Cryptic Lineage and Genetic Structure of Acanthopagrus pacificus Populations in a Natural World Heritage Site Revealed by Population Genetic Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
2.2. Amplification and Sequencing
2.3. Genetic Diversity Analysis
2.4. Population Genetic Structure
2.5. Pattern of Historical Demography
3. Results
3.1. DNA Variation and Genetic Variability
3.2. Inferred Population Demographic History
3.3. Population Genetic Structure
4. Discussion
4.1. Genetic Diversity
4.2. Population Demography and Pleistocene Glaciations Impacts
4.3. Population Genetic Structure
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Smouse, P.E.; Peakall, R. Spatial autocorrelation analysis of individual multiallele and multilocus genetic structure. Heredity 1999, 82, 561–573. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Shan, B.; Song, N.; Gao, T. Genetic diversity and population structure of Acanthopagrus schlegelii inferred from mtDNA sequences. Reg. Stud. Mar. Sci. 2021, 41, 101532. [Google Scholar] [CrossRef]
- Conover, D.O.; Clarke, L.M.; Munich, S.B.; Wagner, G.N. Spatial and temporal scales of adaptive divergence in marine fishes and the implications for conservation. J. Fish Biol. 2006, 69, 21–47. [Google Scholar] [CrossRef]
- Cano, J.M.; Shikano, T.; Kuparinen, A.; Merilä, J. Genetic differentiation, effective population size and gene flow in marine fishes: Implications for stock management. J. Integr. Field Biol. 2008, 5, 1–10. [Google Scholar]
- Zhang, B.-D.; Li, Y.-L.; Xue, D.-X.; Liu, J.-X. Population genomics reveals shallow genetic structure in a connected and ecologically important fish from the Northwestern Pacific Ocean. Front. Mar. Sci. 2020, 5, 374. [Google Scholar] [CrossRef]
- Treml, E.A.; Halpin, P.N.; Urban, D.L.; Pratson, L.F. Modeling population connectivity by ocean currents, a graph-theoretic approach for marine conservation. Landsc. Ecol. 2008, 23, 19–36. [Google Scholar] [CrossRef]
- Priest, M.A.; Halford, A.R.; Mcllwain, J.L. Evidence of stable genetic structure across a remote island archipelago through self-recruitment in a widely dispersed coral reef fish. Ecol. Evol. 2012, 2, 3195–3213. [Google Scholar] [CrossRef]
- Iwamoto, K.; Chang, C.-W.; Imai, H. Spatial genetic structuring and demographic history of the little spinefoot Siganus spinus in the Western Pacific. Biogeography 2020, 22, 26–34. [Google Scholar]
- Salmenkova, E.A. New view on the population genetic structure of Marine Fish. Russ. J. Genet. 2011, 47, 1279–1287. [Google Scholar] [CrossRef]
- Hess, J.E.; Campbell, N.R.; Close, D.A.; Docker, M.F.; Narum, S.R. Population genomics of Pacific lamprey: Adaptive variation in a highly dispersive species. Mol. Ecol. 2013, 22, 2898–2916. [Google Scholar] [CrossRef]
- Guo, B.; Li, Z.; Merilä, J. Population genomic evidence for adaptive differentiation in the Baltic Sea herring. Mol. Ecol. 2016, 25, 2833–2852. [Google Scholar] [CrossRef] [PubMed]
- Rohfritsch, A.; Borsa, P. Genetic structure of Indian scad mackerel Decapterus russelli: Pleistocene vicariance and secondary contact in the central Indo-West Pacific seas. Heredity 2005, 95, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Jones, G.; Milicich, M.; Emslie, M.J.; Lunow, C. Self-recruitment in a coral reef fish population. Nature 1999, 402, 802–804. [Google Scholar] [CrossRef]
