DNA Barcoding of Lepidoptera Species from the Maltese Islands: New and Additional Records, with an Insight into Endemic Diversity
Abstract
1. Introduction
2. Materials and Methods
2.1. Specimen Collection and Morphological Identification
2.2. DNA Extraction, Amplification, and Sequencing
2.3. Molecular Identification
3. Results
3.1. Taxonomic Coverage and General Overview
3.2. Endemic Diversity
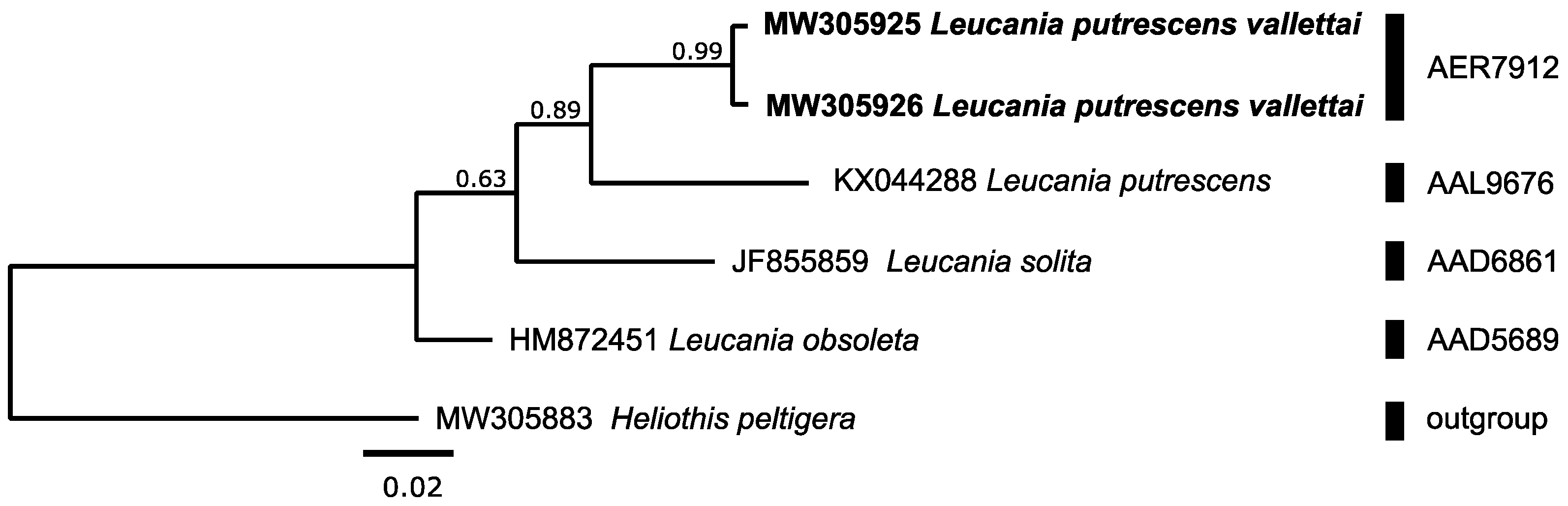
3.2.1. Noctuidae: Leucania putrescens vallettai Boursin, 1952
3.2.2. Noctuidae: Nyctobrya segunai Fibiger, Steiner, & Ronkay, 2009
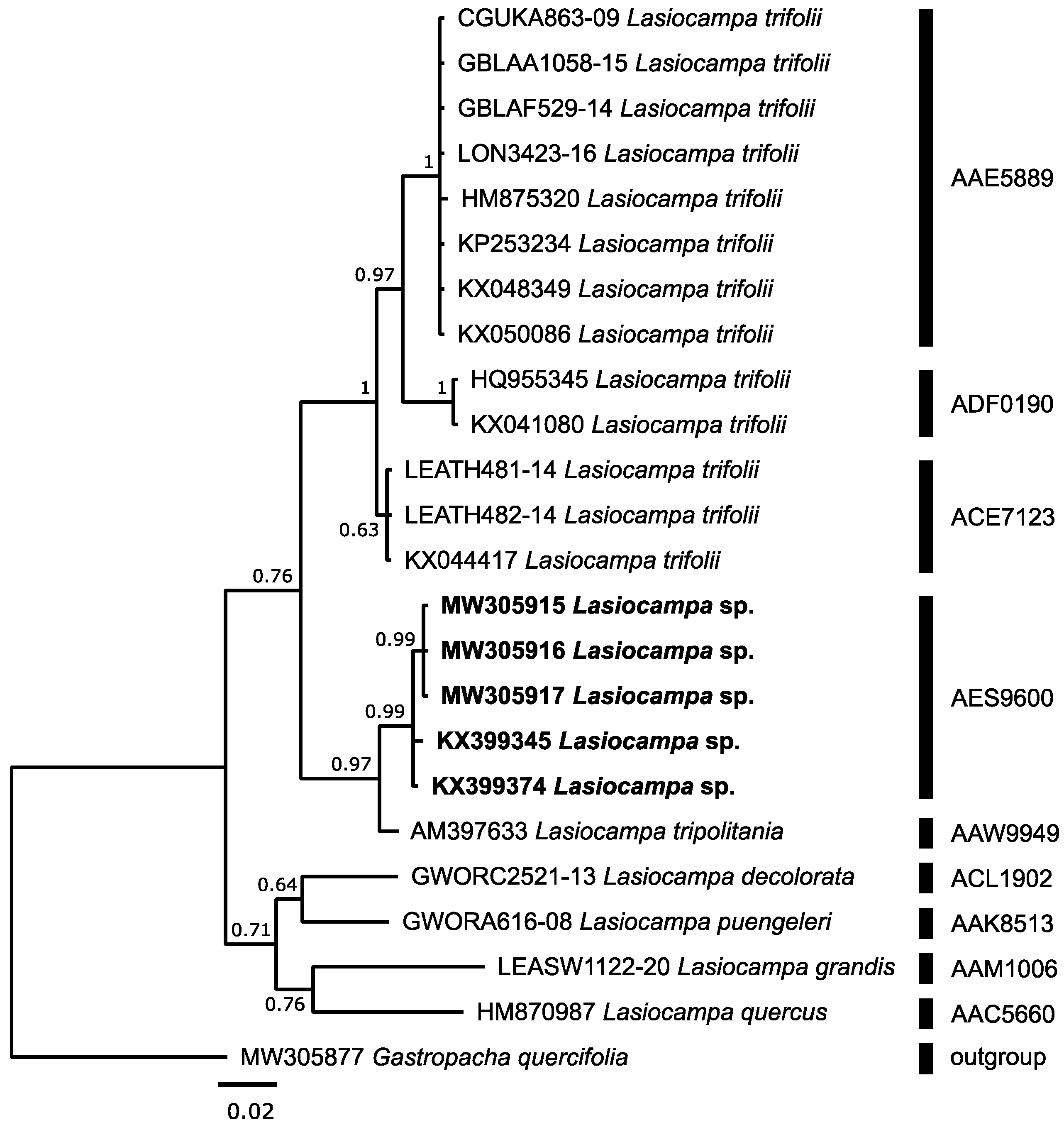
3.2.3. Lasiocampa sp.
3.3. New Additions to the Entomofauna of Malta
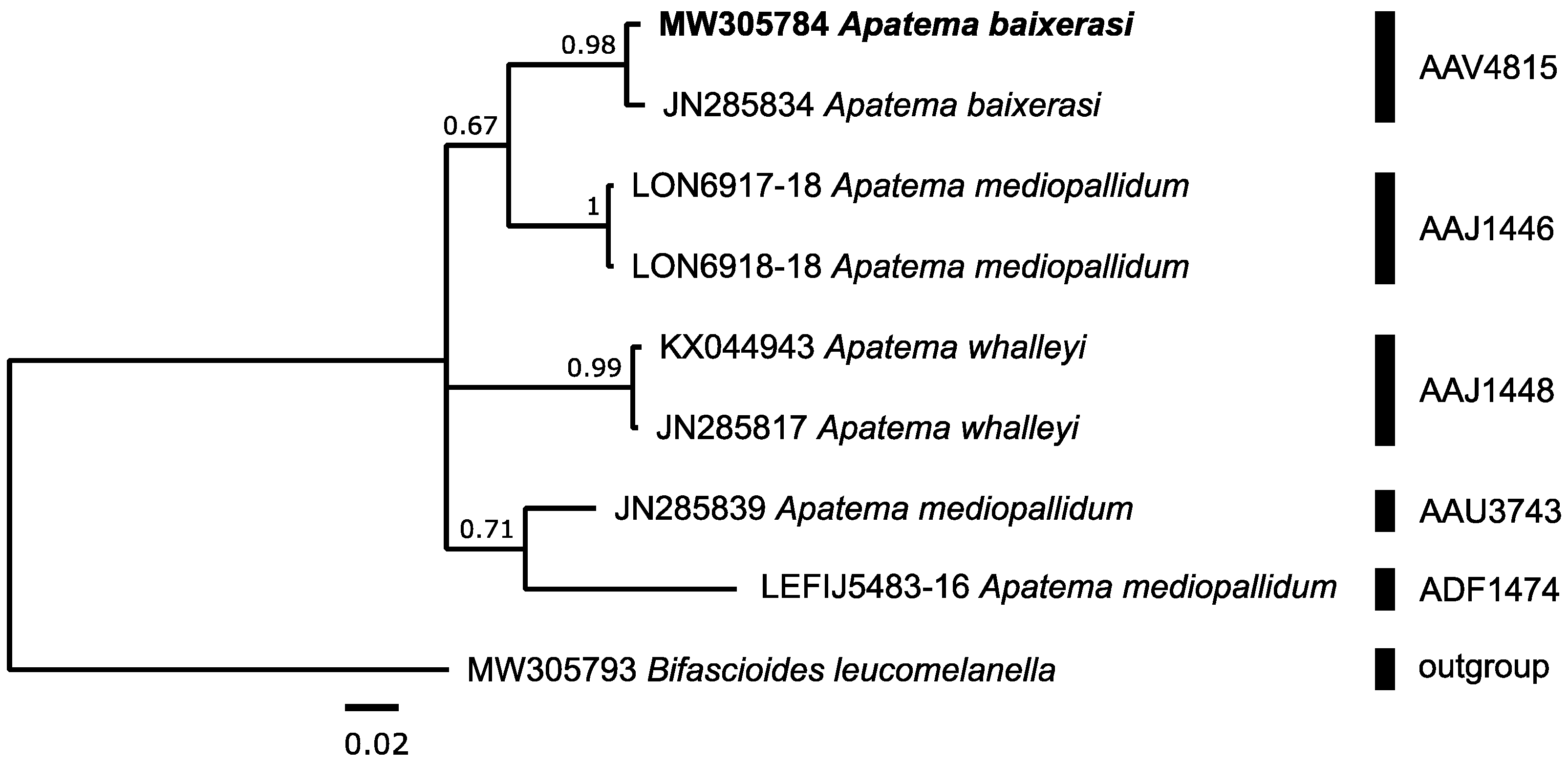
3.3.1. Autostichidae: Apatema baixerasi Vives, 2001
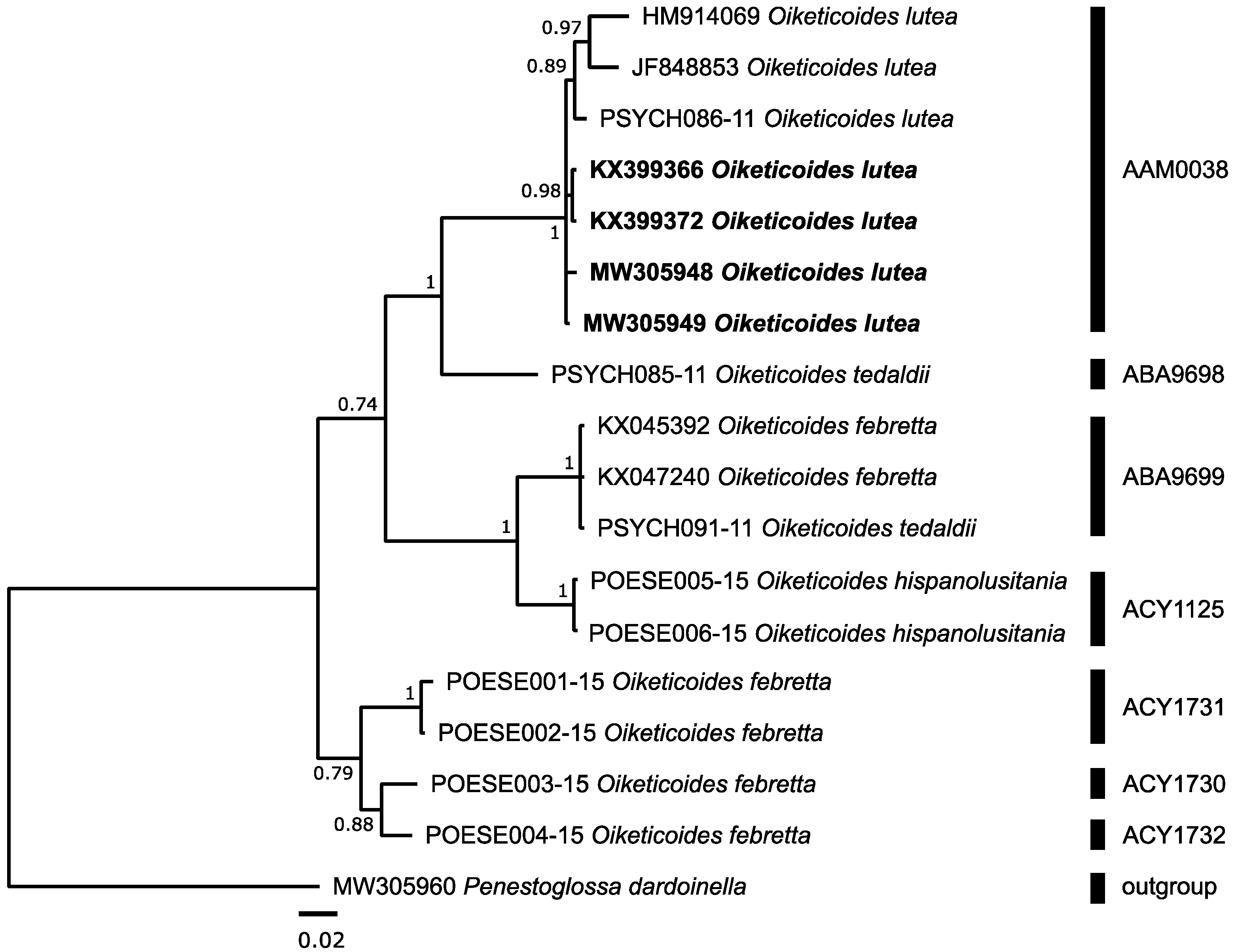
3.3.2. Psychidae: Oiketicoides lutea (Staudinger, 1870)
3.3.3. Pyralidae: Bostra dipectinialis Hampson, 1906
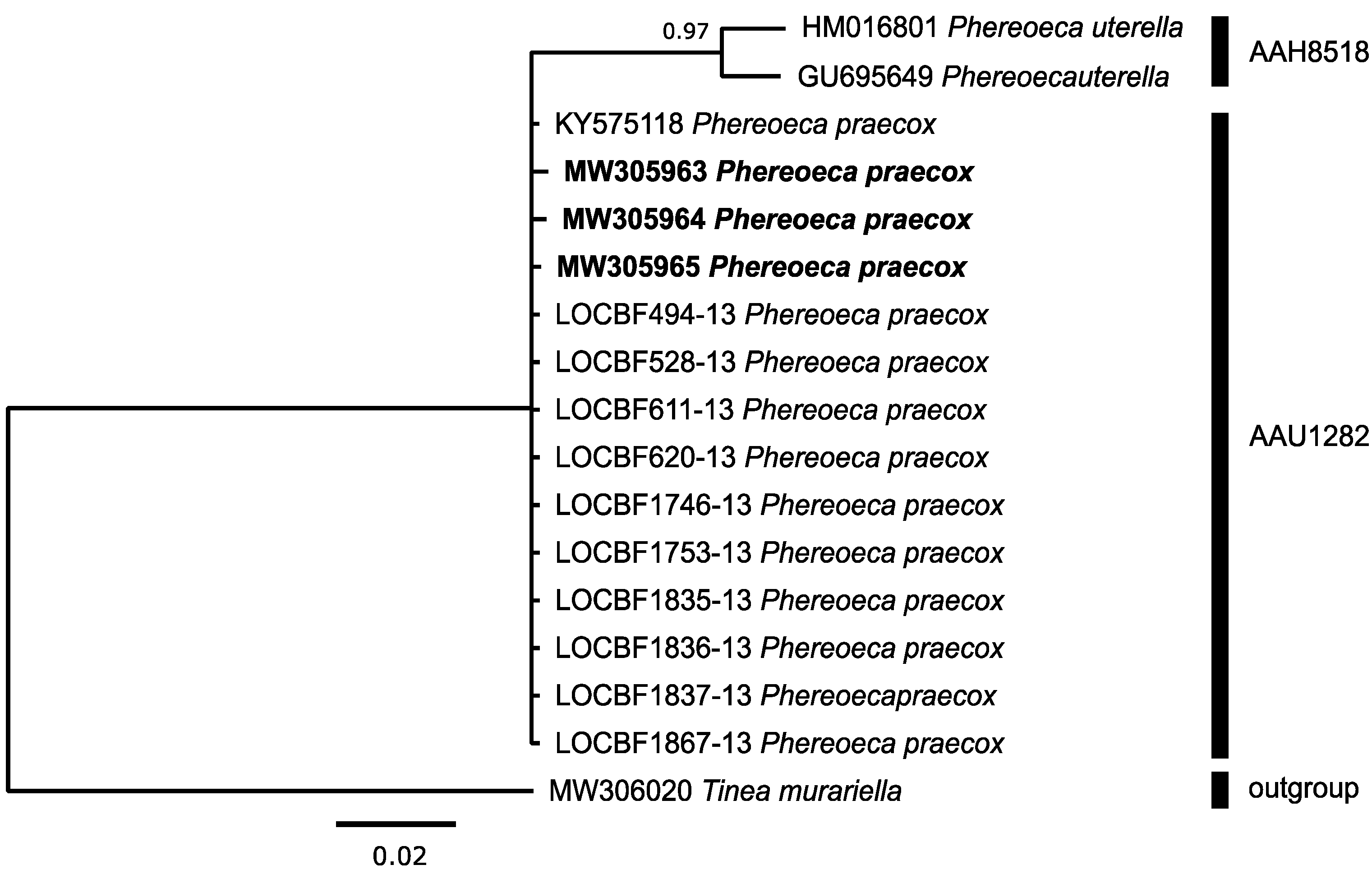
3.3.4. Tineidae: Phereoeca praecox (Gozmany & Vari, 1973)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jinbo, U.; Kato, T.; Ito, M. Current Progress in DNA Barcoding and Future Implications for Entomology. Entomol. Sci. 2011, 14, 107–124. [Google Scholar] [CrossRef]
- Packer, L.; Gibbs, J.; Sheffield, C.; Hanner, R. DNA Barcoding and the Mediocrity of Morphology. Mol. Ecol. Resour. 2009, 9, 42–50. [Google Scholar] [CrossRef]
- Köhler, F. From DNA Taxonomy to Barcoding—How a Vague Idea Evolved into a Biosystematic Tool. Zoosyst. Evol. 2007, 83, 44–51. [Google Scholar] [CrossRef]
- Pires, A.C.; Marinoni, L. DNA Barcoding and Traditional Taxonomy Unified through Integrative Taxonomy: A View That Challenges the Debate Questioning Both Methodologies. Biota Neotrop. 2010, 10, 339–346. [Google Scholar] [CrossRef]
- Schmidt, S.; Taeger, A.; Moriniere, J.; Liston, A.; Blank, S.M.; Kramp, K.; Kraus, M.; Schmidt, O.; Heibo, E.; Prous, M.; et al. Identification of Sawflies and Horntails (Hymenoptera, ‘Symphyta’) through DNA Barcodes: Successes and Caveats. Mol. Ecol. Resour. 2017, 17, 670–685. [Google Scholar] [CrossRef] [PubMed]
- Bickford, D.; Lohman, D.J.; Sodhi, N.S.; Ng, P.K.L.; Meier, R.; Winker, K.; Ingram, K.K.; Das, I. Cryptic Species as a Window on Diversity and Conservation. Trends Ecol. Evol. 2007, 22, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; DeWaard, J.R. Biological Identifications through DNA Barcodes. Proc. R. Soc. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Hajibabaei, M.; Janzen, D.H.; Burns, J.M.; Hallwachs, W.; Hebert, P.D.N. DNA Barcodes Distinguish Species of Tropical Lepidoptera. Proc. Natl. Acad. Sci. USA 2006, 5, 6103. [Google Scholar] [CrossRef]
- Wilson, J.; Sing, K.; Sofian-Azirun, M. Building a DNA Barcode Reference Library for the True Butterflies (Lepidoptera) of Peninsula Malaysia: What about the Subspecies? PLoS ONE 2013, 8, e79969. [Google Scholar] [CrossRef]
- Zhang, J.; Kapli, P.; Pavlidis, P.; Stamatakis, A. A General Species Delimitation Method with Applications to Phylogenetic Placements. Bioinformatics 2013, 29, 2869–2876. [Google Scholar] [CrossRef]
- Kekkonen, M.; Hebert, P.D.N. DNA Barcode-Based Delineation of Putative Species: Efficient Start for Taxonomic Workflows. Mol. Ecol. Resour. 2014, 14, 706–715. [Google Scholar] [CrossRef] [PubMed]
- Park, D.S.; Suh, S.J.; Oh, H.W.; Hebert, P.D.N. Recovery of the Mitochondrial COI Barcode Region in Diverse Hexapoda through tRNA-Based Primers. BMC Genom. 2010, 11, 423. [Google Scholar] [CrossRef] [PubMed]
