Abstract
A new octopus species, Callistoctopus tenuipes sp. nov., was formally described from the southeastern coastal waters of China using morphological description and molecular analysis methods. C. tenuipes sp. nov. is a small- to moderate-sized octopus, which is characterized by very narrow and long arms. Although it was previously misidentified as the juvenile of Octopus minor (Sasaki, 1920), it can be recognised by spots, gill lamellae count, funnel organ shape, enlarged suckers, and ligula shape. C. tenuipes sp. nov. differs from the small-sized octopus Callistoctopus xiaohongxu, mainly in the gill lamellae count, funnel organ shape, and arm-length index. In the molecular analysis, sequences obtained from the cytochrome c-oxidase subunit I (COI) gene of eight specimens were 590 bp in length. The pairwise Kimura 2-parameter (K2P) genetic distances between Octopodidae species ranged from 8.58 to 23.79% based on the COI gene. The phylogenetic analyses suggested that C. tenuipes sp. nov. belonged to the Callistoctopus clade and may have a close affinity with C. xiaohongxu and O. minor. Moreover, three species delimitation methods all strongly supported C. tenuipes as a separate species.
1. Introduction
The total length of China’s coastline is over 32,000 km, of which the mainland coastline is approximately 18,000 km and the island coastline is about 14,000 km. The East China Sea and the South China Sea have a high biological diversity and productivity as a consequence of the warm current flowing from the tropics [1]. In the past 20 years, five new species [2,3,4,5], a newly recorded squid [6], and a newly recorded and redescribed octopus Amphioctopus ovulum (Sasaki, 1917) [7] have been reported from Chinese waters. In 2012, Lu, et al. [8] recorded 134 species of cephalopods in Chinese waters. In 2022, Xu, et al. [9], using DNA barcoding analysis, found that there was an underestimation of species diversity in cephalopods in Chinese waters. Therefore, there may be far more than 134 species of cephalopods in Chinese waters, and it is important to continuously review and describe the cephalopods of China.
The genus Callistoctopus is characterized by a reddish body with white spots or stripes and long arms, often regarded as “Octopus macropus group” in the past [10]. Currently, this genus contains 14 members, including Callistoctopus alpheus (Norman, 1993), Callistoctopus aspilosomatis (Norman, 1993), Callistoctopus bunurong (Stranks, 1990), Callistoctopus dierythraeus (Norman, 1993), Callistoctopus graptus (Norman, 1993), Callistoctopus lechenaultii (d’Orbigny, 1826), Callistoctopus luteus (Sasaki, 1929), Callistoctopus macropus (Risso, 1826), Callistoctopus nocturnus (Norman and Sweeney, 1997), Callistoctopus ornatus (Gould, 1852), Callistoctopus rapanui (Voss, 1979), Callistoctopus xiaohongxu Zheng, Xu and Li, 2022, Callistoctopus furvus (Gould, 1852), and Callistoctopus taprobanensis (Robson, 1926), with the status of C. furvus and C. taprobanensis unresolved [11]. In the last few decades, C. furvus has been considered a potential name for the “macropus” of the West Atlantic. However, Jesus, et al. [12] recently separated this species from C. macropus and used integrative methods of ethnoknowledge, classic taxonomy, and molecular analysis to describe a neotype. Thus, two species of Callistoctopus are now known to exist in the Mediterranean Sea and the Atlantic Ocean, whereas the other members of the genus are scattered throughout the Indo-Pacific region. In the Indo-Pacific waters, several informally described species of this genus have been reported in recent years. Three unverified species of Callistoctopus from the coast of Vietnam were briefly described by Kaneko, et al. [13] and labeled as Callistoctopus sp.1, Callistoctopus sp.2, and Callistoctopus sp.3. Meanwhile, Sreeja [14] found two cryptic Callistoctopus species along the coast of Kerala, India.
The diversity and taxonomy of cephalopods in China have been studied over the last 20 years [9,15,16,17]. The new cryptic Callistoctopus species described in this study was first collected in Xiamen, Fujian Province, and then reported in a DNA-barcoding study of cephalopods from Chinese waters in 2011 [15]. In subsequent sample collections, we were fortunate to have collected this octopus species several times on Dongshan Island in Fujian Province. This new species and another similar octopus, C. xiaohongxu, which was reported in this area, are both small in size, with reddish-orange skin and long arms. Octopus minor (Sasaki, 1920) is a common economic species with red skin and long arms in East Asia, and these two small octopuses were misidentified as juveniles of O. minor on Dongshan Island. This new species and C. xiaohongxu were called Xiaohongxu by the locals in Chinese, which means small and red-skinned octopus.
In this study, this new octopod species is formally described and named Callistoctopus tenuipes sp. nov. Phylogenetic trees were reconstructed using COI gene sequences to analyse the phylogenetic position of C. tenuipes sp. nov in the family Octopodidae.
2. Materials and Methods
2.1. Specimen Collection
All samples were collected from Dongshan Fish Market, Fujian Province, China. After collecting, samples were covered with ice, then fixed in 10% formalin for 7 days, and then preserved in 70% alcohol. The type specimens are deposited in the Specimen Room, Fisheries College, Ocean University of China (OUC), China.