- Swearer, S.E.; Caselle, J.E.; Lea, D.W.; Warner, R.R. Larval retention and recruitment in an island population of a coral-reef fish. Nature 1999, 402, 799–802. [Google Scholar] [CrossRef]
- Barber, P.H.; Palumbi, S.R.; Erdmann, M.V.; Moosa, M.K. Sharp genetic breaks among populations of Haptosquilla pulchella (Stomatopoda) indicates limits to larval transport: Patterns, causes, and consequences. Mol. Ecol. 2002, 11, 659–674. [Google Scholar] [CrossRef]
- Hellberg, M.E.; Burton, R.S.; Neigel, J.E.; Palumbi, S.R. Genetic assessment of connectivity among marine populations. Bull. Mar. Sci. 2002, 70, 273–290. [Google Scholar]
- Taylor, M.S.; Hellberg, M.E. Genetic evidence for local retention of pelagic larvae in a Caribbean reef fish. Science 2003, 299, 107–109. [Google Scholar] [CrossRef]
- Cowen, R.K.; Lwiza, K.M.M.; Sponaugle, S.; Paris, C.B.; Olsen, D.B. Connectivity of marine populations: Open or closed? Science 2000, 287, 857–859. [Google Scholar] [CrossRef]
- Strathmann, R.R.; Hughes, T.R.; Kuris, A.M.; Lindeman, K.C.; Morgan, S.G.; Pandolfi, J.M.; Warner, R.R. Evolution of local recruitment and its consequences for marine populations. Bull. Mar. Sci. 2002, 70, 377–396. [Google Scholar]
- Iwamoto, K.; Chang, C.W.; Takemura, A.; Imai, H. Genetically structured population and demographic history of the goldlined spinefoot Siganus guttatus in the northwestern Pacific. Fish. Sci. 2012, 78, 249–257. [Google Scholar] [CrossRef]
- Imai, H.; Hanamura, Y.; Cheng, J.H. Genetic and morphological differentiation in the Sakura shrimp (Sergia lucens) between Japanese and Taiwanese populations. Contrib. Zool. 2013, 82, 123–130. [Google Scholar] [CrossRef]
- Iwamoto, K.; Abdullah, M.F.; Chang, C.W.; Yoshino, T.; Imai, H. Genetic isolation of the mottled spinefoot Siganus fuscescens Ryukyu Archipelago population. Biogeography 2015, 17, 61–85. [Google Scholar]
- Kuriiwa, K.; Chiba, S.N.; Motomura, H.; Matsuura, K. Phylogeography of Blacktip Grouper, Epinephelus fasciatus (Perciformes: Serranidae), and influence of the Kuroshio Current on cryptic lineages and genetic population structure. Ichthyol. Res. 2014, 61, 361–374. [Google Scholar] [CrossRef]
- Tokuyama, T.; Shy, J.Y.; Lin, H.C.; Henmi, Y.; Mather, P.; Hughes, J.; Tsuchiya, M.; Imai, H. Genetic population structure of the fiddler crab Austruca lactea (De Haan, 1835) based on mitochondrial DNA control region sequences. Crustacean Res. 2020, 49, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Teske, P.R.; Sandoval-Castillo, J.; van Sebille, E.; Waters, J.; Beheregaray, L.B. On-shelf larval retention limits population connectivity in a coastal broadcast spawner. Mar. Ecol. Prog. Ser. 2015, 532, 1–12. [Google Scholar] [CrossRef][Green Version]
- Hellberg, M.E. Genetic approaches to understanding marine metapopulation dynamics. In Marine Metapopulations; Kritzer, J.P., Sale, P.F., Eds.; Elsevier Academic Press: Amsterdam, The Netherlands, 2006; pp. 431–449. [Google Scholar]
- Kume, M.; Yoshino, T. Acanthopagrus chinshira, a new sparid fish (Perciformes: Sparidae) from the East Asia. Bull. Natl. Mus. Nat. Sci. Ser. A 2008, 2, 47–57. [Google Scholar]
- Iwatsuki, Y.; Kume, M.; Yoshino, T. A new species, Acanthopagrus pacificus from the Western Pacific (Pisces, Sparidae). Bull. Natl. Mus. Nat. Sci. Ser. A 2010, 36, 115–130. [Google Scholar]
- Allen, G.R.; Midgley, S.H.; Allen, M. Field Guide to the Freshwater Fishes of Australia; Western Australian Museum: Perth, Australia, 2002; 394p.
