Dietary Variation Is Driven by Landscape Heterogeneity in an Insular Omnivorous Endemic Lizard, Revealed by DNA Metabarcoding
Abstract
1. Introduction
2. Materials and Methods
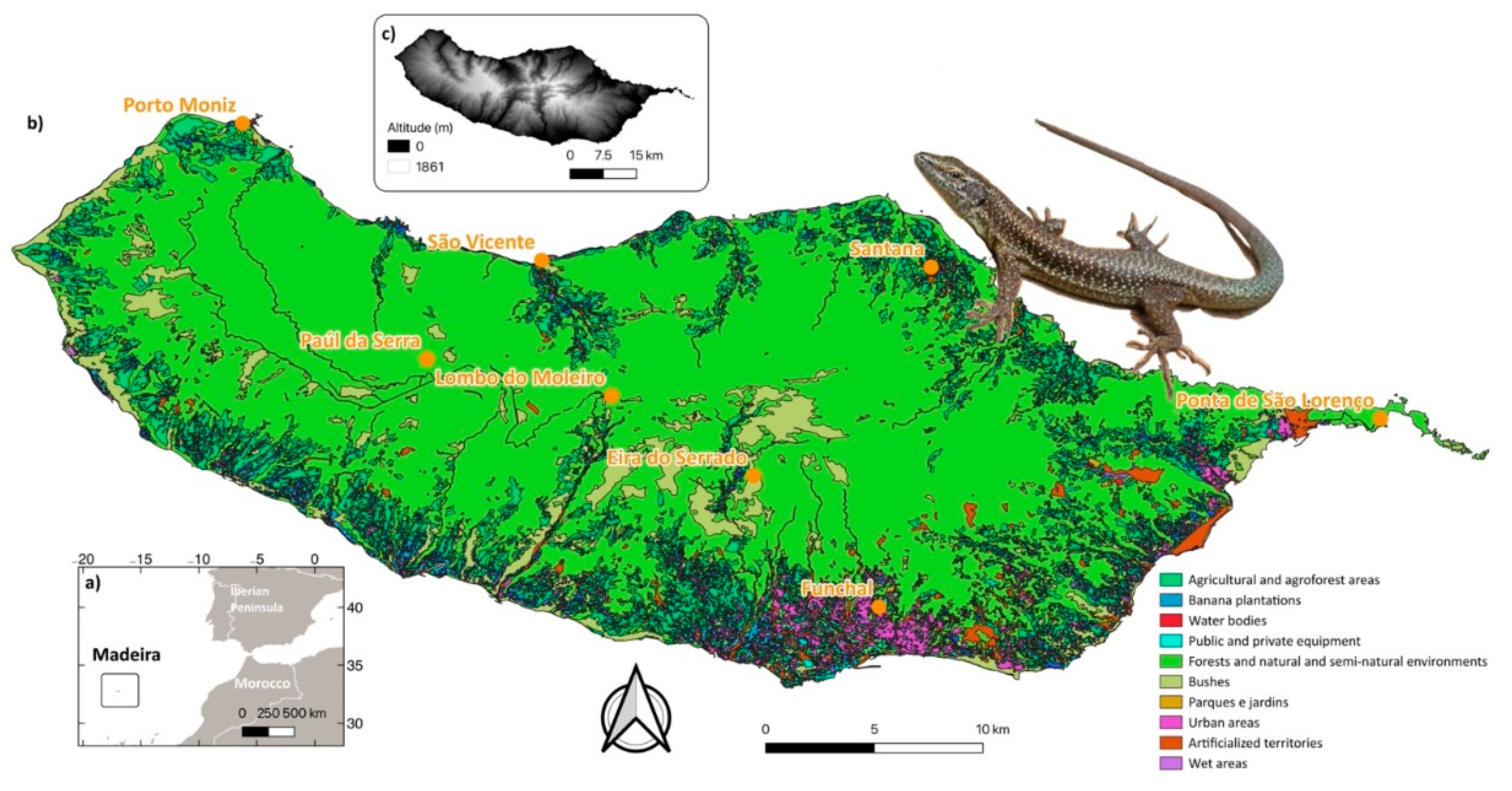
2.1. Study Area
2.2. Field Sampling
2.3. Molecular Analysis
2.4. Analyses of DNA Sequence Data
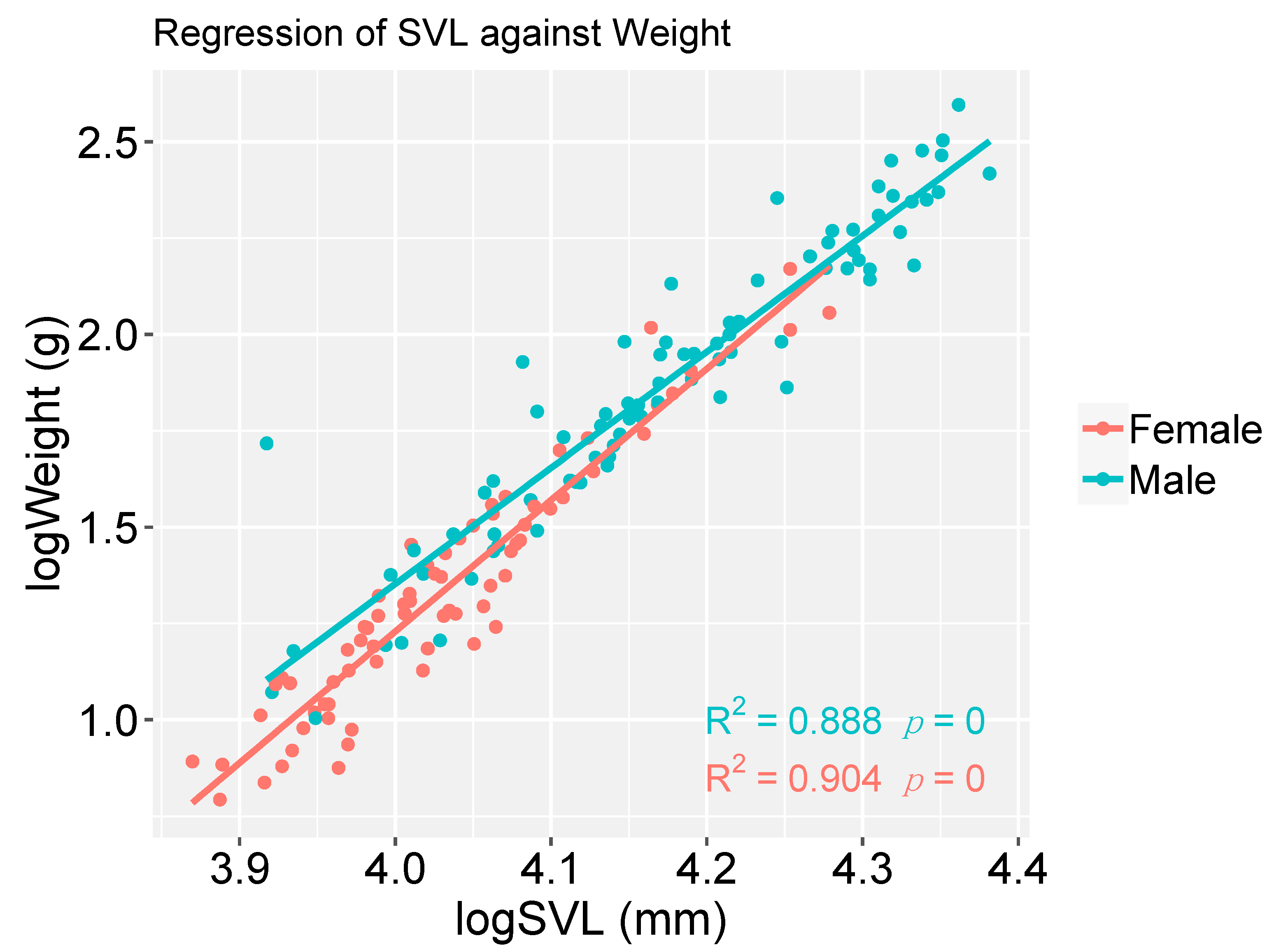
2.5. Statistical Analyses
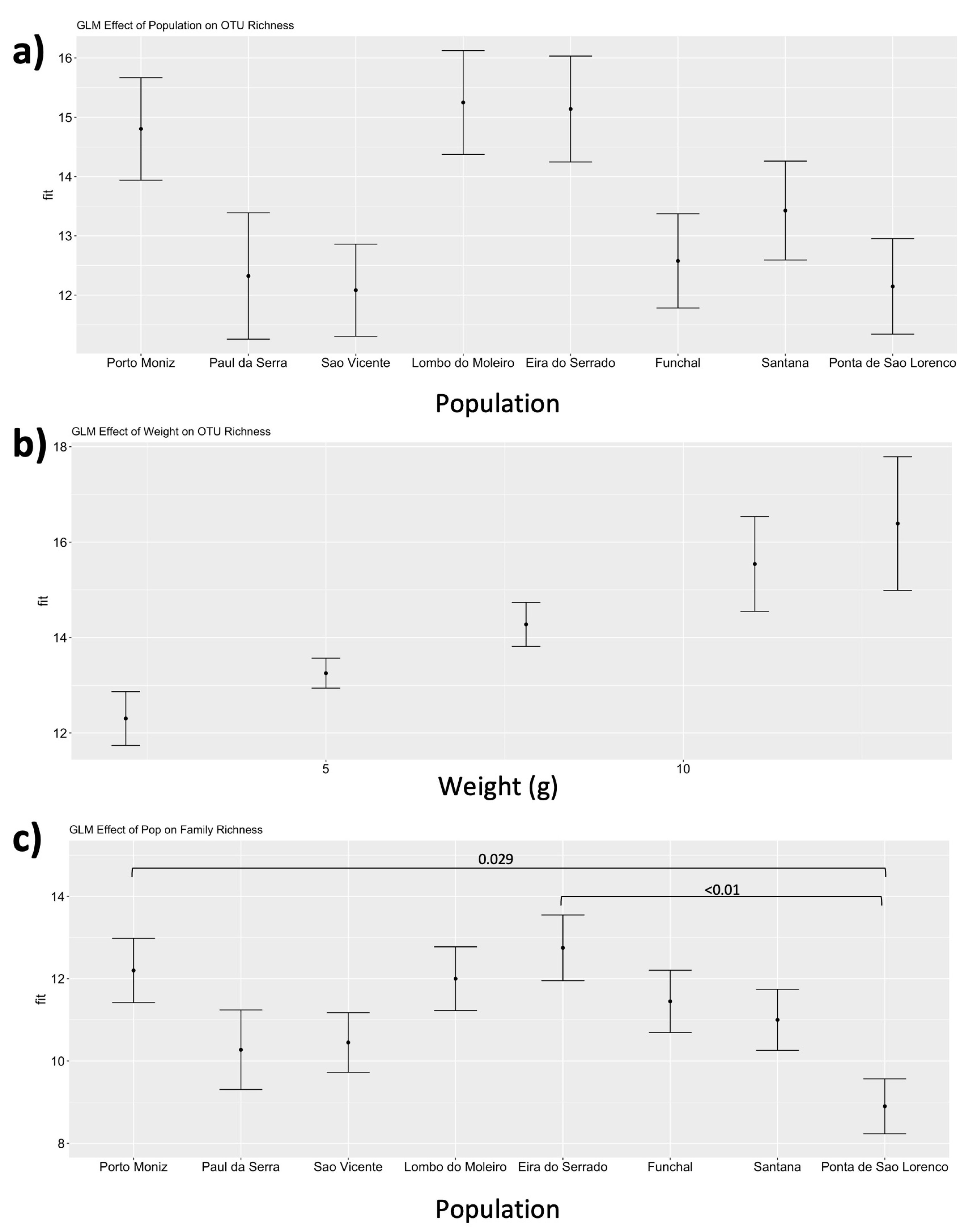
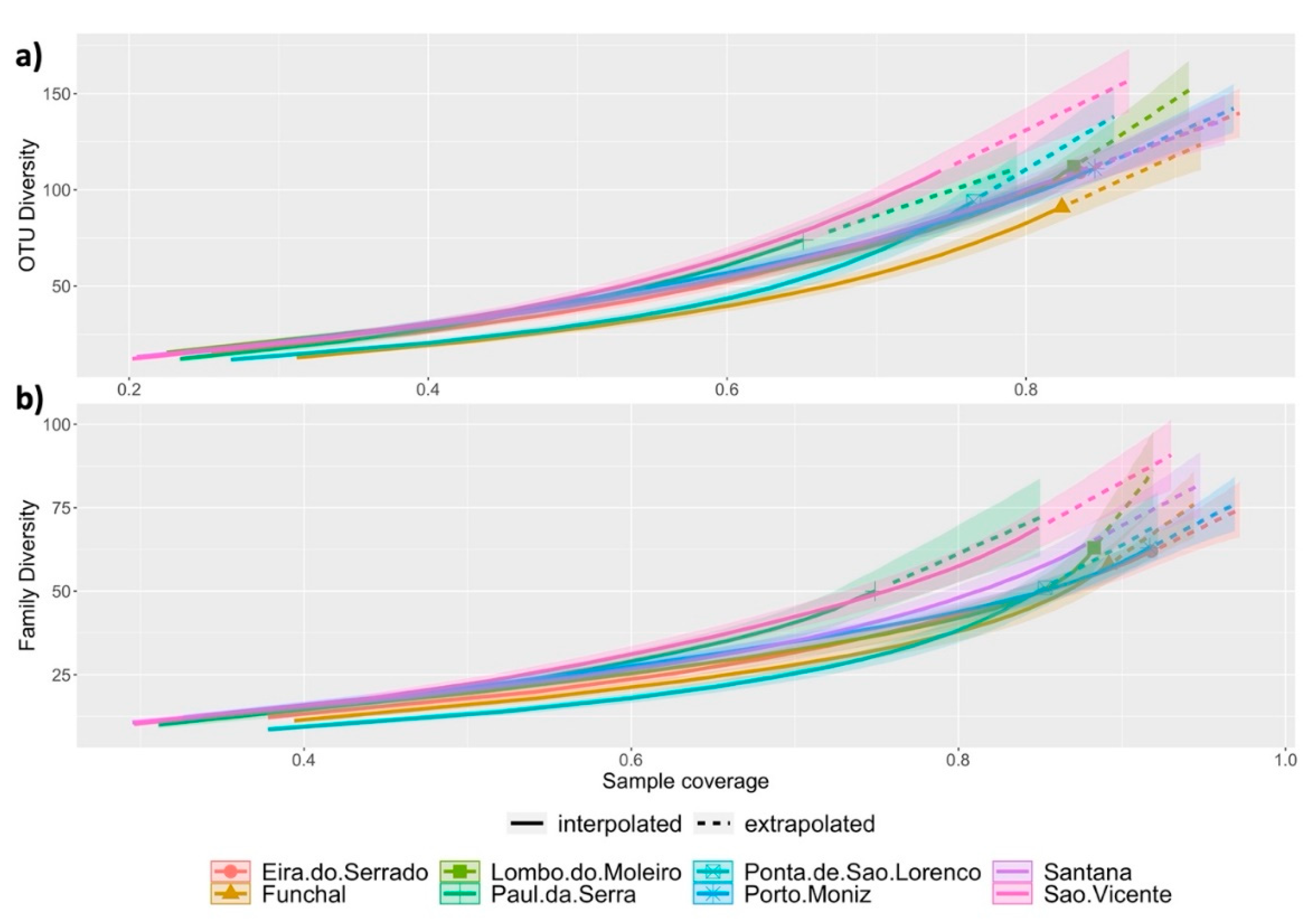
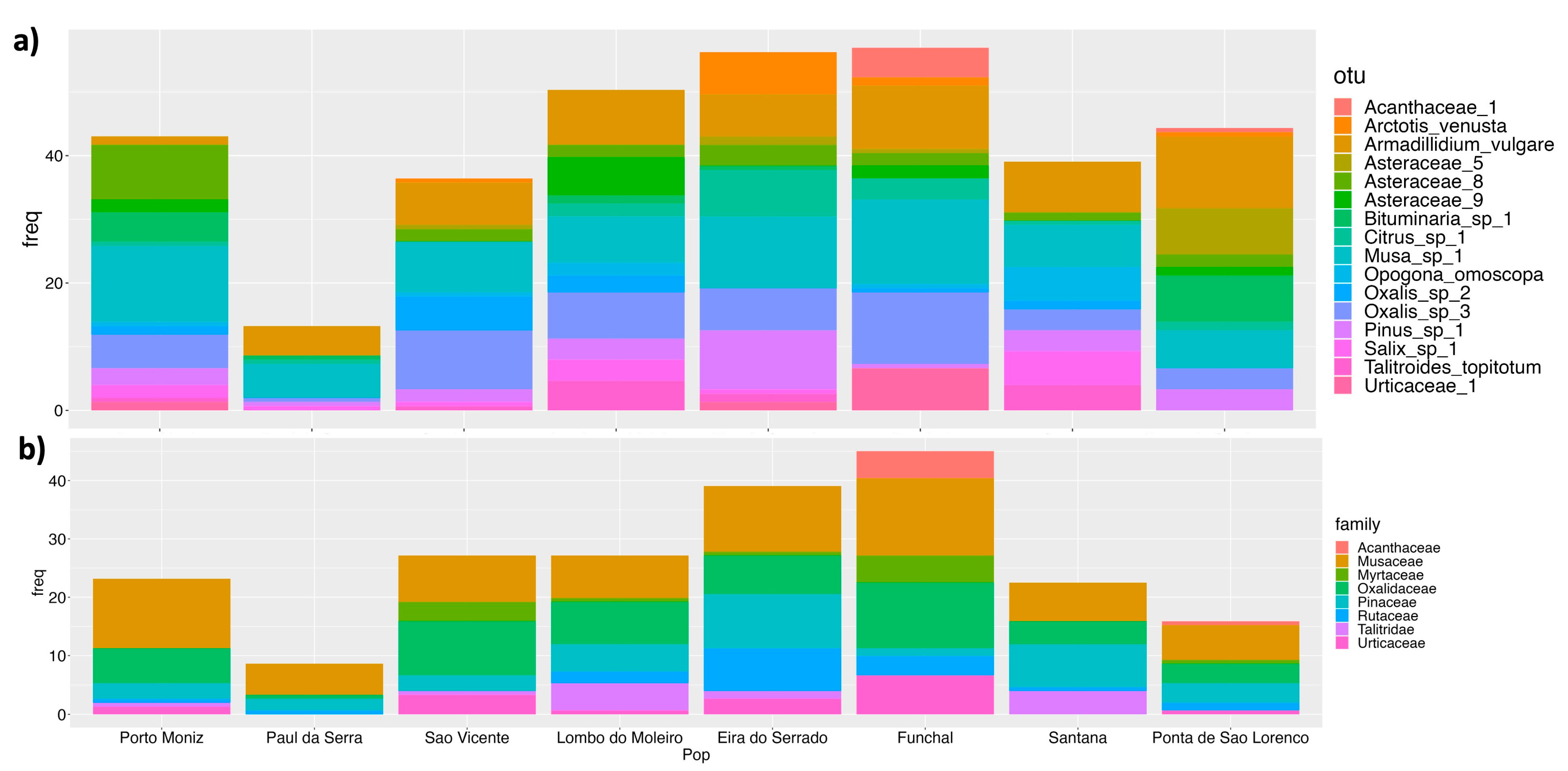
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Crawley, M.J. Herbivory. The Dynamics of Animal–Plant Interactions; Blackwell Scientific Publications: Hoboken, NJ, USA, 1983. [Google Scholar]
- King, G.M. Reptiles and Herbivory; University of California Press: Berkeley, CA, USA, 1996. [Google Scholar]
- Owen-Smith, R.N. Adaptive Herbivore Ecology: From Resources to Populations in Variable Environments; Cambridge University Press: Cambridge, UK, 2002. [Google Scholar]
- Pough, F.H.; Janis, C.M.; Heiser, J.B. Vertebrate Life; Prentice Hall: Upper Saddle River, NJ, USA, 1999. [Google Scholar]
- Sues, H.-D. Evolution of Herbivory in Terrestrial Vertebrates; Cambridge University Press: Cambridge, UK, 2000. [Google Scholar]
- Krebs, C.J. Ecological Methodology, 2nd ed.; Benjamin Cummings: Menlo Park, CA, USA, 1999. [Google Scholar]