2.2. Morphological Analysis
Measurements, counts, and indices follow Roper and Voss [18], Norman and Sweeney [19], and Huffard and Hochberg [20]. Abbreviations and definitions are as follows: ALI—arm length index (arm length/ML × 100); AWI—arm width index (arm width/ML × 100); CaLI—calamus length index (calamus length/ligula length × 100); DWDI—the deepest web depth index (the deepest web depth/the longest arm × 100); EgC—egg count; EgL—egg length; EgW—egg width; FFLI—free funnel length index (free funnel length/funnel length × 100); FLI—funnel length index (funnel length/ML × 100); GC—gill count (number of gill lamellae per outer demibranch, excluding the terminal lamella); HAMI—hectocotylised arm index (hectocotylised arm length/ML × 100); HASC—hectocotylised arm sucker count; HWI—head width index (head width/ML × 100); LLI—ligula length index (ligula length/hectocotylised arm length × 100); ML—dorsal mantle length; MWI—mantle width index (mantle width/ML × 100); OAI—opposite arm index (hectocotylised arm length/normal third arm length × 100); PAI—pallial aperture index (pallial aperture length/ML × 100); SC—sucker count on normal arms; SDIn—normal sucker diameter index (normal sucker diameter/ML × 100); SpC—spermatophore count; SpL—spermatophore length; SpW—spermatophore width; SWDI—the shallowest web depth index (the shallowest web depth/the longest arm × 100); TL—total length; TW—total wet weight.
The beaks and radulae were removed from the buccal mass of some specimens, then cleaned and stored in 75% ethanol. The radulae were cleaned with 10% NaOH, air dried, covered with gold, and then scanned using a VEGA3 scanning electron microscope.
2.3. DNA Extraction, PCR Amplification, and Sequencing
Total genomic DNA was extracted from muscle tissue using CTAB (Hexadecyl trimethyl ammonium bromide) method [21]. DNA was dissolved in TE buffer (10 mM Tris-HCI, 1 mM EDTA, pH 8.0). The COI fragments were amplified by primers referenced by Folmer, et al. [22]. The amplification was carried out in 50 µL reactions, containing 1 µL template DNA of 100 ng, 2 µL of each primer (10 µM), 25 µL of 2 × Hieff® PCR Master Mix (With Dye) (Yeasen), and 20 µL sterile distilled water. The PCRs were performed under the following conditions: denaturation at 94 °C for 5 min, 34 cycles at 94 °C for 10 s, annealing at 55 °C for 20 s, and extension at 72 °C for 30 s, and a final extension of 72 °C for 5 min. PCR products were sequenced with the Sanger sequencing method.
2.4. Molecular and Phylogenetic Analyses
SeqMan v.7.2 (DNASTAR package) was used to assemble sequences, which were then deposited in GenBank under accession number OP184666-OP184673. The other sequences were downloaded from GenBank (Table 1). All sequences were aligned using the Clustal W in MEGA X [23]. Vampyroteuthis infernalis Chun, 1903 was used as an outgroup. The Kimura 2-parameter (K2P) model was used to generate K2P distances in MEGA X [23] as well.
The nucleotide substitution model was selected based on the Bayesian information criterion (BIC) in ModelFinder [24]. The best-fit model GTR + G4 + I + F was selected to construct the BI tree in MrBayes 3.2.6 [25]. The Bayesian analysis was started with four default heat chains, running 1.2 million generations of the Markov Chain Monte Carlo (MCMC) until the standard deviation of split frequencies value was lower than 0.01. The first 25% of generations were discarded as burn-in while the remaining 75% was used to construct the phylogenetic tree. Maximum likelihood phylogenies were inferred using IQ-TREE [26] under the model automatically selected for 1000 ultrafast [27] bootstraps, as well as the Shimodaira–Hasegawa-like approximate-likelihood ratio test [28].

Table 1.
Information of specimens for the COI gene used in this study.
Table 1.
Information of specimens for the COI gene used in this study.
