 2002; ISBN 978-0-521-63985-9. [Google Scholar]
- Riva, F.; Fahrig, L. The Disproportionately High Value of Small Patches for Biodiversity Conservation. Conserv. Lett. 2022, 15, e12881. [Google Scholar] [CrossRef]
- Wright, S. Evolution in Mendelian Populations. Genetics 1931, 16, 97–159. [Google Scholar] [CrossRef]
- Allendorf, F.W.; Luikart, G.; Aitken, S.N. Conservation and the Genetics of Populations, 2nd ed.; John Wiley & Sons: Hoboken, NJ, USA, 2013; ISBN 978-0-470-67146-7. [Google Scholar]
- Fahrig, L. Effects of Habitat Fragmentation on Biodiversity. Annu. Rev. Ecol. Evol. Syst. 2003, 34, 487–515. [Google Scholar] [CrossRef]
- Haddad, N.M.; Brudvig, L.A.; Clobert, J.; Davies, K.F.; Gonzalez, A.; Holt, R.D.; Lovejoy, T.E.; Sexton, J.O.; Austin, M.P.; Collins, C.D.; et al. Habitat Fragmentation and Its Lasting Impact on Earth’s Ecosystems. Sci. Adv. 2015, 1, e1500052. [Google Scholar] [CrossRef]
- Harrisson, K.A.; Pavlova, A.; Amos, J.N.; Takeuchi, N.; Lill, A.; Radford, J.Q.; Sunnucks, P. Fine-Scale Effects of Habitat Loss and Fragmentation despite Large-Scale Gene Flow for Some Regionally Declining Woodland Bird Species. Landsc. Ecol. 2012, 27, 813–827. [Google Scholar] [CrossRef]
- Stevens, K.; Harrisson, K.A.; Hogan, F.E.; Cooke, R.; Clarke, R.H. Reduced Gene Flow in a Vulnerable Species Reflects Two Centuries of Habitat Loss and Fragmentation. Ecosphere 2018, 9, e02114. [Google Scholar] [CrossRef]
- Dale, S. Female-Biased Dispersal, Low Female Recruitment, Unpaired Males, and the Extinction of Small and Isolated Bird Populations. Oikos 2001, 92, 344–356. [Google Scholar] [CrossRef]
- Willi, Y.; Kristensen, T.N.; Sgrò, C.M.; Weeks, A.R.; Ørsted, M.; Hoffmann, A.A. Conservation Genetics as a Management Tool: The Five Best-Supported Paradigms to Assist the Management of Threatened Species. Proc. Natl. Acad. Sci. USA 2022, 119, e2105076119. [Google Scholar] [CrossRef] [PubMed]
- Duncan, C.; Böhm, M.; Turvey, S.T. Identifying the Possibilities and Pitfalls of Conducting IUCN Red List Assessments from Remotely Sensed Habitat Information Based on Insights from Poorly Known Cuban Mammals. Conserv. Biol. 2021, 35, 1598–1614. [Google Scholar] [CrossRef] [PubMed]
- Carroll, E.L.; Bruford, M.W.; DeWoody, J.A.; Leroy, G.; Strand, A.; Waits, L.; Wang, J. Genetic and Genomic Monitoring with Minimally Invasive Sampling Methods. Evol. Appl. 2018, 11, 1094–1119. [Google Scholar] [CrossRef] [PubMed]
- Bruford, M.W.; Davies, N.; Dulloo, M.E.; Faith, D.P.; Walters, M. (Eds.) Monitoring changes in genetic diversity. In The GEO Handbook on Biodiversity Observation Networks; Springer: Cham, Switzerland, 2017; pp. 107–128. [Google Scholar]
- Schlaepfer, D.R.; Braschler, B.; Rusterholz, H.-P.; Baur, B. Genetic Effects of Anthropogenic Habitat Fragmentation on Remnant Animal and Plant Populations: A Meta-Analysis. Ecosphere 2018, 9, e02488. [Google Scholar] [CrossRef]
- Schwartz, M.K.; Luikart, G.; Waples, R.S. Genetic Monitoring as a Promising Tool for Conservation and Management. Trends Ecol. Evol. 2007, 22, 25–33. [Google Scholar] [CrossRef]
- Ripperger, S.P.; Tschapka, M.; Kalko, E.K.V.; Rodriguez-Herrera, B.; Mayer, F. Life in a Mosaic Landscape: Anthropogenic Habitat Fragmentation Affects Genetic Population Structure in a Frugivorous Bat Species. Conserv. Genet. 2013, 14, 925–934. [Google Scholar] [CrossRef]