- IUCN. IUCN World Heritage Evaluations 2020–2021. 2021. Available online: https://www.iucn.org/theme/world-heritage/about/world-heritage-committee/iucn-44whc/nominations-2020-and-2021 (accessed on 8 September 2021).
- Imai, H.; Hamasaki, K.; Cheng, J.H.; Numachi, K. Identification of four mud crab species (genus Scylla) using ITS-1 and 16s rDNA markers. Aquat. Living Resour. 2004, 17, 31–34. [Google Scholar] [CrossRef]
- Tomita, S.; Matsuzaki, S.; Oka, S.; Toda, M.; Imai, H. High levels of genetic diversity and gene flow in the endangered goby Hypseleotris cyprinoides on Okinawajima island, Ishigakijima island and Luzon island. Jpn. J. Ichthyol. 2016, 63, 27–32. [Google Scholar]
- Palumbi, S.; Martin, A.; Romano, S.; MacMillan, W.O.; Stice, L.; Grabowski, G. The Simple Fool’s Guide to PCR, Version 2.0; Department of Zoology and Kewalo Marine Laboratory, University of Hawaii: Honolulu, HI, USA, 1991. [Google Scholar]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The Clustal_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger data sets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Phylogenet. Evol. 1993, 10, 512–526. [Google Scholar]
- Tajima, F. Evolutionary relationship of DNA sequences in finite populations. Genetics 1983, 105, 437–460. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Leigh, J.W.; Bryant, D. POPART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Hsu, T.H.; Guillén Madrid, A.G.; Burridge, C.P.; Cheng, H.Y.; Gao, J.C. Resolution of the Acanthopagrus black seabream complex based on mitochondrial and amplified fragment-length polymorphism analysis. J. Fish Biol. 2011, 79, 1182–1192. [Google Scholar] [CrossRef]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Fu, Y.X. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 1997, 147, 915–925. [Google Scholar] [CrossRef]
- Harpending, H.C. Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Hum. Biol. 1994, 66, 591–600. [Google Scholar] [PubMed]
- Harpending, H.C.; Sherry, S.T.; Rogers, A.R.; Stoneking, M. Genetic structure of ancient human populations. Curr. Anthropol. 1993, 34, 483–496. [Google Scholar] [CrossRef]
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007, 7, 214. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.; Rambaut, A.; Shapiro, B.; Pybus, O.G. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol. Biol. Evol. 2005, 22, 1185–1192. [Google Scholar] [CrossRef] [PubMed]
- Donaldson, K.A.; Wilson, R.R., Jr. Amphi-panamic geminates of snook (Percoidei: Centropomidae) provide a calibration of the divergence rate in the mitochondrial DNA control region of fishes. Mol. Phylogenet. Evol. 1999, 13, 208–213. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, E.B.; Umino, T.; Nagasawa, K. Stock enhancement programme for black sea bream, Acanthopagrus schlegelii (Bleeker), in Hiroshima Bay, Japan: A review. Aquaculture 2008, 39, 1307–1315. [Google Scholar] [CrossRef]
- Rogers, A.R.; Harpending, H. Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 1992, 9, 552–569. [Google Scholar]
- Rambaut, A.; Drummond, A.J. Tracer v1.5. 2009. Available online: http://beast.bio.edu.ac.UK/Tracer (accessed on 13 August 2021).