- Carretero, M.A. From set menu to a la carte. Linking issues in trophic ecology of Mediterranean lacertids. Ital. J. Zool. 2004, 71, 121–133. [Google Scholar] [CrossRef]
- Tirado, C.; Cortés, A.; Miranda-Urbina, E.; Carretero, M. Trophic preferences in an assemblage of mammal herbivores from Andean Puna (Northern Chile). J. Arid Environ. 2012, 79, 8–12. [Google Scholar] [CrossRef]
- Cooper, W.E., Jr.; Vitt, L.J. Distribution, extent, and evolution of plant consumption by lizards. J. Zool. 2002, 257, 487–517. [Google Scholar] [CrossRef]
- Van Damme, R. Evolution of herbivory in lacertid lizards: Effects of insularity and body size. J. Herpetol. 1999, 33, 663–674. [Google Scholar] [CrossRef]
- Pough, F.H. Lizard energetics and diet. Ecology 1973, 54, 837–844. [Google Scholar] [CrossRef]
- Losos, J.B.; Ricklefs, R.E. Adaptation and diversification on islands. Nature 2009, 457, 830–836. [Google Scholar] [CrossRef]
- Hofman, C.A.; Rick, T.C.; Hawkins, M.T.; Funk, W.C.; Ralls, K.; Boser, C.L.; Collins, P.W.; Coonan, T.; King, J.L.; Morrison, S.A. Mitochondrial genomes suggest rapid evolution of dwarf California Channel Islands foxes (Urocyon littoralis). PLoS ONE 2015, 10, e0118240. [Google Scholar] [CrossRef]
- Buglione, M.; Petrelli, S.; Maselli, V.; Trapanese, M.; Salvemini, M.; Aceto, S.; Di Cosmo, A.; Fulgione, D. Fixation of genetic variation and optimization of gene expression: The speed of evolution in isolated lizard populations undergoing Reverse Island Syndrome. PLoS ONE 2019, 14, e0224607. [Google Scholar] [CrossRef]