- Peeters, B.; Le Moullec, M.; Raeymaekers, J.A.M.; Marquez, J.F.; Røed, K.H.; Pedersen, Å.Ø.; Veiberg, V.; Loe, L.E.; Hansen, B.B. Sea Ice Loss Increases Genetic Isolation in a High Arctic Ungulate Metapopulation. Glob. Change Biol. 2020, 26, 2028–2041. [Google Scholar] [CrossRef]
- Barker, J.S.F. Effective Population Size of Natural Populations of Drosophila Buzzatii, with a Comparative Evaluation of Nine Methods of Estimation. Mol. Ecol. 2011, 20, 4452–4471. [Google Scholar] [CrossRef]
- Crispo, E.; Chapman, L.J. Temporal Variation in Population Genetic Structure of a Riverine African Cichlid Fish. J. Hered. 2010, 101, 97–106. [Google Scholar] [CrossRef]
- Charlier, J.; Laikre, L.; Ryman, N. Genetic Monitoring Reveals Temporal Stability over 30 Years in a Small, Lake-Resident Brown Trout Population. Heredity 2012, 109, 246–253. [Google Scholar] [CrossRef]
- Hagen, S.B.; Kopatz, A.; Aspi, J.; Kojola, I.; Eiken, H.G. Evidence of Rapid Change in Genetic Structure and Diversity during Range Expansion in a Recovering Large Terrestrial Carnivore. Proc. R. Soc. B Biol. Sci. 2015, 282, 20150092. [Google Scholar] [CrossRef] [PubMed]
- Kunz, F.; Kohnen, A.; Nopp-Mayr, U.; Coppes, J. Past, Present, Future: Tracking and Simulating Genetic Differentiation over Time in a Closed Metapopulation System. Conserv. Genet. 2021, 22, 355–368. [Google Scholar] [CrossRef]
- Gómez-Catasús, J.; Garza, V.; Morales, M.B.; Traba, J. Hierarchical Habitat-Use by an Endangered Steppe Bird in Fragmented Landscapes Is Associated with Large Connected Patches and High Food Availability. Sci. Rep. 2019, 9, 19010. [Google Scholar] [CrossRef] [PubMed]
- Reverter, M.; Gómez-Catasús, J.; Barrero, A.; Traba, J. Crops Modify Habitat Quality beyond Their Limits. Agric. Ecosyst. Environ. 2021, 319, 107542. [Google Scholar] [CrossRef]
- Traba, J.; Pérez Granados, C. Extensive Sheep Grazing Is Associated with Trends in Steppe Birds in Spain: Recommendations for the Common Agricultural Policy. PeerJ 2022, 10, e12870. [Google Scholar] [CrossRef]
- García, J.T.; Suárez, F.; Garza, V.; Calero-Riestra, M.; Hernández, J.; Pérez-Tris, J. Genetic and Phenotypic Variation among Geographically Isolated Populations of the Globally Threatened Dupont’s Lark Chersophilus Duponti. Mol. Phylogenet. Evol. 2008, 46, 237–251. [Google Scholar] [CrossRef]
- Suárez LA ALONDRA RICOTI (CHERSOPHILUS DUPONTI)|VV.AA.|Casa del Libro. Available online: https://www.casadellibro.com/libro-la-alondra-ricoti-chersophilus-duponti/9788480147736/1769714 (accessed on 22 November 2022).
- García-Antón, A.; Garza, V.; Hernández Justribó, J.; Traba, J. Factors Affecting Dupont´s Lark Distribution and Range Regression in Spain. PLoS ONE 2019, 14, e0211549. [Google Scholar] [CrossRef]
- Vögeli, M.; Serrano, D.; Pacios, F.; Tella, J.L. The Relative Importance of Patch Habitat Quality and Landscape Attributes on a Declining Steppe-Bird Metapopulation. Biol. Conserv. 2010, 143, 1057–1067. [Google Scholar] [CrossRef]
- Gómez-Catasús, J.; Barrero, A.; Garza, V.; Traba, J. Alondra ricotí—Chersophilus duponti. In Enciclopedia Virtual de los Vertebrados Españoles; Salvador, A., Morales, M.B., Eds.; Museo Nacional de Ciencias Naturales: Madrid, Spain, 2016; p. 25. Available online: http://www.vertebradosibericos.org/ (accessed on 21 November 2022).