- Xia, J.-H.; Huang, J.-H.; Gong, J.-B.; Jiang, S.-G. Significant population genetic structure of yellowfin seabream Acanthopagrus latus in China. J. Fish Biol. 2008, 73, 1979–1992. [Google Scholar] [CrossRef]
- Grant, W.S.; Bowen, B.W. Shallow population histories in deep evolutionary lineages of marine fishes: Insights from sardines and anchovies and lessons for conservation. J. Hered. 1998, 89, 415–426. [Google Scholar] [CrossRef]
- Rogers, A.R. Genetic evidence for a Pleistocene population explosion. Evolution 1995, 49, 608–615. [Google Scholar]
- Fontanella, F.M.; Feldman, C.R.; Siddall, M.E.; Burbrink, F.T. Phylogeography of Diadophis punctatus: Extensive lineage diversity and repeated patterns of historical demography in a trans-contintal snake. Mol. Phylogenet. Evol. 2008, 46, 1049–1070. [Google Scholar] [CrossRef] [PubMed]
- Pillans, B.; Chappell, J.; Naish, T.R. A review of the Milankovitch climatic beat: Template for Plio-Pleistocene sea-level changes and sequence stratigraphy. Sediment Geol. 1998, 122, 5–21. [Google Scholar] [CrossRef]
- Inoue, K.; Monroe, E.M.; Elderkin, C.L.; Berg, D.J. Phylogeographic and population genetic analyses reveal Pleistocene isolation followed by high gene flow in a wide ranging, but endangered, freshwater mussel. Heredity 2014, 112, 282–290. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, G.M. The genetic legacy of the Quaternary ice ages. Nature 2000, 405, 907–913. [Google Scholar] [CrossRef] [PubMed]
- Lambeck, K.; Esat, T.M.; Potter, E.K. Links between climate and sea levels for the past three million years. Nature 2002, 419, 199–206. [Google Scholar] [CrossRef]
- Bernatchez, L. Mitochondrial DNA analysis confirms the existence of two glacial races of rainbow smelt Osmerus mordax and their reproductive isolation in the St Lawrence River estuary (Quebec, Canada). Mol. Ecol. 1997, 6, 73–83. [Google Scholar] [CrossRef]
- Liu, J.X.; Gao, T.X.; Yokogawa, K.; Zhang, Y.P. Differential population structuring and demographic history of two closely related fish species, Japanese sea bass (Lateolabrax japonicus) and spotted sea bass (Lateolabrax maculates) in Northwestern Pacific. Mol. Phylogenet. Evol. 2006, 39, 799–811. [Google Scholar] [CrossRef]