| Species | GenBank Accession No. | Locality | References |
|---|---|---|---|
| Amphioctopus aegina | NC_029702 | Haikou, Hainan, China | [29] |
| Amphioctopus fangsiao | HQ846126 | Xiamen, Fujian, China | [15] |
| Amphioctopus marginatus | NC_036351 | Haikou, Hainan, China | [30] |
| Amphioctopus neglectus | NC_049899 | Nanning, Guangxi, China | [31] |
| Amphioctopus rex | MN987271 | Coastal water of India | [7] |
| Callistoctopus aspilosomatis | AB430525 | Miyagi Island, Okinawa, Japan | [32] |
| Callistoctopus furvus | MT892962 | Morro de São Paulo, Bahia, Brazil | [12] |
| Callistoctopus luteus | MT214050 | Zhangzhou, Fujian, China | [9] |
| Callistoctopus macropus | MN933634 | Mediterranean Sea | [33] |
| Callistoctopus ornatus | HM104257 | - | [34] |
| Callistoctopus tenuipes | OP184666-OP184673 | Zhangzhou, Fujian, China | This study |
| Callistoctopus xiaohongxu | OP135961 | Zhangzhou, Fujian, China | [5] |
| Callistoctopus sp.1 | AB385875 | Nha Trang, Vietnam | [32] |
| Callistoctopus sp.2 | AB385876 | Nha Trang, Vietnam | [32] |
| Callistoctopus sp.3 | AB385877 | Nha Trang, Vietnam | [32] |
| Callistoctopus sp.4 | KF489435 | Kerala, India | [14] |
| Callistoctopus sp.5 | MN933632 | Coastal water of Brazil | [33] |
| Cistopus chinensis | KF017606 | Xiamen, Fujian, China | [35] |
| Cistopus taiwanicus | HQ846142 | Xiamen, Fujian, China | [15] |
| Octopus cyanea | NC_039847 | Xisha Islands, China | Unpublished |
| Octopus minor 1 | NC_015896 | Weihai, Shandong, China | [36] |
| Octopus minor 2 | AB191275 | Akashi, Osaka, Japan | [37] |
| Octopus minor 3 | MF631967 | Penghu Islands, China | [16] |
| Octopus sinensis | OK001740 | Wenzhou, Zhejiang, China | [38] |
| Octopus vulgaris | KU525762 | Galicia, Spain | [39] |
| Vampyroteuthis infernalis | NC_009689 | Ogasawara Island, Japan | [40] |
2.5. Species Delimitation
We implemented three widely used molecular methods for species delimitation, including Automatic Barcode Gap Discovery (ABGD), Generalized Mixed Yule Coalescent model (GMYC), and Bayesian Poisson Tree Processes (bPTP). The ABGD analysis was performed on the online server (https://bioinfo.mnhn.fr/abi/public/abgd/) using Kimura (K80) distance model with the relative gap width (X) set to 1.0 and the other parameters maintained as default [41]. The GMYC model was conducted in the split package in R using single-threshold and multiple-threshold analyses (https://www.R-project.org/) [42], respectively. GTR + G4 + I + F was selected as the substitution model, which was evaluated by ModelFinder [24]. The ultrametric tree was generated in BEAST 2.7.1 with a Yule model and strict clock as an assumption [43], running 30 million generations and sampling every 10,000 generations. Then Tracer v1.7.2 was used to assess the convergence by effective sample size (ESS) of each value [44]. The bPTP analysis was carried out on the bPTP online server (https://species.h-its.org/ptp/) with 10,000 generations and 0.1 burn-in [45].
3. Results
3.1. Molecular and Phylogenetic Analyses
Fragments obtained from eight specimens were 590 bp in length. The K2P genetic distances between C. tenuipes sp. nov. and the other species in Octopodidae based on the COI gene are shown in Table 2, ranging from 8.58 to 23.79%.

Table 2.
Pairwise Kimura 2-parameter (K2P) genetic distance (%) between Octopodidae species analysed in this study based on the COI gene.
The COI gene dataset represented four genera of Octopodidae. The Maximum-Likelihood tree (Figure 1) showed that Octopodidae were divided into two clades. One clade comprised the Callistoctopus species and O. minor, with a bootstrap probability (BP) of 84%. The other clade included Octopus, Amphioctopus, and Cistopus. Eight specimens formed a highly supported monophyletic clade, with a BP of 95%. C. tenuipes sp. nov. had a close relationship with C. xiaohongxu and O. minor group, whereas the BP value was ≤ 50%. The topology of the Bayesian Inference analysis is presented in Figure 2. Similarly, C. tenuipes sp. nov. fell into a monophyletic branch with a posterior probability (PP) of 1. C. tenuipes sp. nov., C. xiaohongxu, and O. minor group converged into a single clade. In both phylogenetic trees, species of the genus Amphioctopus and Cistopus formed a monophyletic clade, respectively. However, Octopus was observed to be polyphyletic.
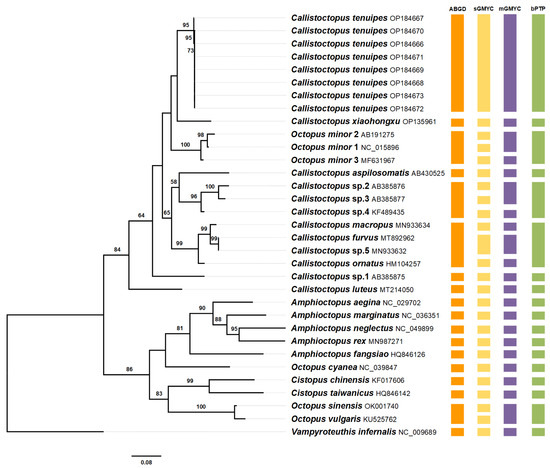
Figure 1.
Phylogenetic tree based on Maximum Likelihood of the mitochondrial COI gene. The values (%) at each node show the bootstrap probability (BP) of ML analyses (only values ≥ 50% are shown). The coloured rectangles on the right present results of species delimitation, including ABGD, sGMYC, mGMYC, and bPTP.