- Gómez-Catasús, J.; Pérez-Granados, C.; Barrero, A.; Bota, G.; Giralt, D.; López-Iborra, G.M.; Serrano, D.; Traba, J. European Population Trends and Current Conservation Status of an Endangered Steppe-Bird Species: The Dupont’s Lark Chersophilus Duponti. PeerJ 2018, 6, e5627. [Google Scholar] [CrossRef]
- BirdLife International. Chersophilus Duponti. IUCN Red List Threat Spec 2020. Available online: https://www.birdlife.org (accessed on 15 November 2022).
- García-Antón, A.; Traba, J. Population Viability Analysis of the Endangered Dupont’s Lark Chersophilus Duponti in Spain. Sci. Rep. 2021, 11, 19947. [Google Scholar] [CrossRef]
- Tella, J.L.; Vögeli, M.; Serrano, D.; Carrete, M. Current Status of the Threatened Dupont’s Lark Chersophilus Duponti in Spain: Overestimation, Decline, and Extinction of Local Populations. Oryx 2005, 39, 90–94. [Google Scholar] [CrossRef]
- Méndez, M.; Tella, J.; Godoy, J. Restricted Gene Flow and Genetic Drift in Recently Fragmented Populations of an Endangered Steppe Bird. Biol. Conserv. 2011, 144, 2515–2622. [Google Scholar] [CrossRef]
- Pérez-Granados, C.; López-Iborra, G.M. Census of Breeding Birds and Population Trends of the Dupont’s Lark Chersophilus Duponti in Eastern Spain. Ardeola 2013, 60, 143–150. [Google Scholar] [CrossRef]
- Laiolo, P.; Vögeli, M.; Serrano, D.; Tella, J.L. Testing Acoustic versus Physical Marking: Two Complementary Methods for Individual-Based Monitoring of Elusive Species. J. Avian Biol. 2007, 38, 672–681. [Google Scholar] [CrossRef]
- Pérez-Granados, C.; Sáez-Gómez, P.; López-Iborra, G.M. Breeding Dispersal Movements of Dupont’s Lark Chersophilus Duponti in Fragmented Landscape. Bird Conserv. Int. 2022, 32, 53–63. [Google Scholar] [CrossRef]
- Pérez-Granados, C.; López-Iborra, G.M.; Seoane, J. A Multi-Scale Analysis of Habitat Selection in Peripheral Populations of the Endangered Dupont’s Lark Chersophilus Duponti. Bird Conserv. Int. 2017, 27, 398–413. [Google Scholar] [CrossRef]
- García-Antón, A.; Garza, V.; Traba, J. Connectivity in Spanish Metapopulation of Dupont’s Lark May Be Maintained by Dispersal over Medium-Distance Range and Stepping Stones. PeerJ 2021, 9, e11925. [Google Scholar] [CrossRef]
- García, J.T.; Suárez, F.; Garza, V.; Justribó, J.H.; Oñate, J.J.; Hervás, I.; Calero, M.; García de la Morena, E.L. Assessing the Distribution, Habitat, and Population Size of the Threatened Dupont’s Lark Chersophilus Duponti in Morocco: Lessons for Conservation. Oryx 2008, 42, 592. [Google Scholar] [CrossRef]
- Ceballos, G.; Ehrlich, P.R.; Dirzo, R. Biological Annihilation via the Ongoing Sixth Mass Extinction Signaled by Vertebrate Population Losses and Declines. Proc. Natl. Acad. Sci. USA 2017, 114, E6089–E6096. [Google Scholar] [CrossRef]
- Boughalmi, A.; Araba, A.; Yessef, M. Dynamics of Extensive Sheep Production Systems in Morocco. Livest. Res. Rural Dev. 2015, 27, 221. [Google Scholar]
- Gaouar, S.B.S.; Lafri, M.; Djaout, A.; El-Bouyahiaoui, R.; Bouri, A.; Bouchatal, A.; Maftah, A.; Ciani, E.; Da Silva, A.B. Genome-Wide Analysis Highlights Genetic Dilution in Algerian Sheep. Heredity 2017, 118, 293–301. [Google Scholar] [CrossRef] [PubMed]
- Laiolo, P.; Tella, J.L. Fate of Unproductive and Unattractive Habitats: Recent Changes in Iberian Steppes and Their Effects on Endangered Avifauna. Environ. Conserv. 2006, 33, 223–232. [Google Scholar] [CrossRef]
- Íñigo Íñigo, A.; Garza, V.; Tella, J.L.; Laiolo, P.; Suárez, F.; Barov, B. Action Plan for the Dupont’s Lark Chersophilus Duponti in the European Union; SEO/BirdLife/BirdLife International: Madrid, Spain, 2008. [Google Scholar]