- Shui, B.N.; Han, Z.Q.; Gao, T.X.; Miao, Z.Q.; Yanagimoto, T. Mitochondrial DNA variation in the East China Sea and Yellow Sea populations of Japanese Spanish mackerel Scomberomorus niphonius. Fish Sci. 2009, 75, 593–600. [Google Scholar] [CrossRef]
- Wee, A.K.S.; Takayama, K.; Asakawa, T.; Thompson, B.; Onrizal, S.S.; Tung, N.X.; Nazre, M.; Soe, K.K.; Tan, H.T.W.; Watano, Y.; et al. Oceanic currents, not land masses, maintain the genetic structure of the mangrove Rhizophora mucronata Lam. (Rhizophoraceae) in Southeast Asia. J. Biogeogr. 2014, 41, 954–964. [Google Scholar] [CrossRef]
- Kojima, S.; Kamimura, S.; Iijima, A.; Kimura, T.; Kurozumi, T.; Furota, T. Molecular phylogeny and population structure of tideland snails in the genus Cerithidea around Japan. Mar. Biol. 2006, 149, 525–535. [Google Scholar] [CrossRef]
- Kobayashi, G. Small-scale population genetic structure of the sand bubbler crab Scopimera ryukyuensis in the Ryukyu Islands, Japan. Mol. Biol. Rep. 2020, 47, 2619–2626. [Google Scholar] [CrossRef] [PubMed]
- Swearer, S.E.; Shima, J.S.; Hellberg, M.E.; Thorrold, S.R.; Jones, G.P.; Robertson, D.R.; Morgan, S.G.; Selkoe, K.A.; Ruiz, G.M.; Warner, R.R. Evidence of self-recruitment in demersal marine populations. Bull. Mar. Sci. 2002, 70, 251–271. [Google Scholar]
- Thorrold, S.R.; Jones, G.P.; Hellberg, M.E.; Burton, R.S.; Swearer, S.E.; Neigel, J.E.; Morgan, S.G.; Warner, R.R. Quantifying larval retention and connectivity in marine populations with artificial and natural markers. Bull. Mar. Sci. 2002, 70, 291–308. [Google Scholar]
- Rocha, L.A.; Robertson, D.R.; Roman, J.; Bowen, B.W. Ecological speciation in tropical reef fishes. Proc. R. Soc. Lond. B 2005, 272, 573–579. [Google Scholar] [CrossRef]
- Macpherson, E.; Raventos, N. Relationships between pelagic larval duration and geographic distribution in Mediterranean littoral fishes. Mar. Ecol. Prog. Ser. 2006, 327, 257–265. [Google Scholar] [CrossRef]
- Carreras-Carbonell, J.; Macpherson, E.; Pascual, M. High self-recruitment levels in a Mediterranean littoral fish population revealed by microsatellite markers. Mar. Biol. 2007, 151, 719–727. [Google Scholar] [CrossRef]
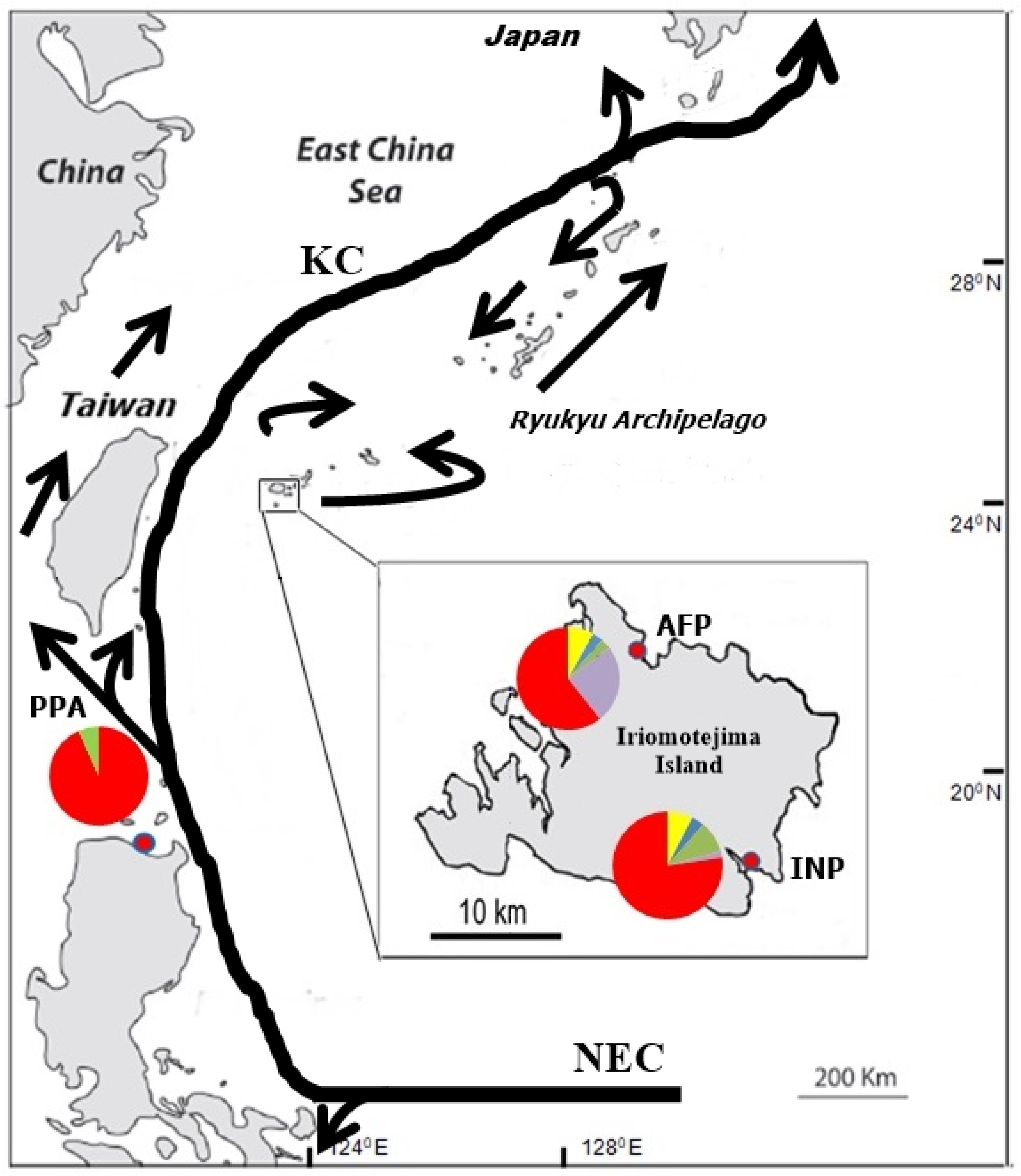




| Localities | Code | Geographic Coordinate | N | Year | Nh | h (SD) | k (SD) | π (SD) in % |
|---|---|---|---|---|---|---|---|---|
| Funaura Bay, Iriomotejima Island, Japan | AFP | 24°24′17.6″ N 123°48′40.3″ E | 61 | 2020 | 21 | 0.915 (0.229) | 5.608 (2.729) | 0.80(0.40) |