- Brown, R.; Pérez-Mellado, V. Ecological energetics and food acquisition in dense Menorcan islet populations of the lizard Podarcis lilfordi. Funct. Ecol. 1994, 8, 427–434. [Google Scholar] [CrossRef]
- Pafilis, P.; Foufopoulos, J.; Poulakakis, N.; Lymberakis, P.; Valakos, E. Digestive performance in five Mediterranean lizard species: Effects of temperature and insularity. J. Comp. Physiol. B 2007, 177, 49–60. [Google Scholar] [CrossRef]
- Pérez-Mellado, V.; Corti, C. Dietary adaptations and herbivory in lacertid lizards of the genus Podarcis from western Mediterranean islands (Reptilia: Sauria). Bonn. Zool. Beitr 1993, 44, 193–220. [Google Scholar]
- Janzen, D.H. Sweep samples of tropical foliage insects: Effects of seasons, vegetation types, elevation, time of day, and insularity. Ecology 1973, 54, 687–708. [Google Scholar] [CrossRef]
- Olesen, J.M.; Valido, A. Lizards as pollinators and seed dispersers: An island phenomenon. Trends Ecol. Evol. 2003, 18, 177–181. [Google Scholar] [CrossRef]
- Perez-Cembranos, A.; Leon, A.; Perez-Mellado, V. Omnivory of an insular lizard: Sources of variation in the diet of Podarcis lilfordi (Squamata, Lacertidae). PLoS ONE 2016, 11, e0148947. [Google Scholar] [CrossRef] [PubMed]