- Strauss, W.M. Preparation of Genomic DNA from Mammalian Tissue. Curr. Protoc. Mol. Biol. 1998, 42, 2.2.1–2.2.3. [Google Scholar] [CrossRef] [PubMed]
- Méndez, M.; Prats, L.; Tella, J.L.; Godoy, J.A. Isolation and Characterization of Twenty Polymorphic Microsatellite Markers for the Endangered Dupont′s Lark (Chersophilus Duponti) and Cross-Amplification in Crested Lark (Galerida Cristata) and Thekla Lark (Galerida Theklae). Conserv. Genet. Resour. 2010, 3, 107–111. [Google Scholar] [CrossRef]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. Micro-Checker: Software for Identifying and Correcting Genotyping Errors in Microsatellite Data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Raymond, M.; Rousset, F. An Exact Test for Population Differentiation. Evolution 1995, 49, 1280–1283. [Google Scholar] [CrossRef]
- Keenan, K.; McGinnity, P.; Cross, T.F.; Crozier, W.W.; Prodöhl, P.A. DiveRsity: An R Package for the Estimation and Exploration of Population Genetics Parameters and Their Associated Errors. Methods Ecol. Evol. 2013, 4, 782–788. [Google Scholar] [CrossRef]
- Kalinowski, S.T. Counting Alleles with Rarefaction: Private Alleles and Hierarchical Sampling Designs. Conserv. Genet. 2004, 5, 539–543. [Google Scholar] [CrossRef]
- Leberg, P.L. Estimating Allelic Richness: Effects of Sample Size and Bottlenecks. Mol. Ecol. 2002, 11, 2445–2449. [Google Scholar] [CrossRef]
- Do, C.; Waples, R.S.; Peel, D.; Macbeth, G.M.; Tillett, B.; Ovenden, J.; Do, C.; Waples, R.S.; Peel, D.; Macbeth, G.M.; et al. NeEstimator v2: Re-Implementation of Software for the Estimation of Contemporary Effective Population Size (Ne) from Genetic Data. Mol. Ecol. Resour. 2014, 14, 209–214. [Google Scholar] [CrossRef]
- Waples, R.S. A Generalized Approach for Estimating Effective Population Size from Temporal Changes in Allele Frequency. Genetics 1989, 121, 379–391. [Google Scholar] [CrossRef] [PubMed]
- Jorde, P.E.; Ryman, N. Unbiased Estimator for Genetic Drift and Effective Population Size. Genetics 2007, 177, 927–935. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Tajima, F. Genetic Drift and Estimation of Effective Population Size. Genetics 1981, 98, 625–640. [Google Scholar] [CrossRef] [PubMed]
- Tessier, N.; Bernatchez, L. Stability of Population Structure and Genetic Diversity across Generations Assessed by Microsatellites among Sympatric Populations of Landlocked Atlantic Salmon (Salmo Salar L.). Mol. Ecol. 1999, 8, 169–179. [Google Scholar] [CrossRef]
- Ozerov, M.Y.; Veselov, A.E.; Lumme, J.; Primmer, C.R. Temporal Variation of Genetic Composition in Atlantic Salmon Populations from the Western White Sea Basin: Influence of Anthropogenic Factors? BMC Genet. 2013, 14, 88. [Google Scholar] [CrossRef]
- Luikart, G.; Cornuet, J.-M. Empirical Evaluation of a Test for Identifying Recently Bottlenecked Populations from Allele Frequency Data. Conserv. Biol. 1998, 12, 228–237. [Google Scholar] [CrossRef]
- Piry, S.; Luikart, G.; Cornuet, J.-M. Computer Note. BOTTLENECK: A Computer Program for Detecting Recent Reductions in the Effective Size Using Allele Frequency Data. J. Hered. 1999, 90, 502–503. [Google Scholar] [CrossRef]
- Di Rienzo, A.; Peterson, A.C.; Garza, J.C.; Valdes, A.M.; Slatkin, M.; Freimer, N.B. Mutational Processes of Simple-Sequence Repeat Loci in Human Populations. Proc. Natl. Acad. Sci. USA 1994, 91, 3166–3170. [Google Scholar] [CrossRef]
- Weir, B.S.; Cockerham, C.C. Estimating F-Statistics for the Analysis of Population Structure. Evolution 1984, 38, 1358–1370. [Google Scholar] [CrossRef]
- Nei Molecular Evolutionary Genetics. Available online: https://www.degruyter.com/document/doi/10.7312/nei-92038/html (accessed on 22 November 2022).