- Hansen, J.A.; Klingeman, W.E.; Moulton, J.K.; Oliver, J.B.; Windham, M.T.; Zhang, A.; Trigiano, R.N. Molecular Identification of Synanthedonini Members (Lepidoptera: Sesiidae) Using Cytochrome Oxidase I. Ann. Entomol. Soc. Am. 2012, 105, 520–528. [Google Scholar] [CrossRef]
- Hausmann, A.; Charles, H.; Godfray, J.; Huemer, P.; Mutanen, M.; Rougerie, R.; Van Nieukerken, E.J.; Ratnasingham, S.; Hebert, P.D.N. Genetic Patterns in European Geometrid Moths Revealed by the Barcode Index Number (BIN) System. PLoS ONE 2013, 8, e84518. [Google Scholar] [CrossRef]
- Raupach, M.J.; Astrin, J.J.; Hannig, K.; Peters, M.K.; Stoeckle, M.Y.; Wägele, J.W. Molecular Species Identification of Central European Ground Beetles (Coleoptera: Carabidae) Using Nuclear RDNA Expansion Segments and DNA Barcodes. Front. Zool. 2010, 7, 26. [Google Scholar] [CrossRef]
- Hausmann, A.; Haszprunar, G.; Hebert, P.D.N. DNA Barcoding the Geometrid Fauna of Bavaria (Lepidoptera): Successes, Surprises, and Questions. PLoS ONE 2011, 6, e17134. [Google Scholar] [CrossRef]
- Gossner, M.; Hausmann, A. DNA Barcoding Enables the Identification of Caterpillars Feeding on Native and Alien Oak. Mitt. Münch. Ent. Ges. 2009, 99, 135–140. [Google Scholar]
- Vella, A.; Vella, N.; Schembri, S. A Molecular Approach towards Taxonomic Identification of Elasmobranch Species from Maltese Fisheries Landings. Mar. Genom. 2017, 36, 17–23. [Google Scholar] [CrossRef]
- Beng, K.C.; Tomlinson, K.W.; Shen, X.H.; Surget-Groba, Y.; Hughes, A.C.; Corlett, R.T.; Slik, J.W.F. The Utility of DNA Metabarcoding for Studying the Response of Arthropod Diversity and Composition to Land-Use Change in the Tropics. Sci. Rep. 2016, 6, 24965. [Google Scholar] [CrossRef]
- Ball, S.L.; Armstrong, K.F. DNA Barcodes for Insect Pest Identification: A Test Case with Tussock Moths (Lepidoptera: Lymantriidae). Can. J. For. Res. 2006, 36, 337–350. [Google Scholar] [CrossRef]
- Madden, M.J.L.; Young, R.G.; Brown, J.W.; Miller, S.E.; Frewin, A.J.; Hanner, R.H. Using DNA Barcoding to Improve Invasive Pest Identification at U.S. Ports-of-Entry. PLoS ONE 2019, 14, e0222291. [Google Scholar] [CrossRef]
- Wu, Y.; Trepanowski, N.F.; Molongoski, J.J.; Reagel, P.F.; Lingafelter, S.W.; Nadel, H.; Myers, S.W.; Ray, A.M. Identification of Wood-Boring Beetles (Cerambycidae and Buprestidae) Intercepted in Tradeassociated Solid Wood Packaging Material Using DNA Barcoding and Morphology. Sci. Rep. 2017, 7, 40316. [Google Scholar] [CrossRef]
- Pons, J.; Barraclough, T.G.; Gomez-zurita, J.; Cardoso, A.; Duran, D.P.; Hazell, S.; Kamoun, S.; Sumlin, W.; Vogler, A.P. Sequence-Based Species Delimitation for the DNA Taxonomy of Undescribed Insects. Syst. Biol. 2006, 55, 595–609. [Google Scholar] [CrossRef]
- Derkarabetian, S.; Starrett, J.; Hedin, M. Using Natural History to Guide Supervised Machine Learning for Cryptic Species Delimitation with Genetic Data. Front. Zool. 2022, 22, 8. [Google Scholar] [CrossRef]
- Vitecek, S.; Ku, M.; Previ, A.; Ivana, Ž.; Stojanovi, K.; Keresztes, L.; Bálint, M.; Hoppeler, F.; Waringer, J.; Graf, W.; et al. Integrative Taxonomy by Molecular Species Delimitation: Multi-Locus Data Corroborate a New Species of Balkan Drusinae Micro-Endemics. BMC Ecol. Evol. 2017, 17, 129. [Google Scholar] [CrossRef]
- Vishnevskaya, M.S.; Saifitdinova, A.F.; Lukhtanov, V.A. Karyosystematics and Molecular Taxonomy of the Anomalous Blue Butterflies (Lepidoptera, Lycaenidae) from the Balkan Peninsula. Comp. Cytogenet. 2016, 10, 1–85. [Google Scholar] [CrossRef]