- Herrel, A.; Vanhooydonck, B.; Van Damme, R. Omnivory in lacertid lizards: Adaptive evolution or constraint? J. Evol. Biol. 2004, 17, 974–984. [Google Scholar] [CrossRef]
- Roca, V. Relación entre las faunas endoparásitas de reptiles y su tipo de alimentación. Rev. Española De Herpetol. 1999, 13, 101–121. [Google Scholar]
- Buglione, M.; Ricca, E.; Petrelli, S.; Baccigalupi, L.; Troiano, C.; Saggese, A.; Rivieccio, E.; Fulgione, D. Gut microbiota plasticity in insular lizards under reversed island syndrome. Sci. Rep. 2022, 12, 12682. [Google Scholar] [CrossRef]
- Kohl, K.D.; Brun, A.; Magallanes, M.; Brinkerhoff, J.; Laspiur, A.; Acosta, J.C.; Bordenstein, S.R.; Caviedes-Vidal, E. Physiological and microbial adjustments to diet quality permit facultative herbivory in an omnivorous lizard. J. Exp. Biol. 2016, 219, 1903–1912. [Google Scholar] [CrossRef]
- Vitt, L.; Caldwell, J.; Sartorius, S.; Cooper, W., Jr.; Baird, T.; Baird, T.; Pérez-Mellado, V. Pushing the edge: Extended activity as an alternative to risky body temperatures in a herbivorous teiid lizard (Cnemidophorus murinus: Squamata). Funct. Ecol. 2005, 19, 152–158. [Google Scholar] [CrossRef]
- Sagonas, K.; Pafilis, P.; Valakos, E.D. Effects of insularity on digestion: Living on islands induces shifts in physiological and morphological traits in island reptiles. Sci. Nat. 2015, 102, 55. [Google Scholar] [CrossRef]