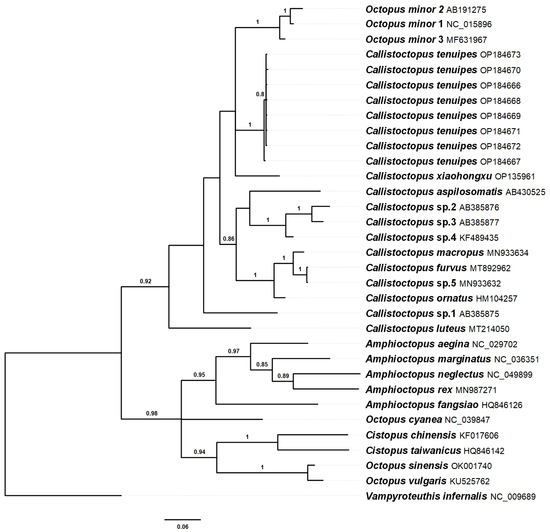
Figure 2.
Phylogenetic tree based on Bayesian Inference of the mitochondrial COI gene. The values (%) at each node show the posterior probability (PP) of BI analyses (only values ≥ 0.8 are shown).
3.2. Species Delimitation
The results of the three species delimitation methods of ABGD, GMYC, and bPTP are shown in Figure 1. Eight specimens of C. tenuipes sp. nov. were recognised as a separate molecular operational taxonomic unit (MOTU) by all three methods. ABGD and bPTP methods showed similar results, with a total of 18 MOTUs identified. GMYC retrieved 25 and 22 MOTUs for the single-threshold model and multiple-threshold model, respectively.
3.3. Taxonomy
Order Octopoda Leach, 1818.
Family Octopodidae d’Orbigny, 1840.
Genus Callistoctopus Taki, 1964.
Type species: Callistoctopus ornatus (Gould, 1852).

Figure 3.
Callistoctopus tenuipes sp. nov. (OUC-201812050307). (A). Dorsal view; (B). ventral view. Scale bar = 10 mm.
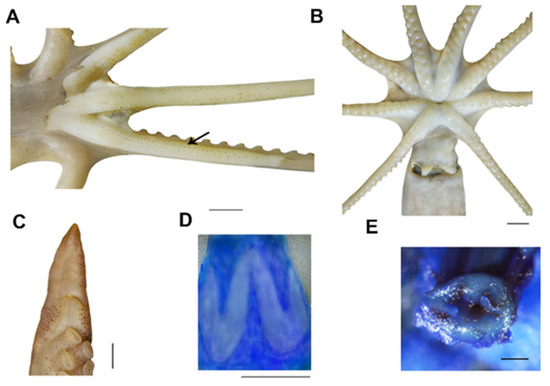
Figure 4.
Callistoctopus tenuipes sp. nov. (A). Chromatophores under the arm skin (OUC-201812050311); (B). oral view (OUC-201812050311); (C). hectocotylus (OUC-201812050309); (D). funnel organ (OUC-201812050311); (E). anal flaps (OUC-201812050311). Scale bars: (A–D) = 10 mm, (E) = 200 μm.
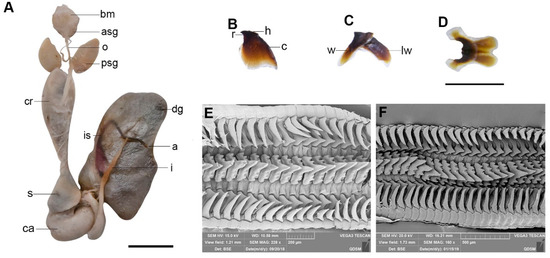
Figure 5.
Callistoctopus tenuipes sp. nov. (A). Digestive system (OUC-201812050311); (B). upper beak (OUC-201808200302), lateral view; (C). lower beak (OUC-201808200302), lateral view; (D). lower beak (OUC-201808200302), ventral view; (E,F). radulae (OUC-201812050308). Scale bars: (A–D) = 10 mm; (E,F) = 200 μm. Abbreviations: a, anus; asg, anterior salivary gland; bm, buccal mass; c, crest; ca, caecum; cr, crop; dg, digestive gland; h, hood; I, intestine; is, ink sac; o, oesophagus; psg, posterior salivary gland; r, rostrum; s, stomach; w, wing; lw, lateral wall.
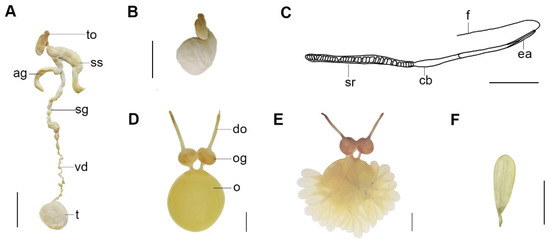
Figure 6.
Callistoctopus tenuipes sp. nov. (A,B). Male reproductive tract (OUC-201812050310); (C). spermatophore (OUC-201812050310), (D). female reproductive tract (OUC-201812050312), (E,F). eggs (OUC-201812050312). Scale bars: (A–F) = 10 mm. Abbreviations: ag, accessory gland; cb, cement body; do, distal oviduct; ea, ejaculatory apparatus; o, ovary; og, oviducal gland; sg, spermatophore gland; sr, sperm reservoir; ss, spermatophore storage sac; t, testis; f, filament; to, terminal organ (penis); vd, vas deferens.