- Marconi, M.; Modesti, A.; Alvarez, L.P.; Ogoña, P.V.; Vecco-giove, C.D.; Luna, J.O.; Giulio, A. Di DNA Barcoding of Stingless Bees (Hymenoptera: Meliponini) in Northern Peruvian Forests: A Plea for Integrative Taxonomy. Diversity 2022, 14, 632. [Google Scholar] [CrossRef]
- Sigut, M.; Kostovćik, M.; Sigutova, H.; Hulcr, J.; Drozd, P.; Hrcek, J. Performance of DNA Metabarcoding, Standard Barcoding, and Morphological Approach in the Identification of Hostparasitoid Interactions. PLoS ONE 2017, 12, e0187803. [Google Scholar] [CrossRef] [PubMed]
- Hausmann, A.; Miller, S.E.; Holloway, J.D.; Dewaard, J.R.; Pollock, D.; Prosser, S.W.J.; Hebert, P.D.N. Calibrating the Taxonomy of a Megadiverse Insect Family: 3000 DNA Barcodes from Geometrid Type Specimens (Lepidoptera, Geometridae). Genome 2016, 59, 671–684. [Google Scholar] [CrossRef] [PubMed]
- Grant, D.M.; Brodnicke, O.B.; Evankow, A.M.; Ferreira, A.O.; Fontes, J.T.; Hansen, A.K.; Jensen, M.R.; Kalaycı, T.E.; Leeper, A.; Patil, S.K.; et al. The Future of DNA Barcoding: Reflections from Early Career Researchers. Diversity 2021, 13, 313. [Google Scholar] [CrossRef]
- Stork, N.E.; McBroom, J.; Gely, C.; Hamilton, A.J. New Approaches Narrow Global Species Estimates for Beetles, Insects, and Terrestrial Arthropods. Proc. Natl. Acad. Sci. USA 2015, 112, 7519–7523. [Google Scholar] [CrossRef] [PubMed]
- Rkristensen, N.; Scoble, M.J.; Karsholt, O. Lepidoptera Phylogeny and Systematics: The State of Inventorying Moth and Butterfly Diversity. Zootaxa 2007, 747, 699–747. [Google Scholar] [CrossRef]
- Pogue, M.G. Biodiversity of Lepidoptera. In Insect Biodiversity: Science and Society; Foottit, R.G., Adler, P.H., Eds.; Blackwell Publishing Ltd.: Hoboken, NJ, USA, 2009; pp. 325–355. ISBN 9781405151429. [Google Scholar]
- Ratnasingham, S.; Hebert, P.D.N. Bold: The Barcode of Life Data System: Barcoding. Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef]
- Ratnasingham, S.; Hebert, P.D.N. A DNA-Based Registry for All Animal Species: The Barcode Index Number (BIN) System. PLoS ONE 2013, 8, e66213. [Google Scholar] [CrossRef]
- Ortiz, A.S.; Rubio, R.M.; Guerrero, J.J.; Garre, M.J.; Serrano, J.; Hebert, P.D.N.; Hausmann, A. Close Congruence between Barcode Index Numbers (Bins) and Species Boundaries in the Erebidae (Lepidoptera: Noctuoidea) of the Iberian Peninsula. Biodivers. Data J. 2017, 5, e19840. [Google Scholar] [CrossRef]
- Lopez-Vaamonde, C.; Kirichenko, N.; Cama, A.; Doorenweerd, C.; Godfray, H.C.J.; Guiguet, A.; Gomboc, S.; Huemer, P.; Landry, J.F.; Laštůvka, A.; et al. Evaluating DNA Barcoding for Species Identification and Discovery in European Gracillariid Moths. Front. Ecol. Evol. 2021, 9, 626752. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Penton, E.H.; Burns, J.M.; Janzen, D.H.; Hallwachs, W. Ten Species in One: DNA Barcoding Reveals Cryptic Species in the Neotropical Skipper Butterfly Astraptes Fulgerator. Proc. Natl. Acad. Sci. USA 2004, 101, 14812–14817. [Google Scholar] [CrossRef] [PubMed]
- Silva-Brandão, K.L.; Lyra, M.L.; Freitas, A.V.L. Barcoding Lepidoptera: Situação Atual e Perspectivas Sobre a Utilidade de Uma Técnica Controversa. Neotrop. Entomol. 2009, 38, 441–451. [Google Scholar] [CrossRef]
- Yang, Z.; Landry, J.F.; Handfield, L.; Zhang, Y.; Alma Solis, M.; Handfield, D.; Scholtens, B.G.; Mutanen, M.; Nuss, M.; Hebert, P.D.N. DNA Barcoding and Morphology Reveal Three Cryptic Species of Anania (Lepidoptera: Crambidae: Pyraustinae) in North America, All Distinct from Their European Counterpart. Syst. Entomol. 2012, 37, 686–705. [Google Scholar] [CrossRef]