- Jost, L. G(ST) and Its Relatives Do Not Measure Differentiation. Mol. Ecol. 2008, 17, 4015–4026. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. Genalex 6: Genetic Analysis in Excel. Population Genetic Software for Teaching and Research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic Analysis in Excel. Population Genetic Software for Teaching and Research—An Update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Lischer, H.E.L. Arlequin Suite Ver 3.5: A New Series of Programs to Perform Population Genetics Analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Smouse, P.E.; Peakall, R. Spatial Autocorrelation Analysis of Individual Multiallele and Multilocus Genetic Structure. Heredity 1999, 82, 561–573. [Google Scholar] [CrossRef] [PubMed]
- Alcala, N.; Goudet, J.; Vuilleumier, S. On the Transition of Genetic Differentiation from Isolation to Panmixia: What We Can Learn from GST and D. Theor. Popul. Biol. 2014, 93, 75–84. [Google Scholar] [CrossRef]
- Sundqvist, L.; Keenan, K.; Zackrisson, M.; Prodöhl, P.; Kleinhans, D. Directional Genetic Differentiation and Relative Migration. Ecol. Evol. 2016, 6, 3461–3475. [Google Scholar] [CrossRef]
- Hoban, S.; Bruford, M.; D’Urban Jackson, J.; Lopes-Fernandes, M.; Heuertz, M.; Hohenlohe, P.A.; Paz-Vinas, I.; Sjögren-Gulve, P.; Segelbacher, G.; Vernesi, C.; et al. Genetic Diversity Targets and Indicators in the CBD Post-2020 Global Biodiversity Framework Must Be Improved. Biol. Conserv. 2020, 248, 108654. [Google Scholar] [CrossRef]
- Frankham, R. Effective Population Size/Adult Population Size Ratios in Wildlife: A Review. Genet. Res. 1995, 66, 95–107. [Google Scholar] [CrossRef]
- Palstra, F.P.; Ruzzante, D.E. Genetic Estimates of Contemporary Effective Population Size: What Can They Tell Us about the Importance of Genetic Stochasticity for Wild Population Persistence? Mol. Ecol. 2008, 17, 3428–3447. [Google Scholar] [CrossRef]
- Nei, M.; Maruyama, T.; Chakraborty, R. The Bottleneck Effect and Genetic Variability in Populations. Evolution 1975, 29, 1–10. [Google Scholar] [CrossRef]
- Barrandeguy, M.E.; García, M.V. The Sensitiveness of Expected Heterozygosity and Allelic Richness Estimates for Analyzing Population Genetic Diversity; IntechOpen: London, UK, 2021; ISBN 978-1-83881-097-9. [Google Scholar]
- Schilling, J.; Freier, K.P.; Hertig, E.; Scheffran, J. Climate Change, Vulnerability and Adaptation in North Africa with Focus on Morocco. Agric. Ecosyst. Environ. 2012, 156, 12–26. [Google Scholar] [CrossRef]
- del Barrio, G.; Sanjuan, M.; Hirche, A.; Yassin, M.; Ruiz, A.; Ouessar, M.; Martinez Valderrama, J.; Essifi, B.; Puigdefabregas, J. Land Degradation States and Trends in the Northwestern Maghreb Drylands, 1998–2008. Remote Sens. 2016, 8, 603. [Google Scholar] [CrossRef]