- Rocha, C.F.D. Ontogenetic shift in the rate of plant consumption in a tropical lizard (Liolaemus lutzae). J. Herpetol. 1998, 32, 274–279. [Google Scholar] [CrossRef]
- Fuentes, E.R.; Di Castri, F. Ensayo de Herbivoría Experimental en Especies de Liolaemus (Iguanidae) Chilenos. Anales del Museo de Historia Natural de Valparaíso (Chile) 2015, 8, 66–75. [Google Scholar]
- Schluter, D. Body size, prey size and herbivory in the Galapagos lava lizard, Tropidurus. Oikos 1984, 43, 291–300. [Google Scholar] [CrossRef]
- Cunningham, P.L. Notes on the diet, survival rate, and burrow specifics of Uromastyx aegyptius. Asiat. Herpetol. Res. 2001, 9, 30–33. [Google Scholar]
- Espinoza, R.E.; Wiens, J.J.; Tracy, C.R. Recurrent evolution of herbivory in small, cold-climate lizards: Breaking the ecophysiological rules of reptilian herbivory. Proc. Natl. Acad. Sci. USA 2004, 101, 16819–16824. [Google Scholar] [CrossRef]
- Brehm, A.; Jesus, J.; Spınola, H.; Alves, C.; Vicente, L.; Harris, D. Phylogeography of the Madeiran endemic lizard Lacerta dugesii inferred from mtDNA sequences. Mol. Phylogenetics Evol. 2003, 26, 222–230. [Google Scholar] [CrossRef]
- Paulo Sá-Sousa, P.; Sindaco, R. Teira dugesii (errata version published in 2017). In The IUCN Red List of Threatened Species 2009; 2009; e.T61521A121720531. Available online: https://www.iucnredlist.org/ (accessed on 17 November 2022).