- Mutanen, M.; Kivelä, S.M.; Vos, R.A.; Doorenweerd, C.; Ratnasingham, S.; Hausmann, A.; Huemer, P.; Dinča, V.; Van Nieukerken, E.J.; Lopez-Vaamonde, C.; et al. Species-Level Para- and Polyphyly in DNA Barcode Gene Trees: Strong Operational Bias in European Lepidoptera. Syst. Biol. 2016, 65, 1024–1040. [Google Scholar] [CrossRef] [PubMed]
- Cusser, S.; Haddad, N.M.; Jha, S. Unexpected Functional Complementarity from Non-Bee Pollinators Enhances Cotton Yield. Agric. Ecosyst. Environ. 2021, 314, 107415. [Google Scholar] [CrossRef]
- Macgregor, C.J.; Pocock, M.J.O.; Fox, R.; Evans, D.M. Pollination by Nocturnal Lepidoptera, and the Effects of Light Pollution: A Review. Ecol. Entomol. 2015, 40, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Ferracini, C.; Pogolotti, C.; Mancardi, P.; Miglio, M.; Bonelli, S.; Barbero, F. The Box Tree Moth: An Invasive Species Severely Threatening Buxus Natural Formation in NW Italy. Forests 2022, 13, 178. [Google Scholar] [CrossRef]
- Kravchenko, V.D.; Müller, G.C.; Allan, S.A.; Yefremova, Z.A. Seven Invasive Owlet Moths (Lepidoptera: Noctuidae) in Israel and Their Potential Parasitoids (Hymenoptera: Chalcidoidea). Phytoparasitica 2014, 42, 333–339. [Google Scholar] [CrossRef]
- Karlsson, F.M.; Rachidatou, S.; Sahadatou, M.S.; Joseph, Z.A.; Georg, G. First Report of Tuta Absoluta Meyrick (Lepidoptera: Gelechiidae) in the Republic of Benin. BioInvasions Rec. 2018, 7, 463–468. [Google Scholar] [CrossRef]
- Legal, L.; Valet, M.; Dorado, O.; de Jesus-Almonte, J.M.; López, K.; Céréghino, R. Lepidoptera Are Relevant Bioindicators of Passive Regeneration in Tropical Dry Forests. Diversity 2020, 12, 231. [Google Scholar] [CrossRef]
- Ismail, N.; Rahman, A.A.A.; Mohamed, M.; Bakar, M.F.A.; Tokiman, L. Butterfly as Bioindicator for Development of Conservation Areas in Bukit Reban Kambing, Bukit Belading and Bukit Tukau, Johor, Malaysia. Biodiversitas 2020, 21, 334–344. [Google Scholar] [CrossRef]
- Valtonen, A.; Jantunen, J.; Saarinen, K. Flora and Lepidoptera Fauna Adversely Affected by Invasive Lupinus polyphyllus along Road Verges. Biol. Conserv. 2006, 133, 389–396. [Google Scholar] [CrossRef]
- Sabbour, M.M. Control of Leopard Zeuzera pyrina (L.) (Lepidoptera: Cossidae), by Imidaclorprid in Olive Trees. Egypt. Acad. J. Biol. Sci. F. Toxicol. Pest Control 2017, 9, 191–195. [Google Scholar] [CrossRef]
- Fekrat, L.; Farashi, A. Impacts of Climatic Changes on the Worldwide Potential Geographical Dispersal Range of the Leopard Moth, Zeuzera pyrina (L.) (Lepidoptera: Cossidae). Glob. Ecol. Conserv. 2022, 34, e02050. [Google Scholar] [CrossRef]
- Eurostat. Demography of Europe: Statistics Visualised; European Union: Brussel, Belgium, 2021.
- Seguna, A. Malta Lepidoptera. Available online: http://seguna4.wix.com/maltalepidoptera (accessed on 25 October 2022).
- Sammut, P. Il-Lepidoptera; Pin Publishers: Pieta’, Malta, 2000; 246p. [Google Scholar]
- de Jong, Y.; Verbeek, M.; Michelsen, V.; de Place Bjørn, P.; Los, W.; Steeman, F.; Bailly, N.; Basire, C.; Chylarecki, P.; Stloukal, E.; et al. Fauna Europaea—All European Animal Species on the Web. Biodivers. Data J. 2014, 2, 4034. [Google Scholar] [CrossRef]
- Mazzei, P.; Morel, D.; Panfili, R. Moths and Butterflies of Europe and North Africa. Available online: https://www.leps.it (accessed on 25 October 2022).