Holotype. OUC-201812050307, mature male, 37.6 mm ML, 23.42° N, 117.85° E, 5 December 2018, coll.
Paratypes. OUC-201812050308, mature male, 41.4 mm ML, 23.42° N, 117.85° E, 5 December 2018, coll. OUC-201812050309, mature male, 42.8 mm ML, 23.42° N, 117.85° E, 5 December 2018, coll. OUC-201812050310, mature male, 41.1 mm ML, 23.42° N, 117.85° E, 5 December 2018, coll. OUC-201808200302, mature male, 30.5 mm ML, 23.42° N, 117.85° E, 20 August 2018, coll. OUC-201812050311, mature female, 54.7 mm ML, 23.42° N, 117.85° E, 5 December 2018, coll. OUC-201812050312, mature female, 57.3 mm ML, 23.42° N, 117.85° E, 5 December 2018, coll. OUC-201812050313, immature female, 34.6 mm ML, 23.42° N, 117.85° E, 5 December 2018, coll.
Diagnosis: Small- to medium-sized species. Mantle ovoid to elongate. Stylets absent. Arms long and slender, with dorsal arms longest (arm formula 1 > 2 > 3 > 4). Web shallow, web formula typically A > B > C > D > E. Suckers small and biserial, enlarged suckers absent. Hectocotylised arm with 67–73 suckers. Ligula moderate size (around 5% of hectocotylised arm length), triangular with shallow groove. Funnel organ W-shaped. Gills with 8 to 10 lamellae per demibranch. Colour typically reddish-orange to reddish-brown in fresh specimens. Lateral margins of arms start with two lines of chromatophores under the skin, then increase to three rows and end in a single line.
Description: The following description is based on five mature males and three females (two mature, one immature). Counts, measurements, and indices are presented in Table 3. Small- to moderate-sized species (ML 30.5–57.3 mm). Total length 146.5–318.8 mm. Body weight to 27.3 g. Skin smooth. Mantle ovoid to elongate. Mantle opening length moderate (PAI 63.1–101.1). Head width narrower than mantle width (HWI 25.6–40.7). Funnel short (FLI 27.7–38.9), free funnel length around 33–58% funnel length (FFLI 33.3–58.4). Funnel organ W-shaped (Figure 4D), outer limbs approximately equal in length with medial ones. Arms long (ALI 148.9–635.7) and slender (AWI 11.4–20.7), dorsal arms longest (typical arm formula 1 > 2 > 3 > 4). Webs shallow (WDI 3.5–13.8) (Figure 4B). Dorsal webs deepest, ventral webs shallowest (web formula A > B > C > D > E).

Table 3.
Measurements (mm), counts, and morphometric indices of Callistoctopus tenuipes sp. nov.
Suckers in two rows, small-sized (SDIn 3.3–6.2) (Figure 4B), enlarged suckers absent. The third right arm of mature males hectocotylised with 67–73 suckers, approximately 50% length of the opposite arm (OAI 45.9–52.6). Ligula small, triangular with shallow groove (Figure 4C), about 5% of arm length (LLI 4.4–7.2). Calamus of moderate size (CaLI 24.4–44.4). Gills with 8–10 lamellae per demibranch.
Digestive tract (Figure 5A): Buccal mass moderate size. Anterior salivary glands small. Posterior salivary glands large, triangular, and as long as buccal mass. Crop well developed. Stomach about as large as caecum. Caecum with about one whorl. Intestine long. Digestive gland well developed, dark grey. Ink sac present, embedded in digestive gland, and attached to the intestine posteriorly, then opening into anus. Anal flaps small (Figure 4E).
Chitinous beaks dark brown. Upper beak (Figure 5B) with short hood, sharp and small rostrum. Crest curved and longer than total wing length. Lower break (Figure 5C,D) with a medium-sized hood, moderate broad wings, and short lateral wings. Radula (Figure 5E,F) with seven teeth and two marginal plates per transverse row. Rachidian teeth with two or three lateral cups on each side. First lateral teeth small and sharp. Second lateral teeth larger than the first, sharp. Third lateral teeth long, curved, sharply pointed, longer than second lateral teeth. Marginal plates flat.
Male reproductive tract (Figure 6A): Terminal organ ‘—’ shaped (Figure 6B) in mature males. Spermatophore gland long. Accessory gland curved, almost same length as the Needham’s sac. Spermatophore storage sac long and slender. Vas deferens short, narrow. Testis roundish and large. Spermatophores narrow (Figure 6C), average length 46.4 mm, few (4–6 in storage sac).
Female reproductive tract (Figure 6D–F): Ovary large and round in mature females. Proximal oviducts short and narrow. Distal oviducts long and thin. About 110 eggs in ovary of a single mature female. Largest mature ovarian eggs approximately 15.1 mm.