- Barakat, A.; Ouargaf, Z.; Khellouk, R.; El Jazouli, A.; Touhami, F. Land Use/Land Cover Change and Environmental Impact Assessment in Béni-Mellal District (Morocco) Using Remote Sensing and GIS. Earth Syst. Environ. 2019, 3, 113–125. [Google Scholar] [CrossRef]
- Heshmati, G.A.; Squires, V.R. (Eds.) Combating Desertification in Asia, Africa and the Middle East: Proven Practices; Springer: Dordrecht, The Netherlands, 2013; ISBN 978-94-007-6651-8. [Google Scholar]
- Le Cuziat, J.L.; Vidal, E.; Roche, P.; Lacroix, F. Human Acitivities Affect the Potential Distribution of the Houbara Bustard. Ardeola 2005, 52, 21–30. [Google Scholar]
- Frankham, R. Genetic Rescue of Small Inbred Populations: Meta-Analysis Reveals Large and Consistent Benefits of Gene Flow. Mol. Ecol. 2015, 24, 2610–2618. [Google Scholar] [CrossRef]
- Jangjoo, M.; Matter, S.F.; Roland, J.; Keyghobadi, N. Connectivity Rescues Genetic Diversity after a Demographic Bottleneck in a Butterfly Population Network. Proc. Natl. Acad. Sci. USA 2016, 113, 10914–10919. [Google Scholar] [CrossRef]
- Couvet, D. Deleterious Effects of Restricted Gene Flow in Fragmented Populations. Conserv. Biol. 2002, 16, 369–376. [Google Scholar] [CrossRef]
- Keller, I.; Largiadèr, C.R. Recent Habitat Fragmentation Caused by Major Roads Leads to Reduction of Gene Flow and Loss of Genetic Variability in Ground Beetles. Proc. R. Soc. B Biol. Sci. 2003, 270, 417–423. [Google Scholar] [CrossRef]
- Chan, J.; Pan, B.; Geng, D.; Zhang, Q.; Zhang, S.; Guo, J.; Xu, Q. Genetic Diversity and Population Structure Analysis of Three Deep-Sea Amphipod Species from Geographically Isolated Hadal Trenches in the Pacific Ocean. Biochem. Genet. 2020, 58, 157–170. [Google Scholar] [CrossRef]
- Smith, M.M.; Gilbert, J.H.; Olson, E.R.; Scribner, K.T.; Van Deelen, T.R.; Van Stappen, J.F.; Williams, B.W.; Woodford, J.E.; Pauli, J.N. A Recovery Network Leads to the Natural Recolonization of an Archipelago and a Potential Trailing Edge Refuge. Ecol. Appl. 2021, 31, e02416. [Google Scholar] [CrossRef]
- Boudjemadi, K.; Lecomte, J.; Clobert, J. Influence of Connectivity on Demography and Dispersal in Two Contrasting Habitats: An Experimental Approach. J. Anim. Ecol. 1999, 68, 1207–1224. [Google Scholar] [CrossRef]
- Hastings, A.; Botsford, L.W. Persistence of Spatial Populations Depends on Returning Home. Proc. Natl. Acad. Sci. USA 2006, 103, 6067–6072. [Google Scholar] [CrossRef] [PubMed]
| Morocco | Spain | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AA | ABM | MID | REKK | EV | IM | NP | SP | SS | ||
| N | P | 7 | 57 | 43 | 19 | 17 | 57 | 25 | 26 | 9 |
| C | 5 | 25 | 21 | 9 | 53 | 272 | 28 | 74 | 5 | |
| A | P | 43 | 109 | 97 | 72 | 74 | 95 | 75 | 81 | 64 |