- Lewandowski, S.; Fischer, H. Revision Der Artengruppen von Lasiocampa trifolii und L. Serrula Der Gattung Lasiocampa von Paula Schrank, 1802 (Lepidoptera: Lasiocampidae). Nachrichten Entomol. Vereins Apollo 2005, 26, 183–196. [Google Scholar]
- Sammut, P.; Sammut, A.; Catania, A.; Seguna, A.; Magro, D. New Records of Noctuidae (Lepidoptera) from the Maltese Islands. Cent. Mediterr. Nat. 2003, 4, 51–54. [Google Scholar]
- Fibiger, M.; Sammut, P.; Seguna, A.; Catania, A. Recent Records of Noctuidae from Malta, with Five Species New to the European Fauna, and a New Subspecies. Nota Lepidopterol. 2006, 29, 193–213. [Google Scholar]
- Waring, P.; Townsend, M. Field Guide to the Moths of Great Britain and Ireland, 2nd ed.; Bloomsbury: London, UK, 2009. [Google Scholar]
- Brock, P.D. A Photographic Guide to Insects of Southern Europe and the Mediterranean; Piscespublications: Newbury, UK, 2017; ISBN 989-1-874357-79-7. [Google Scholar]
- Sammut, P. Die Geometriden Der Maltesischen Inseln (Lepidoptera: Geometridae). Neue Entomol. Nachrichten 1983, 6, 61–64. [Google Scholar]
- Sammut, P.; Borg, J. An Annotated Catalogue of the Lepidoptera Collection of Guido Lanfranco at the National Museum of Natural History in Malta. Bull. Entomol. Soc. Malta 2008, 1, 67–78. [Google Scholar]
- Sammut, P.; Seguna, A.; Catania, A. Notes on Geometridae of the Maltese Islands with New Records (Lepidoptera: Geometridae). Shil. Rev. Lepidopterol. 2008, 36, 105–111. [Google Scholar]
- Fibiger, M.; Hacker, H.H. Systematic List of the Noctuoidea of Europe; Esperiana: Bad Staffelstein, Germany, 2005; Volume 11, ISBN 0147-5185. [Google Scholar]
- Koster, S.; Sammut, P. Faunistic Notes on Momphidae, Batrachedridae, Stathmopodidae and Cosmopterigidae from the Maltese Islands. Nota Lepidopterol. 2006, 29, 49–63. [Google Scholar]
- Folmer, O.; BLACK, M.; HOEH, W.; Lutz, R.; Vrijenhoek, R. DNA Primers for Amplification of Mitochondrial Cytochrome c Oxidase Subunit I from Diverse Metazoan Invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar] [CrossRef]
- Mifsud, C.M.; Vella, N.; Vella, A. Contribution to the Knowledge of the Beetle Fauna (Insecta, Coleoptera) of Malta: New Records of Seven Species with Supporting DNA Barcodes. Check List 2021, 17, 1443–1449. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: A Multiple Sequence Alignment Method with Reduced Time and Space Complexity. BMC Bioinf. 2004, 5, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2013, 41, 36–42. [Google Scholar] [CrossRef]
- Zhang, Z.; Schwartz, S.; Wagner, L.; Miller, W. A Greedy Algorithm for Aligning DNA Sequences. J. Comput. Biol. 2000, 7, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Morgulis, A.; Coulouris, G.; Raytselis, Y.; Madden, T.L.; Agarwala, R.; Schäffer, A.A. Database Indexing for Production MegaBLAST Searches. Bioinformatics 2008, 24, 1757–1764. [Google Scholar] [CrossRef] [PubMed]
- Posada, D. JModelTest: Phylogenetic Model Averaging. Mol. Biol. Evol. 2008, 25, 1253–1256. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Lerget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian Inference of Phylogenetic Trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef]
- Falck, P.; Karsholt, O.; Simonsen, T. The Genus Apatema Walsingham, 1900 in the Canary Islands and Madeira, with Description of 13 New Species (Lepidoptera, Autostichidae, Oegoconiinae). Shil. Revta. Lepid. 2021, 49, 273–318. [Google Scholar]
- Ashfaq, M.; Akhtar, S.; Rafi, M.A.; Mansoor, S.; Hebert, P.D.N. Mapping Global Biodiversity Connections with DNA Barcodes: Lepidoptera of Pakistan. PLoS ONE 2017, 12, e174749. [Google Scholar] [CrossRef]
- Huemer, P.; Mutanen, M.; Sefc, K.M.; Hebert, P.D.N. Testing DNA Barcode Performance in 1000 Species of European Lepidoptera: Large Geographic Distances Have Small Genetic Impacts. PLoS ONE 2014, 9, e115774. [Google Scholar] [CrossRef] [PubMed]
- Boursin, C. A New Maltese Subspecies of Leucania Putrescens Hübner. Entomology 1952, 85, 132. [Google Scholar]
- Lewandowski, S.; Fischer, H. Zweiter Nachtrag Zu Den Revisionen Der Trifolii- Und Serrula-Artengruppen Der Gattung Lasiocampa Sowie Neue Erkenntnisse Zum Typenfundort von L. Decolorata (Lepidoptera: Lasiocampidae). Nachr. Entomol. Ver. Apollo 2012, 32, 97–104. [Google Scholar]
- Arnscheid, W.R.; Sobczyk, T.; Zerafa, M. Notes on the Identity of Oiketicoides tedaldii (Heylaerts, 1882) (Psychidae, Oiketicinae). Nota Lepidopterol. 2021, 44, 1–15. [Google Scholar] [CrossRef]
- Heckford, R.J. A Further Record of Phereoeca lodli Vives, 2001, from Spain, with Illustrations of the Adult and Female Genitalia (Lepidoptera: Tineidae). Shil. Rev. Lepidopterol. 2012, 40, 465–468. [Google Scholar]
- Bippus, M. Praeacedes atomosella (Walker, 1863) and Phereoeca praecox Gozmány & Vári, 1973—Two Case-Bearing Moths New to the Fauna of La Réunion (Lepidoptera: Tineidae). Contrib. Entomol. 2016, 66, 159–163. [Google Scholar] [CrossRef]
- Mifsud, C.M.; Magro, D.; Vella, A. First Record and DNA Barcode of the Clearwing Moth Tinthia tineiformis (Esper, 1789) from Malta, Central Mediterranean. Check List 2019, 15, 595–599. [Google Scholar] [CrossRef]
- Furlani, S.; Antonioli, F.; Biolchi, S.; Gambin, T.; Gauci, R.; Lo Presti, V.; Anzidei, M.; Devoto, S.; Palombo, M.; Sulli, A. Holocene Sea Level Change in Malta. Quat. Int. 2013, 288, 146–157. [Google Scholar] [CrossRef]
- Dapporto, L.; Dennis, R.L.H. Conservation Biogeography of Large Mediterranean Islands. Butterfly Impoverishment, Conservation Priorities and Inferences for an Ecological “Island Paradigm.” Ecography (Cop). 2009, 32, 169–179. [Google Scholar] [CrossRef]
- Medail, F.; Quezel, P. Biodiversity Hotspots in the Mediterranean Basin: Setting Global Conservation Priorities. Conserv. Biol. 1999, 13, 1510–1513. [Google Scholar] [CrossRef]
- Dapporto, L.; Dennis, R.L.H. Island Size Is Not the Only Consideration. Ranking Priorities for the Conservation of Butterflies on Italian Offshore Islands. J. Insect Conserv. 2008, 12, 237–249. [Google Scholar] [CrossRef]
- Gillespie, R.G.; Will, K. Biodiversity of Arthropods on Islands. Insect Biodivers. 2018, II, 81–104. [Google Scholar] [CrossRef]
- Emerson, B.C.; Borges, P.A.V.; Cardoso, P.; Convey, P.; deWaard, J.R.; Economo, E.P.; Gillespie, R.G.; Kennedy, S.; Krehenwinkel, H.; Meier, R.; et al. Collective and Harmonized High Throughput Barcoding of Insular Arthropod Biodiversity: Toward a Genomic Observatories Network for Islands. Mol. Ecol. 2022, 16, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Özden, Ö.; Ciesla, W.M.; Fuller, W.J.; Hodgson, D.J. Butterfly Diversity in Mediterranean Islands and in Pentadaktylos Pinus brutia Forests of Cyprus. Biodivers. Conserv. 2008, 17, 2821–2832. [Google Scholar] [CrossRef]