- Jesus, J. Lacerta dugesii. In Atlas dos Anfíbios e Répteis de Portugal; Loureiro, A., Ferrand de Almeida, N., Carretero, M.A., Paulo, O.S., Eds.; Instituto de Conservação da Natureza e da Biodiversidade: Lisboa, Portugal, 2008; pp. 190–191. [Google Scholar]
- Ulfstrand, S. On the vertebrate fauna of the Azores. Bol. Mus. Munic. Funchal 1961, 14, 75–86. [Google Scholar]
- Malkmus, R. Zur Verbreitung von Rana perezi und Lacerta dugesii auf den Azoren. Nachr. Des Naturwissen. Mus. Aschaffenbg. 1985, 92, 37–69. [Google Scholar]
- Sá-Sousa, P. The introduced Madeiran lizard, Lacerta (Teira) dugesii in Lisbon. Amphib.-Reptil. 1995, 16, 211–214. [Google Scholar] [CrossRef]
- Ferreira, A.I.; Vasconcelos, D.S.; Harris, D.J. Origins of an introduced Teira dugesii (Lacertidae) population in Porto. Herpetol Notes, 2022; in press. [Google Scholar]
- Mateo, J.A.; Ayres, C.; López-Jurado, L.F. Los anfibios y reptiles naturalizados en España: Historia y evolución de una problemática creciente. Historia y evolución de una problemática creciente. Boletín Asoc. Herpetol. Esp. 2011, 22, 2–42. [Google Scholar]
- Santos, L.-D.; López-Jurado, L.F.; Hernández-Peñate, A.; Mateo, J.A. Una nueva población de lagartija (Teira dugesii) en Las Palmas de Gran Canaria. Boletín Asoc. Herpetol. Esp. 2013, 24, 102–103. [Google Scholar]
- Sadek, R.A. The diet of the Madeiran lizard Lacerta dugesii. Zool. J. Linn. Soc. 1981, 73, 313–341. [Google Scholar] [CrossRef]
- Matias, R.; Rebelo, R.; Granadeiro, J.; Catry, P. Predation by Madeiran wall lizards Teira dugesii on Cory’s shearwater Calonectris diomedea hatchlings at Selvagem Grande, North Atlantic. Waterbirds 2009, 32, 600–603. [Google Scholar] [CrossRef]
- Neves, V.C.; Nava, C.; Monteiro, E.V.; Monteiro, P.R.; Bried, J. Depredation of Monteiro’s Storm-petrel (Hydrobates monteiroi) chicks by Madeiran wall lizards (Lacerta dugesii). Waterbirds 2017, 40, 82–86. [Google Scholar] [CrossRef]
- Neves, V.; Rund, D.; Pinho, C.J.; Vasconcelos, R.; Bustamante, P.; Quillfeldt, P. Diet of the exotic Madeiran wall lizard: First insights into trophic interactions in an Atlantic seabird sanctuary. Herpetozoa 2022, 35, 107–113. [Google Scholar] [CrossRef]
- Elvers, I. Flower-visiting lizards on Madeira. Bot. Not. 1977, 130, 231–234. [Google Scholar]
- Lopez-Darias, M.; Vanhooydonck, B.; Cornette, R.; Herrel, A. Sex-specific differences in ecomorphological relationships in lizards of the genus Gallotia. Funct. Ecol. 2015, 29, 506–514. [Google Scholar] [CrossRef]
- Carretero, M.A.; Jorge, F.; Llorente, G.A.; Roca, V. Relationships between helminth communities and diet in Canarian lizards: The evidence from Gallotia atlantica (Squamata: Lacertidae). J. Nat. Hist. 2014, 48, 1199–1216. [Google Scholar] [CrossRef]
- Martin, J.; Llorente, G.; Roca, V.; Carretero, M.; Montori, A.; Santos, X.; Romeu, R. Relationship between diet and helminths in Gallotia caesaris (Sauria: Lacertidae). Zoology 2005, 108, 121–130. [Google Scholar] [CrossRef]
- Carretero, M.A.; Llorente, G.A.; Santos, X.; Montori, A. The diet of an introduced population of Podarcis pityusensis. ls herbivory fixed? In Mediterranean Basin Lacertid Lizards. A Biological Approach; Vicente, L., Crespo, E.G., Eds.; ICN: Lisboa, Portugal, 2001; pp. 113–124. [Google Scholar]
- Gil, V.; Pinho, C.J.; Aguiar, C.A.; Jardim, C.; Rebelo, R.; Vasconcelos, R. Questioning the proverb ‘more haste, less speed’: Classic versus metabarcoding approaches for the diet study of a remote island endemic gecko. PeerJ 2020, 8, e8084. [Google Scholar] [CrossRef] [PubMed]
- Tercel, M.P.T.G.; Symondson, W.O.C.; Cuff, J.P. The problem of omnivory: A synthesis on omnivory and DNA metabarcoding. Mol. Ecol. 2021, 30, 2199–2206. [Google Scholar] [CrossRef] [PubMed]
- Taberlet, P.; Coissac, E.; Pompanon, F.; Brochmann, C.; Willerslev, E. Towards next-generation biodiversity assessment using DNA metabarcoding. Mol. Ecol. 2012, 21, 2045–2050. [Google Scholar] [CrossRef] [PubMed]
- Alberdi, A.; Aizpurua, O.; Bohmann, K.; Gopalakrishnan, S.; Lynggaard, C.; Nielsen, M.; Gilbert, M.T.P. Promises and pitfalls of using high-throughput sequencing for diet analysis. Mol. Ecol. Resour. 2019, 19, 327–348. [Google Scholar] [CrossRef] [PubMed]
- Ando, H.; Mukai, H.; Komura, T.; Dewi, T.; Ando, M.; Isagi, Y. Methodological trends and perspectives of animal dietary studies by noninvasive fecal DNA metabarcoding. Environ. DNA 2020, 2, 391–406. [Google Scholar] [CrossRef]
- Geldmacher, J.; van den Bogaard, P.; Hoernle, K.; Schmincke, H.U. The 40Ar/39Ar age dating of the Madeira Archipelago and hotspot track (eastern North Atlantic). Geochem. Geophys. Geosyst. 2000, 1, 1008. [Google Scholar] [CrossRef]