| C | 43 | 94 | 78 | 63 | 87 | 113 | 76 | 87 | 54 | |
| Ho | P | 0.662 | 0.687 | 0.721 | 0.737 | 0.701 | 0.721 | 0.633 | 0.71 | 0.747 |
| C | 0.618 | 0.676 | 0.693 | 0.646 | 0.631 | 0.716 | 0.646 | 0.671 | 0.782 | |
| uHe | P | 0.699 | 0.792 | 0.785 | 0.763 | 0.733 | 0.751 | 0.735 | 0.751 | 0.75 |
| C | 0.697 | 0.779 | 0.759 | 0.746 | 0.721 | 0.748 | 0.721 | 0.738 | 0.741 | |
| AR | P | 3.563 | 5.075 | 4.95 | 4.588 | 4.547 | 4.754 | 4.515 | 4.546 | 4.666 |
| C | 3.909 | 5.085 | 4.553 | 4.596 | 4.472 | 4.652 | 4.381 | 4.421 | 4.909 | |
| HWE | P | 0.621 | 0.000 | 0.000 | 0.001 | 0.009 | 0.031 | 0.000 | 0.078 | 0.736 |
| C | 0.340 | 0.014 | 0.009 | 0.079 | 0.000 | 0.000 | 0.001 | 0.000 | 0.955 | |
| FIS | P | −0.016 | 0.123 * | 0.068 * | −0.01 | 0.009 | 0.036 | 0.127 * | 0.038 | −0.058 |
| C | 0.02 | 0.124 * | 0.07 | 0.08 | 0.108 * | 0.036 * | 0.075 | 0.09 * | −0.191 | |
| Ap a | P | 0 | 12 (11) | 7 (7.2) | 2 (2.8) | 3 (4.1) | 3 (3.2) | 0 | 1 (1.2) | 1 (1.6) |
| C | 1 (2.3) | 7 (7.4) | 1 (1.3) | 2 (3.2) | 1 (1.1) | 8 (7.2) | 1 (1.3) | 1 (1.1) | 0 | |
| Ap_losses | 0 | 7 (5) | 5(2) | 2 (2) | 3 (3) | 1 (1) | 0 | 0 (0) | 1 (0) | |
| Ap_gains | 1 | 3 (1) | 0 (0) | 2 (2) | 1 (0) | 6 (3) | 0 (0) | 1 (0) | 0 | |
| RA b | P | 0 | 45(41.3) | 34 (35) | 10 (13.9) | 14 (18.9) | 33 (34.7) | 19 (25.3) | 24 (29.6) | 0 |
| C | 0 | 29 (30.8) | 28 (35.9) | 0 | 28 (32.2) | 53 (46.9) | 22 (28.9) | 34 (39.1) | 0 | |
| RA_losses | 0 | 23 | 17 | 6 | 5 | 2 | 6 | 3 | 0 | |
| RA_gains | 0 | 4 | 14 | 0 | 8 | 4 | 8 | 9 | 0 | |
| Census size c | 162 | 1079 | 6553 | 3426 | 629 | 2612 | 216 | 300 | 67 | |
| He ≠ Heq (prob) | 0.232 | 0.027 | 0.001 | 0.002 | 0.103 | 0.034 | 0.051 | 0.002 | 0.087 | |
| FS a | FC b | ||||||
|---|---|---|---|---|---|---|---|
| NC | Ne * | Ne † | Ne | Low 95%–High 95% | Ne | Low 95%–High 95% | |
| Morocco | |||||||
| AA | 162 | 68 | 16 | 298 | 23–∞ | 687 | 411–1033 |
| ABM | 1079 | 453 | 108 | 410 | 126–∞ | 274 | 186–378 |
| MID | 6553 | 2752 | 655 | 190 | 74–2147 | 202 | 136–281 |
| REKK | 3426 | 1439 | 343 | ∞ | 81–∞ | ∞ | ∞–∞ |
| Spain | |||||||
| EV | 629 | 264 | 63 | 278 | 87–∞ | 157 | 105–219 |
| IM | 2612 | 1097 | 261 | 765 | 279–∞ | 1592 | 1074–2210 |
| NP | 216 | 91 | 22 | 486 | 110–∞ | 457 | 298–651 |
| SP | 300 | 126 | 30 | 123 | 61–278 | 118 | 78–167 |
| SS | 67 | 28 | 7 | 110 | 26–∞ | 58 | 38–82 |
| Morocco | Spain | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AA | ABM | MID | REKK | EV | IM | NP | SP | SS | ||
| AA | 0.046 | 0.075 * | 0.085 * | 0.098 * | EV | 0.014 | 0.007 * | 0.014 * | 0.012 * | 0.042 |
| ABM | 0.052 * | 0.010 | 0.022 * | 0.023 | IM | 0.014 * | 0.003 | 0.010 * | 0.008 * | 0.040 * |
| MID | 0.050 * | 0.010 * | 0.013 | 0.029 | NP | 0.016 | 0.007 | 0.010 | 0.013 * | 0.040 |