- Malcolm, J.R.; Liu, C.; Neilson, R.P.; Hansen, L.; Hannah, L. Global Warming and Extinctions of Endemic Species from Biodiversity Hotspots. Conserv. Biol. 2006, 20, 538–548. [Google Scholar] [CrossRef]
- Badalamenti, E.; Cusimano, D.; La Mantia, T.; Pasta, S.; Romano, S.; Troia, A.; Ilardi, V. The Ongoing Naturalisation of Eucalyptus spp. in the Mediterranean Basin: New Threats to Native Species and Habitats. Aust. For. 2018, 81, 239–249. [Google Scholar] [CrossRef]
- Casazza, G.; Giordani, P.; Benesperi, R.; Foggi, B.; Viciani, D.; Filigheddu, R.; Farris, E.; Bagella, S.; Pisanu, S.; Mariotti, M.G. Climate Change Hastens the Urgency of Conservation for Range-Restricted Plant Species in the Central-Northern Mediterranean Region. Biol. Conserv. 2014, 179, 129–138. [Google Scholar] [CrossRef]



| FAMILY Species | n | H | BIN BOLD: | nB | AvD (%) | MxD (%) | DNN (%) | NN BIN BOLD: | NN Taxonomy | nBN | NN AvD (%) | NN MxD (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AUTOSTICHIDAE | ||||||||||||
| Apatema baixerasiF | 1 | 1 1 | AAV4815 | 10 | 0.56 | 0.98 | 4.04 | ADR6916 | Apatema sp. | 4 | 0.14 | 0.33 |
BLASTOBASIDAE | ||||||||||||
| Blastobasis phycidella | 3 | 3 3 | AAF0414 | 69 | 0.57 | 2.43 | 2.97 | AAZ8649 | Blastobasis sp. | 2 | 0.46 | 0.46 |
COSMOPTERIGIDAE | ||||||||||||
| Bifascioides leucomelanella | 1 | 1 1 | ABA4555 | 18 | 0.27 | 0.84 | 2.41 | ADU3943 | Lepidoptera sp. | 1 | - | - |
| Pyroderces argyrogrammos | 3 | 3 3 | AAQ0242 | 114 | 0.78 | 3.05 | 6.46 | AEJ1178 | Lepidoptera sp. | 1 | - | - |
COSSIDAE | ||||||||||||
| Zeuzera pyrina | 2 | 1 2 | AET9156 N | 2 | 0.00 | 0.00 | 1.93 | ADC8403 | Zeuzera sp. | 1 | - | - |
CRAMBIDAE | ||||||||||||
| Agriphila trabeatellus | 2 | 2 2 | ACA9410 | 9 | 0.70 | 1.77 | 8.26 | ABA4409 | Catoptria confusellus | 3 | 0.27 | 0.33 |
| Ancylolomia pectinatellus | 1 | 1 1 | ACA9335 | 5 | 0.49 | 0.67 | 7.87 | ACI1349 | Crambidae sp. | 2 | 0.15 | 0.15 |
| Antigastra catalaunalis | 3 | 3 2 | AAE6976 | 38 | 0.61 | 1.53 | 3.22 | AAP5696 | Antigastra catalaunalis | 1 | - | - |
| Aporodes floralis | 3 | 1 0 | AAN7323 | 40 | 0.13 | 0.59 | 2.62 | AAV4122 | Aporodes floralis | 6 | 0.65 | 1.12 |
| Dolicharthria bruguieralis | 1 | 1 1 | AED1102 | 2 | 0.16 | 0.16 | 1.28 | AAO3560 | Dolicharthria bruguieralis | 10 | 0.12 | 0.48 |
| Duponchelia fovealis | 5 | 3 2 | AAD9727 | 106 | 0.16 | 1.99 | 2.25 | ACR2019 | Duponchelia fovealis | 9 | 0.36 | 0.80 |
| Euchromius cambridgei | 4 | 2 2 | ABY3890 | 12 | 1.24 | 2.18 | 7.21 | ADZ9070 | Phycitinae sp. | 1 | - | - |
| Euchromius ocellea | 8 | 4 2 | AAA5671 | 231 | 0.37 | 1.77 | 3.19 | ABA8488 | Euchromius sp. | 1 | - | - |
| Evergestis sp. | 2 | 2 2 | AEU3700 N | 2 | 0.16 | 0.16 | 2.73 | ADL3576 | Evergestis isatidalis | 3 | 0.54 | 0.80 |
| Hellula undalis | 5 | 4 1 | AAC8519 | 78 | 0.48 | 1.61 | 3.45 | AAE6944 | Hellula rogatalis | 54 | 0.06 | 0.55 |
| Herpetogramma licarsisalis | 1 | 1 1 | AAA3965 | 292 | 0.14 | 1.77 | 3.98 | AAA3967 | Herpetogramma licarsisalis | 38 | 0.06 | 0.32 |
| Nomophila noctuella | 3 | 1 0 | AAA7880 | 319 | 0.46 | 3.75 | 3.86 | AAB5466 | Nomophila corticalis | 123 | 0.35 | 1.12 |
| Palpita vitrealis | 1 | 1 0 | AAC1043 | 100 | 0.72 | 2.57 | 2.51 | AAB0733 | Palpita margaritacea | 51 | 0.18 | 0.64 |
| Spoladea recurvalis | 1 | 1 0 | AAA3666 | 364 | 0.72 | 3.19 | 6.18 | ABA0182 | Scoparia paracycla | 1 | - | - |
| Udea ferrugalis | 3 | 2 0 | AAC3729 | 104 | 0.64 | 3.04 | 3.47 | ABA1630 | Udea stellata | 1 | - | - |
| Uresiphita gilvata | 1 | 1 0 | ACF5204 | 43 | 0.50 | 1.77 | 1.10 | AAA3568 | Uresiphita ornithopteralis | 170 | 0.15 | 1.02 |
EREBIDAE | ||||||||||||
| Clytie illunaris | 1 | 1 1 | AAK5589 | 18 | 0.25 | 0.65 | 1.12 | AEH3335 | Clytie sp. | 3 | 0.11 | 0.16 |
| Cymbalophora pudica | 3 | 2 2 | AAG6227 | 12 | 0.74 | 1.50 | 4.81 | ABZ5736 | Turuptiana obliqua | 1 | - | - |
| Dysauxes famula | 2 | 2 1 | AAM0427 | 24 | 0.24 | 0.80 | 1.12 | ACF0669 | Dysauxes famula | 8 | 0.53 | 0.96 |
| Eilema caniola | 2 | 1 0 | AAF6264 | 71 | 0.78 | 2.73 | 4.19 | AAA4503 | Manulea bicolor | 198 | 0.30 | 2.68 |
| Eublemma ostrina | 1 | 1 0 | AAG1829 | 34 | 0.56 | 2.25 | 2.39 | ABW0690 | Eublemma staudingeri | 22 | 0.06 | 0.37 |
| Eublemma parva | 5 | 3 1 | AAM5884 | 43 | 0.25 | 0.64 | 3.52 | ACL9149 | Eublemma saldaitis | 1 | - | - |
| Eublemma scitula | 1 | 1 1 | ACD0717 | 6 | 0.62 | 1.77 | 4.33 | ACN9797 | Noctuidae sp. | 1 | - | - |
| Eublemma sp. | 1 | 1 1 | AAL4752 | 13 | 0.81 | 1.77 | 1.12 | ACL7422 | Eublemma parva | 4 | 0.48 | 0.80 |
| Hypena lividalis | 3 | 2 0 | AAE1121 | 69 | 0.19 | 1.15 | 5.93 | AAA2868 | Chytolita morbidalis | 187 | 0.70 | 1.92 |
| Hypena obsitalis | 10 | 5 3 | AAK3686 | 23 | 0.12 | 0.39 | 3.19 | ACF0234 | Hypena sordidula | 17 | 0.16 | 0.65 |
| Metachrostis velocior | 3 | 2 1 | AAH6931 | 8 | 0.51 | 1.25 | 1.44 | ACK1973 | Metachrostis dardouini | 6 | 0.05 | 0.16 |
| Metachrostis velox | 2 | 2 1 | AAH6930 | 14 | 0.45 | 0.98 | 1.77 | ACK1973 | Metachrostis dardouini | 6 | 0.05 | 0.16 |
| Nodaria nodosalis | 1 | 1 0 | AAK3749 | 50 | 0.76 | 3.00 | 3.10 | AAD1694 | Simplicia cornicalis | 50 | 0.25 | 2.17 |
| Ophiusa tirhaca | 1 | 1 0 | ABZ7648 | 21 | 0.40 | 1.18 | 1.77 | ABZ4334 | Ophiusa sp. | 22 | 0.05 | 0.36 |
| Orgyia trigotephras | 1 | 1 1 | AAM0804 | 4 | 1.03 | 1.44 | 4.17 | ACB6683 | Orgyia sp. | 4 | 0.00 | 0.00 |
| Pechipogo plumigeralis | 1 | 1 0 | AAI4196 | 22 | 0.05 | 0.32 | 3.37 | AAA2868 | Chytolita morbidalis | 187 | 0.70 | 1.92 |
| Phragmatobia fuliginosa | 4 | 1 0 | AAA6178 | 94 | 0.59 | 2.25 | 2.25 | AAN2564 | Phragmatobia fuliginosa | 2 | 0.36 | 0.36 |
| Utetheisa pulchella | 1 | 1 0 | AAF0098 | 61 | 0.17 | 0.64 | 1.42 | ACT3042 | Utetheisa elata | 3 | 0.82 | 1.22 |
| Zebeeba falsalis | 6 | 1 0 | AAJ9181 | 26 | 0.98 | 2.57 | 8.01 | AAN6974 | Elaphria sp. | 1 | - | - |
GELECHIIDAE | ||||||||||||
| Agonopterix olusatri | 1 | 1 1 | ABW7168 | 15 | 0.52 | 1.61 | 1.93 | ADF2495 | Agonopterix sp. | 5 | 0.10 | 0.16 |
| Agonopterix subpropinquella | 1 | 1 1 | AER7434 N | 1 | - | - | 1.93 | AAZ9000 | Agonopterix subpropinquella | 13 | 0.05 | 0.38 |
| Aproaerema sp. | 1 | 1 1 | AET5627 N | 1 | - | - | 1.77 | AEA1472 | Aproaerema sp. | 5 | 0.00 | 0.00 |
| Ornativalva plutelliformis | 1 | 1 1 | ABW9166 | 11 | 0.65 | 1.77 | 2.49 | ABX8241 | Ornativalva plutelliformis | 4 | 0.33 | 0.66 |
| Phthorimaea operculella | 4 | 1 0 | AEL8356 | 95 | 0.18 | 4.31 | 5.54 | AED9067 | Phthorimaea sp. | 1 | - | - |