Colouration: Fresh specimens reddish orange. Skin smooth with no papillae on dorsal mantle. Two or more lines of chromatophores obviously presented on the lateral margins of arms under the skin (Figure 4A).
Etymology: The species name tenuipes is proposed for its very narrow arms.
Distribution: Through the survey of fishermen and our sampling collection, this species is mainly found in the East China Sea to the South China Sea, and potentially south to Hainan Province, including Beibu Gulf.
Remarks: The species reported in this study have differences between males and females. Generally, mature females are larger than males. To determine the maturation times, we collected samples three times from the same sea area and found that males and females mature synchronously.
4. Discussion
As mentioned above, C. tenuipes sp. nov. was misidentified as the juvenile of O. minor. To prove the validity of the nominal taxon C. tenuipes sp. nov., we compared it with the original descriptive features of three unsolved subspecies of O. minor. Table 4 shows that C. tenuipes sp. nov. differs from the other three subspecies, mainly in spots, gill count, funnel organ shape, enlarged suckers, and ligula shape. Meanwhile, C. tenuipes sp. nov. is distinct from C. xiaohongxu in body size (mature individuals to 39.2 g versus 27.3 g), funnel organ shape (W-shaped versus \ ∧ /-shaped), web depth index (WDI 3.5–13.8 versus 15.7–22.9), arm length index (ALI 247.8–635.7 versus 154.9–336.3), and ligula index (LLI 4.4–7.2 versus 7.0–11.6). On the other hand, C. tenuipes sp. nov. resembles C. xiaohongxu in orange skin as in the fish market. However, in mature individuals, C. tenuipes sp. nov. has a smaller body size and longer arms than C. xiaohongxu. Notably, chromatophores on the lateral margins of arms under the skin were also found in O. minor and C. xiaohongxu. However, the arrangement of chromatophores in the three species was not consistent. In addition, small chromatophores of an unresolved octopus Callistoctopus sp.1, collected by Kaneko, et al. [13] from Vietnamese waters, were scattered on the surface of the arms. It remains to be seen whether chromatophores will be found in more species and whether there is a pattern of arrangement of chromatophores in certain taxa.

Table 4.
Comparison of Callistoctopus tenuipes sp. nov., Callistoctopus xiaohongxu, and three subspecies of Octopus minor group.
In the molecular analysis, the pairwise K2P genetic distances displayed in Table 3 indicate that C. tenuipes sp. nov. can be separated as a new species since the genetic distance between other sequences of Octopodidae analysed in this study ranged from 8.58 to 23.79%. Three species delimitation analyses also strongly supported C. tenuipes sp. nov. as a separate new species. Phylogenetic topologies showed that C. tenuipes sp. nov. may have a close relationship with C. xiaohongxu and O. minor. Regarding the taxonomic status of O. minor, Kaneko, et al. [32] placed it into Callistoctopus based on its morphological characteristics and DNA-barcoding analyses. In this study, three O. minor sequences converged to the Callistocotpus clade, which may indicate that O. minor had a closer affinity to the genus Callistocotpus than Octopus. However, the taxonomic classification of O. minor still needs further study.
In summary, we described a new octopod species, C. tenuipes sp. nov., using morphological and molecular methods. The South China Sea coastal current flows from Guangdong Province, and one branch flows southwest along the Hainan coast. Considering the current system, we speculate that this new octopus will be collected in Hainan and Beibu Gulf after a comprehensive resource survey. Our work adds some useful information to the classification of octopuses. Moreover, this study increases the knowledge of species diversity in the Southeast China Sea.
Author Contributions
Conceptualization, X.Z., J.L. and C.X.; methodology, J.L. and C.X.; software, J.L. and C.X.; validation, J.L., X.Z. and C.X.; formal analysis, J.L. and X.Z.; investigation, J.L. and X.Z.; resources, X.Z. and C.X.; data curation, J.L. and X.Z.; writing—original draft preparation, J.L.; writing—review and editing, X.Z.; visualization, J.L., X.Z. and C.X.; supervision, X.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (Project NO. 32170536 and 31672257).
Institutional Review Board Statement
The animal study protocol was approved by the Experimental Animal Ethics Committee of the Ocean University of China.
Data Availability Statement
The molecular data presented in this study are openly available in the National Center for Biotechnology Information under the GenBank accession numbers OP184666-OP184673.