| REKK | 0.058 * | 0.011 | 0.016* | 0.020 | SP | 0.020 * | 0.009 | 0.014 | 0.012 * | 0.048 * |
| SS | 0.025 | 0.018 | 0.019 | 0.016 | 0.051 | |||||
| Source of Variation | df | Variance Components | % of Variation | p-Value |
|---|---|---|---|---|
| Morocco | ||||
| Among groups (regions) | 3 | 0.14 | 3.50 | <0.001 |
| Among temporal samples | 4 | 0.01 | 0.31 | 0.18 |
| Within regions | 178 | 3.95 | 96.19 | <0.001 |
| Spain | ||||
| Among groups (regions) | 4 | 0.02 | 0.53 | 0.047 |
| Among temporal samples | 5 | 0.02 | 0.47 | 0.040 |
| Within regions | 556 | 3.70 | 98.99 | <0.001 |
| Country | Region | AA | ABM | MID | REKK | |
|---|---|---|---|---|---|---|
| Morocco | AA | 3.1 | 2.7 | 2.3 | ||
| ABM | 4.5 | 11.3 | 10.6 | |||
| MID | 4.8 | 24.6 | 8.5 | |||
| REKK | 4.1 | 23.3 | 15.2 | |||
| EV | IM | NP | SP | SS | ||
| Spain | EV | 38.2 | 17.0 | 20.5 | 5.7 | |
| IM | 18.1 | 25.6 | 33.1 | 6.1 | ||
| NP | 15.2 | 34.0 | 18.5 | 6.0 | ||
| SP | 12.1 | 26.7 | 18.1 | 5.0 | ||
| SS | 9.7 | 13.9 | 13.2 | 15.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bustillo-de la Rosa, D.; Traba, J.; Calero-Riestra, M.; Morales, M.B.; Barrero, A.; Viñuela, J.; Pérez-Granados, C.; Gómez-Catasús, J.; Oñate, J.J.; Reverter, M.; et al. Recent Changes in Genetic Diversity, Structure, and Gene Flow in a Passerine Experiencing a Rapid Population Decline, the Dupont’s Lark (Chersophilus duponti). Diversity 2022, 14, 1120. https://doi.org/10.3390/d14121120
Bustillo-de la Rosa D, Traba J, Calero-Riestra M, Morales MB, Barrero A, Viñuela J, Pérez-Granados C, Gómez-Catasús J, Oñate JJ, Reverter M, et al. Recent Changes in Genetic Diversity, Structure, and Gene Flow in a Passerine Experiencing a Rapid Population Decline, the Dupont’s Lark (Chersophilus duponti). Diversity. 2022; 14(12):1120. https://doi.org/10.3390/d14121120
Chicago/Turabian StyleBustillo-de la Rosa, Daniel, Juan Traba, María Calero-Riestra, Manuel B. Morales, Adrián Barrero, Javier Viñuela, Cristian Pérez-Granados, Julia Gómez-Catasús, Juan J. Oñate, Margarita Reverter, and et al. 2022. "Recent Changes in Genetic Diversity, Structure, and Gene Flow in a Passerine Experiencing a Rapid Population Decline, the Dupont’s Lark (Chersophilus duponti)" Diversity 14, no. 12: 1120. https://doi.org/10.3390/d14121120
APA StyleBustillo-de la Rosa, D., Traba, J., Calero-Riestra, M., Morales, M. B., Barrero, A., Viñuela, J., Pérez-Granados, C., Gómez-Catasús, J., Oñate, J. J., Reverter, M., Hervás, I., Hernández Justribó, J., García de la Morena, E. L., López-Iborra, G. M., & García, J. T. (2022). Recent Changes in Genetic Diversity, Structure, and Gene Flow in a Passerine Experiencing a Rapid Population Decline, the Dupont’s Lark (Chersophilus duponti). Diversity, 14(12), 1120. https://doi.org/10.3390/d14121120