| Platyedra subcinerea | 6 | 5 4 | AAD8749 | 49 | 0.54 | 1.50 | 5.36 | AAU3620 | Pexicopia sp. | 2 | 0.00 | 0.00 |
| Ptocheuusa paupella | 1 | 1 0 | AAV2188 | 18 | 0.17 | 0.82 | 3.21 | ACW2460 | Ptocheuusa paupella | 7 | 0.57 | 1.29 |
| Tuta absoluta | 1 | 1 0 | AAJ8033 | 973 | 0.04 | 1.80 | - | - | - | - | - | - |
GEOMETRIDAE | ||||||||||||
| Charissa variegata | 5 | 4 4 | AAC1039 | 35 | 1.62 | 3.49 | 2.75 | AAC4341 | Charissa subtaurica | 18 | 0.35 | 1.50 |
| Cyclophora puppillaria | 1 | 1 0 | AAB2523 | 60 | 0.03 | 0.65 | 3.17 | ACF3607 | Cyclophora albipunctata | 36 | 0.43 | 1.44 |
| Epirrhoe alternata | 2 | 1 0 | ACE4142 | 134 | 0.63 | 2.81 | 1.02 | AAA3371 | Epirrhoe alternata | 85 | 0.85 | 2.09 |
| Eucrostes indigenata | 2 | 2 2 | AAC6469 | 16 | 0.37 | 1.16 | 3.60 | ADF4899 | Eucrostes sp. | 1 | - | - |
| Eupithecia centaureata | 2 | 2 2 | ACE9420 | 67 | 1.76 | 4.33 | 6.06 | ACJ9495 | Eupithecia sp. | 1 | - | - |
| Gymnoscelis rufifasciata | 1 | 1 1 | AAA7404 | 97 | 0.74 | 2.20 | 2.74 | ADL3671 | Gymnoscelis rufifasciata | 2 | 0.00 | 0.00 |
| Idaea elongaria | 1 | 1 1 | AAA8985 | 7 | 0.26 | 0.55 | 2.57 | ACK1747 | Idaea elongaria | 1 | - | - |
| Idaea fractilineata | 4 | 3 2 | AAK4252 | 8 | 0.34 | 0.97 | 2.85 | ACM9078 | Idaea purpurariata | 5 | 0.00 | 0.00 |
| Idaea obsoletaria | 1 | 1 1 | AAB4939 | 6 | 0.11 | 0.33 | 2.30 | ACE4926 | Idaea obsoletaria | 4 | 0.40 | 0.71 |
| Idaea seriata | 3 | 1 1 | AAA9645 | 56 | 0.13 | 0.71 | 1.92 | ABZ4137 | Idaea seriata | 6 | 0.10 | 0.31 |
| Isturgia pulinda | 1 | 1 0 | AAA6139 | 95 | 0.40 | 1.34 | 3.50 | AAU7783 | Isturgia exerraria | 2 | 0.00 | 0.00 |
| Menophra japygiaria | 5 | 1 0 | AAB6706 | 42 | 0.41 | 2.64 | 2.66 | AAC8802 | Menophra berenicidaria | 13 | 0.18 | 0.75 |
| Phaiogramma etruscaria | 1 | 1 0 | ABY4065 | 42 | 0.20 | 0.67 | 1.12 | ACW6537 | Phaiogramma etruscaria | 9 | 0.17 | 0.32 |
| Phaiogramma faustinata | 1 | 1 0 | AAB4914 | 82 | 0.68 | 2.67 | 1.04 | ACW6536 | Phaiogramma stibolepida | 14 | 0.64 | 1.44 |
| Rhodometra sacraria | 6 | 3 1 | AAA8983 | 138 | 1.11 | 5.41 | 5.92 | AAQ1498 | Rhodometra sacraria | 3 | 1.47 | 2.25 |
| Scopula imitaria | 2 | 1 0 | AAB6665 | 56 | 0.23 | 1.44 | 1.80 | ABZ6950 | Scopula imitaria syriacaria | 5 | 0.00 | 0.00 |
| Scopula minorata | 1 | 1 1 | AAA9357 | 125 | 0.94 | 3.86 | 2.12 | AEO1263 | Scopula sp. | 1 | - | - |
| Xanthorhoe disjunctaria | 1 | 1 0 | ABY6341 | 27 | 0.45 | 1.15 | 1.77 | AET6043 | Xanthorhoe sardisjuncta | 11 | 0.23 | 0.64 |
LASIOCAMPIDAE | ||||||||||||
| Gastropacha quercifolia | 1 | 1 1 | AAF4844 | 94 | 0.48 | 1.54 | 8.67 | AAI7018 | Gastropacha sikkima | 50 | 1.08 | 2.09 |
| Lasiocampa sp. | 5 | 3 3 | AES9600 N | 3 | 0.00 | 0.00 | 1.93 | AAW9949 | Lasiocampa tripolitania | 3 | 0.20 | 0.31 |
LYCAENIDAE | ||||||||||||
| Polyommatus celina | 4 | 3 1 | AAA3304 | 275 | 0.92 | 2.41 | 3.71 | AAA3303 | Polyommatus erotides | 976 | 1.28 | 3.88 |
MOMPHIDAE | ||||||||||||
| Mompha subbistrigella | 1 | 1 1 | AAD0702 | 71 | 0.30 | 1.44 | 3.61 | ADB9986 | Mompha glaucella | 3 | 0.10 | 0.15 |
NOCTUIDAE | ||||||||||||
| Acontia lucida | 3 | 2 1 | AAD6258 | 38 | 0.25 | 1.01 | 5.22 | ABV2194 | Lepidoptera sp. | 4 | 0.15 | 0.31 |
| Agrotis biconica | 1 | 1 0 | AAE4276 | 36 | 0.67 | 1.51 | 2.09 | ABZ5220 | Agrotis munda | 28 | 0.28 | 1.02 |
| Agrotis ipsilon | 3 | 2 0 | AAA3364 | 336 | 0.31 | 1.94 | 1.00 | ACE7272 | Agrotis infusa | 120 | 0.02 | 0.37 |
| Agrotis lata | 4 | 4 2 | ACE7288 | 11 | 0.29 | 0.80 | 1.28 | AEH3853 | Agrotis lata | 1 | - | - |
| Agrotis puta | 5 | 1 0 | AAB9164 | 79 | 0.09 | 0.64 | 2.19 | AAB9165 | Lepidoptera sp. | 10 | 0.31 | 0.64 |
| Agrotis segetum | 3 | 2 1 | AAC3884 | 172 | 0.17 | 1.69 | 2.32 | AAB9113 | Agrotis exclamationis | 90 | 0.07 | 0.80 |
| Agrotis trux | 14 | 6 6 | AET6510 | 14 | 0.19 | 0.50 | 1.12 | AAM0539 | Agrotis trux | 13 | 0.98 | 2.09 |
| Anarta trifolii | 1 | 1 0 | ABZ1428 | 201 | 0.58 | 3.33 | 1.71 | AAA9985 | Anarta columbica | 71 | 0.13 | 1.07 |
| Autographa gamma | 3 | 2 1 | AAB4345 | 626 | 0.03 | 2.02 | 2.17 | AAB2628 | Autographa californica | 110 | 0.12 | 0.67 |
| Callopistria latreillei | 1 | 1 1 | AAP2182 | 47 | 0.28 | 1.28 | 3.24 | AAN8804 | Callopistria sp. | 2 | 0.00 | 0.00 |
| Caradrina clavipalpis | 3 | 1 0 | AAB6999 | 83 | 0.36 | 2.17 | 2.41 | ABZ7109 | Caradrina selini | 48 | 0.13 | 0.64 |
| Caradrina flava | 2 | 1 0 | AAK4908 | 8 | 0.12 | 0.48 | 3.21 | AET1610 | Lepidoptera sp. | 117 | 0.03 | 0.64 |
| Caradrina flavirena | 6 | 1 0 | AAB7000 | 95 | 0.67 | 2.73 | 1.18 | ADB8712 | Caradrina flavirena | 9 | 0.04 | 0.16 |
| Chrysodeixis chalcites | 3 | 1 0 | AAB3384 | 354 | 0.56 | 3.16 | 3.05 | AAG0704 | Chrysodeixis kebea | 6 | 0.33 | 0.61 |
| Condica viscosa | 1 | 1 0 | AAN1812 | 13 | 0.34 | 0.72 | 2.09 | ADU4648 | Noctuidae sp. | 2 | 0.00 | 0.00 |
| Cryphia algae | 5 | 2 2 | AAD6780 | 10 | 0.40 | 1.28 | 1.28 | AAD6780 | Lepidoptera sp. | 65 | 0.30 | 0.96 |
| Hadena sancta | 2 | 1 1 | AAY8457 | 10 | 0.48 | 1.04 | 2.09 | ABY4816 | Hadena ruetimeyeri | 2 | 1.07 | 1.07 |
| Heliothis peltigera | 5 | 3 0 | AAC6990 | 60 | 0.14 | 0.64 | 3.53 | AAV6844 | Heliothis saskai | 2 | 0.00 | 0.00 |
| Leucania putrescens vallettaiE | 2 | 2 2 | AER7912 N | 2 | 0.34 | 0.34 | 2.18 | AAK9298 | Leucania putrescens | 6 | 0.61 | 0.96 |
| Mythimna sicula | 1 | 1 1 | AAF8181 | 53 | 0.49 | 1.13 | 2.75 | ABX0055 | Mythimna opaca | 2 | 0.32 | 0.32 |
| Mythimna unipuncta | 1 | 1 0 | AAA2482 | 555 | 0.51 | 4.41 | 2.08 | ACG2559 | Mythimna unipuncta | 3 | 0.00 | 0.00 |
| Noctua pronuba | 2 | 2 0 | AAA2632 | 321 | 0.22 | 1.61 | 3.69 | AAD0229 | Noctua interjecta | 56 | 0.41 | 1.46 |
| Nyctobrya segunai E | 4 | 4 4 | AET0743 N | 4 | 0.53 | 0.68 | 1.68 | AAN0805 | Cryphia muralis | 8 | 0.21 | 0.50 |
| Pseudozarba bipartita | 6 | 6 6 | AAE4331 | 48 | 1.15 | 3.28 | 3.17 | AAE8111 | Pseudozarba orthopetes | 24 | 0.54 | 1.93 |
| Spodoptera cilium | 1 | 1 0 | AAC8279 | 79 | 0.15 | 0.99 | 2.11 | ACE3456 | Spodoptera depravata | 71 | 0.13 | 0.80 |
| Spodoptera exigua | 8 | 3 0 | AAA6644 | 632 | 0.38 | 3.37 | 2.60 | ADB9075 | Spodoptera exigua | 1 | - | - |
| Synthymia fixa | 3 | 1 0 | AAN0137 | 3 | 0.00 | 0.00 | 1.12 | AES6312 | Synthymia fixa | 6 | 0.27 | 0.80 |
| Trichoplusia ni | 1 | 1 0 | AAC3410 | 50 | 0.02 | 0.35 | 4.01 | AAC3409 | Trichoplusia ni | 86 | 0.19 | 2.57 |
| Tyta luctuosa | 5 | 2 0 | AAD5088 | 39 | 0.27 | 0.80 | 5.35 | AEI5594 | Epharmottomena tenera | 1 | - | - |
| Xylena exsoleta | 1 | 1 1 | AAE4735 | 20 | 0.54 | 1.12 | 4.00 | ACD6521 | Xylena formosa | 9 | 0.79 | 1.61 |
NYMPHALIDAE | ||||||||||||