Acknowledgments
We would like to thank Chungcheng Lu and the anonymous reviewers for their helpful comments and advice on the manuscript. We are also grateful to Feige Sunxie for collecting specimens and Poungthong Itsaret for helping to take a few photos of the new species.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Liu, J.Y. Status of marine biodiversity of the China seas. PLoS ONE 2013, 8, e50719. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.-W.; Lu, C.-C. Two new species of Sepia (Doratosepion) (Cephalopoda: Sepiidae) from Taiwan, based on morphological and molecular data. Phuket Mar. Biol. Cent. Res. Bull. 2005, 66, 51–69. [Google Scholar]
- Liao, J.X.; Lu, C.C. A new species of Cistopus (Cephalopoda: Octopodidae) from Taiwan and morphology of mucous pouches. J. Molluscan Stud. 2009, 75, 269–278. [Google Scholar] [CrossRef][Green Version]
- Zheng, X.; Lin, X.; Lu, C.; Ma, R. A new species of Cistopus Gray, 1849 (Cephalopoda: Octopodidae) from the East and South China Seas and phylogenetic analysis based on the mitochondrial COI gene. J. Nat. Hist. 2012, 46, 355–368. [Google Scholar] [CrossRef]
- Zheng, X.; Xu, C.; Li, J. Morphological description and mitochondrial DNA-based phylogenetic placement of a new species of Callistoctopus Taki, 1964 (Cephalopoda, Octopodidae) from the southeast waters of China. ZooKeys 2022, 1121, 1–15. [Google Scholar] [CrossRef]
- Lee, K.-S.; Lu, C.-C. Histioteuthis miranda (Berry, 1918) (Cephalopoda: Histioteuthidae) from the North Pacific Ocean. Collect. Res. 2019, 33, 9–18. [Google Scholar]
- Tang, Y.; Zheng, X.; Li, Q. Redescription of Amphioctopus ovulum (Sasaki, 1917) (Cephalopoda: Octopodidae) and comparative morphological analyses among three species of violet-ringed octopods. Invertebr. Syst. 2020, 34, 823–836. [Google Scholar] [CrossRef]
- Lu, C.C.; Zheng, X.D.; Lin, X.Z. Diversity of Cephalopoda from the waters of the Chinese mainland and Taiwan. In Proceedings of the 1st Taiwan Symposium of Marine Biodiversity Studies; Lin, M., Wang, C.G., Eds.; Ocean: Beijing, China, 2012; pp. 76–87. [Google Scholar]
- Xu, R.; Lü, Y.; Tang, Y.; Chen, Z.; Xu, C.; Zhang, X.; Zheng, X. DNA Barcoding reveals high hidden species diversity of Chinese waters in the Cephalopoda. Front. Mar. Sci. 2022, 9, 830381. [Google Scholar] [CrossRef]
- Norman, M.D. Cephalopods: A World Guide; ConchBooks: Hackenheim, German, 2000; pp. 1–320. [Google Scholar]
- Norman, M.D.; Hochberg, F.G. The current state of octopus taxonomy. Phuket Mar. Biol. Cent. Res. Bull. 2005, 66, 127–154. [Google Scholar]
- Jesus, M.D.; Sales, J.B.; Martins, R.S.; Ready, J.S.; Costa, T.A.S.; Ablett, J.D.; Schiavetti, A. Traditional knowledge aids description when resolving the taxonomic status of unsettled species using classical and molecular taxonomy: The case of the shallow-water octopus Callistoctopus furvus (Gould, 1852) from the western Atlantic Ocean. Front. Mar. Sci. 2021, 7, 595244. [Google Scholar] [CrossRef]
- Kaneko, N.; Kubodera, T.; Dinh, T.; Chung, B.D. Shallow-water benthic octopuses (Cephalopoda, Octopodidae) collected from the coastal waters of Vietnam. Bull. Natl. Mus. Nat. Sci. Ser. A 2008, 34, 105–122. [Google Scholar]
- Sreeja, V. Taxonomy and Diversity of Cephalopods (Mollusca: Cephalopoda) of Kerala Coast. Ph.D. Thesis, University of Kerala, Thiruvananthapuram, India, 2013; pp. 138–141. [Google Scholar]
- Dai, L.; Zheng, X.; Kong, L.; Li, Q. DNA barcoding analysis of Coleoidea (Mollusca: Cephalopoda) from Chinese waters. Mol. Ecol. Resour. 2012, 12, 437–447. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Bo, Q.; Zheng, X. A divergent lineage among Octopus minor (Sasaki, 1920) populations in the Northwest Pacific supported by DNA barcoding. Mar. Biol. Res. 2018, 14, 335–344. [Google Scholar] [CrossRef]
- Tang, Y.; Zheng, X.; Liu, H.; Sunxie, F. Population genetics and comparative mitogenomic analyses reveal cryptic diversity of Amphioctopus neglectus (Cephalopoda: Octopodidae). Genomics 2020, 112, 3893–3902. [Google Scholar] [CrossRef]
- Roper, C.F.E.; Voss, G.L. Guidelines for taxonomic descriptions of cephalopod species. Mem. Natl. Mus. Vic. 1983, 44, 48–63. [Google Scholar] [CrossRef]
- Norman, M.D.; Sweeney, M.J. The shallow-water octopus (Cephalopoda: Octopodidae) of the Philippines. Invertebr. Taxon. 1997, 11, 89–140. [Google Scholar] [CrossRef]
- Huffard, C.L.; Hochberg, F.G. Description of a new species of the genus Amphioctopus (Mollusca: Octopodidae) from the Hawai’ian Islands. Molluscan Res. 2005, 25, 113–128. [Google Scholar]