| Nakamagawa River, Iriomotejima Island, Japan | INP | 24°17′31.6″ N 123°52′00.6″ E | 53 | 2020 | 20 | 0.942(0.014) | 11.022 (5.089) | 1.60(0.80) |
| Aparri fish market, Luzon Island, Philippines | PPA | 18°35′08.26″ N 121°63′78.62″ E | 45 | 2012 | 36 | 0.989 (0.007) | 17.720 (8.017) | 2.60(1.30) |
| Locality | Demographic Parameters | Test of Goodness of Fit | Neutrality Test | |||||
|---|---|---|---|---|---|---|---|---|
| τ | Obs. | θ0 | θ1 | SSD (p) | Ragged (p) | Tajima’s D (p) | Fu’s Fs (p) | |
| Funaura | 3.728 | 5.609 | 0.000 | 14.087 | 0.006 (0.250) | 0.017 (0.740) | −1.346 (0.063) | −3.805 (0.132) |
| Nakamagawa | 0.000 | 11.022 | 10.800 | 6887.570 | 0.0237 (0.980) | 0.007 (1.000) | 0.647 (0.790) | 0.134 (0.590) |
| Philippines | 8.303 | 17.720 | 15.089 | 174.061 | 0.0054 (0.460) | 0.004 (0.590) | −0.119 (0.512) | −11.169 (0.004) |
| Locality | AFP | INP | PPA |
|---|---|---|---|
| AFP | 0.000 | 0.000 | |
| INP | 0.135 | 0.000 | |
| PPA | 0.237 | 0.089 |
| Comparisons | Source of Variance | df | % var. | Fixation Indices |
|---|---|---|---|---|
| All populations | Among populations | 2 | 15.46 | 0.155 |
| Within populations | 156 | 84.54 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Islam, M.R.-U.; Tachihara, K.; Imai, H. Cryptic Lineage and Genetic Structure of Acanthopagrus pacificus Populations in a Natural World Heritage Site Revealed by Population Genetic Analysis. Diversity 2022, 14, 1117. https://doi.org/10.3390/d14121117
Islam MR-U, Tachihara K, Imai H. Cryptic Lineage and Genetic Structure of Acanthopagrus pacificus Populations in a Natural World Heritage Site Revealed by Population Genetic Analysis. Diversity. 2022; 14(12):1117. https://doi.org/10.3390/d14121117
Chicago/Turabian StyleIslam, Md Rakeb-Ul, Katsunori Tachihara, and Hideyuki Imai. 2022. "Cryptic Lineage and Genetic Structure of Acanthopagrus pacificus Populations in a Natural World Heritage Site Revealed by Population Genetic Analysis" Diversity 14, no. 12: 1117. https://doi.org/10.3390/d14121117
APA StyleIslam, M. R.-U., Tachihara, K., & Imai, H. (2022). Cryptic Lineage and Genetic Structure of Acanthopagrus pacificus Populations in a Natural World Heritage Site Revealed by Population Genetic Analysis. Diversity, 14(12), 1117. https://doi.org/10.3390/d14121117