- Santos, J.M.F. Análise e modelação espácio-temporal do mosquito vetor do dengue na ilha da Madeira. Master’s Thesis, University of Lisbon, Lisbon, Portugal, 2018. [Google Scholar]
- Capelo, J.; Menezes de Sequeira, M.; Jardim, R.; Mesquita, S. Biologia e ecologia das florestas das ilhas—Madeira. In Árvores e Florestas de Portugal, Açores e Madeira—A Floresta das Ilhas; Sande Silva, J., Ed.; Fundação Luso Americana para o Desenvolvimento, Público e Liga para a Protecção da Natureza: Lisbon, Portugal, 2007; Volume 6, pp. 81–134. [Google Scholar]
- Borges, P.A.V.; Abreu, C.; Aguiar, A.M.F.; Carvalho, P.; Jardim, R.; Melo, I.; Oliveira, P.; Sérgio, C.; Serrano, A.R.M.; Vieira, P. Listagem dos Fungos, Flora e Fauna Terrestres dos Arquipélagos da Madeira e Selvagens = A List of the Terrestrial Fungi, Flora and Fauna of Madeira and Selvagens Archipelagos; Secretaria Regional do Ambiente e dos Recursos Naturais do Governo Regional da Madeira: Funchal, Madeira, 2008; p. 438. [Google Scholar]
- Galán, P.; Vicente, L. Reproductive characteristics of the insular lacertid Teira dugesii. Herpetol. J. 2003, 13, 149–154. [Google Scholar]
- Báez, M. Observaciones sobre colorido y diseño de Podarcis dugesii en la isla de Madeira (Sauria, Lacertidae). Vieraea 1990, 18, 197–203. [Google Scholar]
- Taberlet, P.; Coissac, E.; Pompanon, F.; Gielly, L.; Miquel, C.; Valentini, A.; Vermat, T.; Corthier, G.; Brochmann, C.; Willerslev, E. Power and limitations of the chloroplast trn L (UAA) intron for plant DNA barcoding. Nucleic Acids Res. 2007, 35, e14. [Google Scholar] [CrossRef]
- Vamos, E.E.; Elbrecht, V.; Leese, F. Short COI markers for freshwater macroinvertebrate metabarcoding. Metabarcoding Metagenomics 2017, e14625. [Google Scholar] [CrossRef]
- Zhang, J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 2014, 30, 614–620. [Google Scholar] [CrossRef] [PubMed]
- Martins, B.; Silva-Rocha, I.; Rocha, R.; Mata, V.A.; Gonçalves, Y.; Rato, C. Trophic interactions of an invasive gecko in an endemic-rich oceanic island: Insights using DNA metabarcoding. Front. Ecol. Evol. 2022; in press. [Google Scholar] [CrossRef]
- Andújar, C.; Arribas, P.; Gray, C.; Bruce, C.; Woodward, G.; Yu, D.W.; Vogler, A.P. Metabarcoding of freshwater invertebrates to detect the effects of a pesticide spill. Mol. Ecol. 2018, 27, 146–166. [Google Scholar] [CrossRef] [PubMed]
- Boyer, F.; Mercier, C.; Bonin, A.; Le Bras, Y.; Taberlet, P.; Coissac, E. obitools: A unix-inspired software package for DNA metabarcoding. Mol. Ecol. Resour. 2016, 16, 176–182. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- Frøslev, T.G.; Kjøller, R.; Bruun, H.H.; Ejrnæs, R.; Brunbjerg, A.K.; Pietroni, C.; Hansen, A.J. Algorithm for post-clustering curation of DNA amplicon data yields reliable biodiversity estimates. Nat. Commun. 2017, 8, 1188. [Google Scholar] [CrossRef]
- Mata, V.A.; Amorim, F.; Corley, M.F.; McCracken, G.F.; Rebelo, H.; Beja, P. Female dietary bias towards large migratory moths in the European free-tailed bat (Tadarida teniotis). Biol. Lett. 2016, 12, 20150988. [Google Scholar] [CrossRef]
- Evans, H.K.; Bunch, A.J.; Schmitt, J.D.; Hoogakker, F.J.; Carlson, K.B. High-throughput sequencing outperforms traditional morphological methods in Blue Catfish diet analysis and reveals novel insights into diet ecology. Ecol. Evol. 2021, 11, 5584–5597. [Google Scholar] [CrossRef]
- Buchner, D.; Leese, F. BOLDigger–a Python package to identify and organise sequences with the Barcode of Life Data systems. Metabarcoding Metagenomics 2020, 4, e53535. [Google Scholar] [CrossRef]
- Drummond, A.J.; Ashton, B.; Buxton, S.; Cheung, M.; Cooper, A.; Duran, C.; Field, M.; Heled, J.; Kearse, M.; Markowitz, S.; et al. Geneious [Computer Software]. 2010. Available online: http://www.geneious.com/ (accessed on 17 November 2022).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: https://www.R-project.org/ (accessed on 17 November 2022).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer-Verlag: New York, NY, USA, 2016; Available online: https://ggplot2.tidyverse.org (accessed on 17 November 2022).
- Hothorn, T.; Bretz, F.; Westfall, P. Simultaneous inference in general parametric models. Biom. J. J. Math. Methods Biosci. 2008, 50, 346–363. [Google Scholar] [CrossRef]
- Fox, J.; Weisberg, S. An R Companion to Applied Regression, 3rd ed.; Sage: Thousand Oaks CA, USA, 2019. [Google Scholar]
- Mazerolle, M.J. AICcmodavg: Model Selection and Multimodel Inference Based on (Q)AIC(c). R Package Version 2.3-1. 2020. Available online: https://cran.r-project.org/package=AICcmodavg (accessed on 17 November 2022).