- Winnepenninckx, B. Extraction of high molecular weight DNA from molluscs. Trends Genet. 1993, 9, 407. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.B.; Vrijenhoek, R.C. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Bui Quang, M.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Nguyen, M.A.T.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zheng, X.; Ma, Y.; Li, Q. Complete mitochondrial genome and phylogenetic relationship analyses of Amphioctopus aegina (Gray, 1849) (Cephalopoda: Octopodidae). Mitochondrial DNA Part A 2017, 28, 17–18. [Google Scholar] [CrossRef]
- Tang, Y.; Zheng, X.; Ma, Y.; Cheng, R.; Li, Q. The complete mitochondrial genome of Amphioctopus marginatus (Cephalopoda: Octopodidae) and the exploration for the optimal DNA barcoding in Octopodidae. Conserv. Genet. Resour. 2017, 10, 115–118. [Google Scholar] [CrossRef]
- Tang, Y.; Zheng, X.; Zhong, H.; Li, Q. Phylogenetics and comparative analysis of the mitochondrial genomes of three violet-ringed octopuses. Zool. Scr. 2019, 48, 482–493. [Google Scholar] [CrossRef]
- Kaneko, N.; Kubodera, T.; Iguchis, A. Taxonomic study of shallow-water octopuses (Cephalopoda: Octopodidae) in Japan and adjacent waters using mitochondrial genes with perspectives on octopus DNA barcoding. Malacologia 2011, 54, 97–108. [Google Scholar] [CrossRef]
- Lima, F.D.; Strugnell, J.M.; Leite, T.S.; Lima, S.M.Q. A biogeographic framework of octopod species diversification: The role of the Isthmus of Panama. PeerJ 2020, 8, e8691. [Google Scholar] [CrossRef]
- Strugnell, J.M.; Norman, M.D.; Vecchione, M.; Guzik, M.; Allcock, A.L. The ink sac clouds octopod evolutionary history. Hydrobiologia 2014, 725, 215–235. [Google Scholar] [CrossRef]
- Cheng, R.; Zheng, X.; Ma, Y.; Li, Q. The complete mitochondrial genomes of two octopods Cistopus chinensis and Cistopus taiwanicus: Revealing the phylogenetic position of the genus Cistopus within the order Octopoda. PLoS ONE 2013, 8, e84216. [Google Scholar] [CrossRef] [PubMed]
- Cheng, R.; Zheng, X.; Lin, X.; Yang, J.; Li, Q. Determination of the complete mitochondrial DNA sequence of Octopus minor. Mol. Biol. Rep. 2011, 39, 3461–3470. [Google Scholar] [CrossRef] [PubMed]
- Takumiya, M.; Kobayashi, M.; Tsuneki, K.; Furuya, H. Phylogenetic relationships among major species of Japanese coleoid cephalopods (Mollusca: Cephalopoda) using three mitochondrial DNA sequences. Zool. Sci. 2005, 22, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chen, S.; Chen, W.; Xie, S.; Ni, X.; Zheng, X. Morphological and genetic diversity analysis of Octopus sinensis in Nanji Islands. Oceanol. Limnol. Sin. 2022, 53, 486–495. (In Chinese) [Google Scholar] [CrossRef]
- Amor, M.D.; Norman, M.D.; Roura, A.; Leite, T.S.; Gleadall, I.G.; Reid, A.; Perales-Raya, C.; Lu, C.C.; Silvey, C.J.; Vidal, E.A.G.; et al. Morphological assessment of the Octopus vulgaris species complex evaluated in light of molecular-based phylogenetic inferences. Zool. Scr. 2016, 46, 275–288. [Google Scholar] [CrossRef]
- Yokobori, S.; Lindsay, D.J.; Yoshida, M.; Tsuchiya, K.; Yamagishi, A.; Maruyama, T.; Oshima, T. Mitochondrial genome structure and evolution in the living fossil vampire squid, Vampyroteuthis infernalis, and extant cephalopods. Mol. Phylogen. Evol. 2007, 44, 898–910. [Google Scholar] [CrossRef]
- Puillandre, N.; Lambert, A.; Brouillet, S.; Achaz, G. ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol. Ecol. 2012, 21, 1864–1877. [Google Scholar] [CrossRef]
- Ezard, T.; Fujisawa, T.; Barraclough, T. Splits: Species’ Limits by Threshold Statistics, version 1.0-20. 2021.
- Bouckaert, R.; Vaughan, T.G.; Barido-Sottani, J.; Duchene, S.; Fourment, M.; Gavryushkina, A.; Heled, J.; Jones, G.; Kuhnert, D.; De Maio, N.; et al. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019, 15, e1006650. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Zhang, J.; Kapli, P.; Pavlidis, P.; Stamatakis, A. A general species delimitation method with applications to phylogenetic placements. Bioinformatics 2013, 29, 2869–2876. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, M. A monograph of dibranchiate cephalopods of the Japanese and adjacent waters. J. Coll. Agric. Hokkaido Imp. Univ. 1929, 20, 90–96. [Google Scholar]
- Sasaki, M. Report on cephalopods collected during 1906 by the United States Bureau of Fisheries steamer “Albatross” in the northwestern Pacific. Proc. United States Natl. Mus. 1920, 57, 181–182. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).