| Coenonympha pamphilus | 1 | 1 0 | AAA7351 | 305 | 0.15 | 1.38 | 1.25 | ADJ7308 | Coenonympha pamphilus | 42 | 0.39 | 1.28 |
| Danaus chrysippus | 1 | 1 0 | ABX5122 | 215 | 0.59 | 2.75 | 1.58 | AAB3216 | Danaus chrysippus | 22 | 0.02 | 0.20 |
| Lasiommata megera | 1 | 1 0 | AAB0123 | 342 | 0.46 | 1.26 | 1.12 | ACE4512 | Lasiommata paramegaera | 55 | 0.05 | 0.50 |
| Vanessa atalanta | 2 | 2 0 | AAA8638 | 271 | 0.22 | 2.71 | 3.85 | AAE5211 | Antanartia abyssinica | 22 | 0.13 | 0.61 |
PAPILIONIDAE | ||||||||||||
| Papilio machaon | 3 | 2 0 | AAA5810 | 440 | 1.01 | 3.58 | 1.77 | ABZ2147 | Papilio machaon | 2 | 0.12 | 0.12 |
PIERIDAE | ||||||||||||
| Colias croceus | 2 | 2 0 | ABZ3039 | 440 | 0.06 | 1.38 | 1.72 | ACF0844 | Colias pelidne | 115 | 1.16 | 2.57 |
| Pieris brassicae | 1 | 1 1 | AAB0552 | 405 | 0.64 | 4.25 | 2.13 | ACN0735 | Pieris brassicae | 1 | - | - |
| Pieris rapae | 1 | 1 1 | AAA2224 | 904 | 0.46 | 3.37 | 3.10 | AAB3783 | Pieris mannii | 128 | 0.24 | 0.80 |
PLUTELLIDAE | ||||||||||||
| Plutella xylostella | 3 | 3 1 | AAA1513 | 3792 | 0.77 | 4.34 | 6.57 | AAC6876 | Plutella australiana | 121 | 0.06 | 0.62 |
PSYCHIDAE | ||||||||||||
| Oiketicoides lutea F | 4 | 3 3 | AAM0038 | 6 | 2.09 | 3.35 | 7.50 | ABU9696 | Oiketicoides sp. | 1 | - | - |
| Penestoglossa dardoinella | 3 | 1 1 | AEU4296 N | 3 | 0.00 | 0.00 | 1.28 | AAL3705 | Penestoglossa dardoinella | 12 | 0.03 | 0.17 |
PTEROPHORIDAE | ||||||||||||
| Agdistis frankeniae | 1 | 1 1 | AED1693 N | 1 | - | - | 3.37 | ABV2042 | Lepidoptera sp. | 2 | 0.16 | 0.16 |
| Emmelina monodactyla | 2 | 1 0 | ACE4862 | 110 | 0.08 | 0.70 | 1.34 | AAA3882 | Emmelina monodactyla | 111 | 0.45 | 1.77 |
| Merrifieldia malacodactylus | 1 | 1 0 | ACS6787 | 17 | 0.28 | 0.64 | 4.49 | ADZ0299 | Pterophoridae sp. | 1 | - | - |
| Pterophoridae sp. | 1 | 1 1 | ADZ0387 | 2 | 0.18 | 0.18 | 6.31 | AAV5270 | Procapperia linariae | 11 | 0.75 | 1.31 |
| Stenoptilia sp. | 1 | 1 1 | ABW6859 | 7 | 0.12 | 0.32 | 4.65 | ACS3431 | Stenoptilia sp. | 6 | 0.80 | 2.14 |
PYRALIDAE | ||||||||||||
| Aglossa caprealis | 1 | 1 1 | ACY8691 | 3 | 1.07 | 1.91 | 6.26 | ADR4870 | Aglossa sp. | 1 | - | - |
| Apomyelois ceratoniae | 2 | 2 0 | AAU4812 | 114 | 0.43 | 1.44 | 3.00 | ACR0358 | Cadra sp. | 5 | 0.32 | 0.80 |
| Bostra dipectinialis F | 7 | 1 0 | AAU4121 | 11 | 0.30 | 0.81 | 2.09 | AEO2408 | Bostra sp. | 15 | 0.76 | 1.61 |
| Cadra abstersella | 7 | 1 0 | AAW5130 | 22 | 0.20 | 1.30 | 5.54 | AAB9605 | Cadra cautella | 167 | 1.24 | 4.21 |
| Cadra cautella | 2 | 2 1 | AAB9605 | 167 | 1.24 | 4.21 | 3.04 | ADV8858 | Cadra sp. | 1 | - | - |
| Cadra figulilella | 6 | 2 0 | AAZ9283 | 81 | 0.20 | 1.62 | 1.61 | ADS7823 | Cadra sp. | 7 | 0.41 | 0.80 |
| Ceutholopha isidis | 2 | 2 1 | ABA4962 | 28 | 0.19 | 0.68 | 2.45 | ABA4962 | Ceutholopha petalocosma | 42 | 0.24 | 0.75 |
| Ephestia elutella | 1 | 1 0 | AAC6157 | 57 | 0.76 | 1.61 | 4.15 | AAD1430 | Ephestia parasitella | 79 | 0.72 | 2.57 |
| Lamoria anella | 11 | 3 3 | ACY8237 | 26 | 0.83 | 1.93 | 4.94 | AAY8816 | Lamoria anella | 2 | 0.48 | 0.48 |
| Oxybia transversella | 1 | 1 1 | ACA9658 | 13 | 1.12 | 2.57 | 5.78 | AAB9775 | Salebriaria roseopunctella | 115 | 1.16 | 2.59 |
| Phycita diaphana | 1 | 1 1 | ACA9652 | 8 | 0.32 | 0.80 | 5.78 | ACB7132 | Phycitinae sp. | 2 | 0.00 | 0.00 |
| Phycitodes saxicola | 2 | 1 1 | AAD9531 | 42 | 0.63 | 2.02 | 6.04 | ABX8977 | Phycitodes sp. | 3 | 0.31 | 0.46 |
| Plodia interpunctella | 2 | 1 0 | AAB2462 | 101 | 0.52 | 4.05 | 6.96 | ADG1988 | Pyralidae sp. | 1 | - | - |
| Psorosa dahliella | 1 | 1 0 | ACA9753 | 3 | 0.55 | 0.84 | 2.09 | AEF6784 | Psorosa ferrugatella | 3 | 0.00 | 0.00 |
| Pyralis farinalis | 2 | 2 2 | AAB3316 | 57 | 0.40 | 2.70 | 2.41 | AAY8728 | Pyralis farinalis | 4 | 0.19 | 0.32 |
| Stemmatophora brunnealis | 3 | 2 1 | AAV6933 | 13 | 0.28 | 0.65 | 5.16 | AEO2608 | Stemmatophora brunnealis | 1 | - | - |
SESIIDAE | ||||||||||||
| Bembecia albanensis | 1 | 1 1 | AAM2453 | 8 | 1.22 | 2.09 | 4.09 | ABX3895 | Bembecia albanensis | 1 | - | - |
SPHINGIDAE | ||||||||||||
| Acherontia atropos | 1 | 1 0 | AAB7886 | 41 | 0.06 | 0.93 | 4.97 | AAD2845 | Acherontia styx | 24 | 0.28 | 1.39 |
| Agrius convolvuli | 1 | 1 0 | AAA2393 | 162 | 0.60 | 2.09 | 3.32 | AAA2392 | Agrius convolvuli | 158 | 0.23 | 2.82 |
| Hippotion celerio | 2 | 2 0 | ABZ5722 | 28 | 0.19 | 0.64 | 1.10 | ACE8834 | Sphingidae sp. | 18 | 0.10 | 0.33 |
TINEIDAE | ||||||||||||
| Niditinea fuscella | 1 | 1 1 | AAF3430 | 59 | 0.90 | 2.87 | 6.37 | AAG3681 | Niditinea truncicolella | 5 | 0.12 | 0.32 |
| Phereoeca praecox F | 3 | 3 2 | AAU1282 | 33 | 0.06 | 0.66 | 3.75 | AAH8518 | Phereoeca uterella | 11 | 0.63 | 2.43 |
| Tinea murariella | 1 | 1 0 | AAE7470 | 8 | 0.19 | 0.93 | 2.90 | AEI9096 | Tinea translucens | 1 | - | - |
TORTRICIDAE | ||||||||||||
| Aethes sp. | 3 | 2 2 | AEU4088 N | 2 | 0.39 | 0.39 | 3.53 | AAP7561 | Aethes sp. | 26 | 0.50 | 1.44 |
| Cacoecimorpha pronubana | 2 | 1 1 | AAD3477 | 33 | 0.26 | 0.96 | 1.96 | ACS9337 | Cacoecimorpha pronubana | 2 | 0.00 | 0.00 |
| Clepsis sp. | 2 | 2 2 | AED2423 N | 2 | 0.50 | 0.50 | 3.34 | ACT3810 | Clepsis consimilana | 4 | 0.08 | 0.16 |
| Eucosma sp. | 4 | 1 0 | ACT0042 | 5 | 0.71 | 1.34 | 2.10 | AAB4296 | Eucosma sp. | 83 | 0.26 | 1.66 |
| Lobesia botrana | 2 | 2 1 | ACH2178 | 77 | 0.57 | 1.70 | 4.24 | AAC9385 | Lobesia reliquana | 37 | 0.11 | 0.64 |
| Pseudococcyx tessulatana | 1 | 1 0 | ACT0606 | 8 | 0.14 | 0.34 | 7.70 | AAH4831 | Retinia sabiniana | 3 | 0.10 | 0.15 |
| Selania capparidana | 1 | 1 1 | AET4374 N | 1 | - | - | 2.50 | ABA4981 | Selania sp. | 1 | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vella, A.; Mifsud, C.M.; Magro, D.; Vella, N. DNA Barcoding of Lepidoptera Species from the Maltese Islands: New and Additional Records, with an Insight into Endemic Diversity. Diversity 2022, 14, 1090. https://doi.org/10.3390/d14121090
Vella A, Mifsud CM, Magro D, Vella N. DNA Barcoding of Lepidoptera Species from the Maltese Islands: New and Additional Records, with an Insight into Endemic Diversity. Diversity. 2022; 14(12):1090. https://doi.org/10.3390/d14121090
Chicago/Turabian StyleVella, Adriana, Clare Marie Mifsud, Denis Magro, and Noel Vella. 2022. "DNA Barcoding of Lepidoptera Species from the Maltese Islands: New and Additional Records, with an Insight into Endemic Diversity" Diversity 14, no. 12: 1090. https://doi.org/10.3390/d14121090
APA StyleVella, A., Mifsud, C. M., Magro, D., & Vella, N. (2022). DNA Barcoding of Lepidoptera Species from the Maltese Islands: New and Additional Records, with an Insight into Endemic Diversity. Diversity, 14(12), 1090. https://doi.org/10.3390/d14121090