- Burnham, K.P.; Andersen, D.R. Model Selection and Multi-Model Inference: A Practical Information-Theoretic Approach; Springer-Verlag: Heidelberg, Germany, 2002. [Google Scholar]
- Fox, J. Effect Displays in R for Generalised Linear Models. J. Stat. Softw. 2003, 8, 1–27. [Google Scholar] [CrossRef]
- Hsieh, T.; Ma, K.; Chao, A. iNEXT: An R package for rarefaction and extrapolation of species diversity (H ill numbers). Methods Ecol. Evol. 2016, 7, 1451–1456. [Google Scholar] [CrossRef]
- Chao, A.; Jost, L. Coverage-based rarefaction and extrapolation: Standardizing samples by completeness rather than size. Ecology 2012, 93, 2533–2547. [Google Scholar] [CrossRef]
- MacGregor-Fors, I.; Payton, M.E. Contrasting diversity values: Statistical inferences based on overlapping confidence intervals. PLoS ONE 2013, 8, e56794. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.; O’hara, R.; Simpson, G.; Solymos, P.; Stevens, M.H.H.; Wagner, H. Vegan: Community ecology package. R Package Version 2013, 2, 321–326. [Google Scholar]
- Anderson, M.J. Distance-based tests for homogeneity of multivariate dispersions. Biometrics 2006, 62, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Naumann, U.; Eddelbuettel, D.; Wilshire, J.; Warton, D.; Byrnes, J.; dos Santos Silva, R.; Niku, J.; Renner, I.; Wright, S. mvabund: Statistical Methods for Analysing Multivariate Abundance Data. R package version 4.2.1. Available online: https://CRAN.R-project.org/package=mvabund (accessed on 17 November 2022).
- Mella, J.; Tirado, C.; Cortés, A.; Carretero, M. Seasonal variation of prey consumption by Liolaemus barbarae, a highland lizard endemic to Northern Chile. Anim. Biol. 2010, 60, 413–421. [Google Scholar]
- Dearing, M.D.; Schall, J.J. Testing models of optimal diet assembly by the generalist herbivorous lizard Cnemidophorus murinus. Ecology 1992, 73, 845–858. [Google Scholar] [CrossRef]
- Stamps, J.; Tanaka, S.; Krishnan, V. The relationship between selectivity and food abundance in a juvenile lizard. Ecology 1981, 62, 1079–1092. [Google Scholar] [CrossRef]
- Rocha, C.F. Selectivity in plant food consumption in the lizard Liolaemus lutzae from southeastern Brazil. Stud. Neotrop. Fauna Environ. 2000, 35, 14–18. [Google Scholar] [CrossRef] [PubMed]
- Região Autónoma da Madeira. Ponta de S. Lourenço; Secretaria Regional do Ambiente e dos Recursos Naturais; Serviço do Parque Natural da Madeira: Madeira, Portugal, 2010. [Google Scholar]
- Nóbrega, H.; Freitas, G.; Zavattieri, M.; Ragonezi, C.; de Carvalho, M.Â.P. Structure and floristic composition associated with an endangered species Beta patula Aiton (Amaranthaceae) in the Islands of Madeira Archipelago. Biodivers. Data J. 2021, 9, e61091. [Google Scholar] [CrossRef] [PubMed]
- Camacho, R.A.P. Influência de Diferentes Dietas em Teira dugesii (Milne-Edwards, 1829); Universidade da Madeira: Funchal, Madeira, 2002. [Google Scholar]
- Carretero, M.A.; Roca, V.; Martín, J.E.; Llorente, G.A.; Montori, A.; Santos, X.; Mateos, J. Diet and helminth parasites in the Gran Canaria giant lizard, Gallotia stehlini. Rev. Esp. Herpetol. 2006, 20, 105–117. [Google Scholar]
- Molina-Borja, M.; Rodríguez-Domínguez, M. Evolution of biometric and life-history traits in lizards (Gallotia) from the Canary Islands. J. Zool. Syst. Evol. Res. 2004, 42, 44–53. [Google Scholar] [CrossRef]
- Meiri, S. Evolution and ecology of lizard body sizes. Glob. Ecol. Biogeogr. 2008, 17, 724–734. [Google Scholar] [CrossRef]
- Ingerson-Mahar, J. Relating diet and morphology in adult carabid beetles. In The Agroecology of Carabid Beetles; Holland, J.M., Ed.; Intercept Limited: Andover, UK, 2002; pp. 111–136. [Google Scholar]
- Carretero, M.; Llorente, G. What are they really eating? Stomach versus intestine as sources of diet information in lacertids. In Mediterranean Basin Lacertid Lizards: A Biological Approach; Vicente, L., Crespo, E.G., Eds.; ICN: Lisboa, Portugal, 2001; pp. 105–112. [Google Scholar]
- Cook, L. Variation in the Madeiran lizard Lacerta dugesii. J. Zool. 1979, 187, 327–340. [Google Scholar] [CrossRef]
- Crisp, M.; Cook, L.; Hereward, F. Color and heat balance in the lizard Lacerta dugesii. Copeia 1979, 1979, 250–257. [Google Scholar] [CrossRef]
- Díaz, J.A.; Carrascal, L.M. Prey size and food selection of Psammodromus algirus (Lacertidae) in central Spain. J. Herpetol. 1990, 24, 342–347. [Google Scholar] [CrossRef]
- Silva-Rocha, I.; Santos, J.M.; Rocha, R.; Rato, C. Bioclimatic and local drivers modulating the expansion of an introduced temperate reptile in a subtropical island. Glob. Ecol. Conserv. 2022, 37, e02164. [Google Scholar] [CrossRef]
- Guarino, F. Diet of a large carnivorous lizard, Varanus varius. Wildl. Res. 2001, 28, 627–630. [Google Scholar] [CrossRef]
- Weatherhead, P.J.; Blouin-Demers, G. Understanding avian nest predation: Why ornithologists should study snakes. J. Avian Biol. 2004, 35, 185–190. [Google Scholar] [CrossRef]
- SRA. Estratégia Marinha para a subdivisão da Madeira. Diretiva Quadro Estratégia Marinha; Secretaria Regional do Ambiente e dos Recursos Naturais: Lisbon, Portugal, 2014.
- Kaliontzopoulou, A.; Adams, D.C.; van der Meijden, A.; Perera, A.; Carretero, M.A. Relationships between head morphology, bite performance and ecology in two species of Podarcis wall lizards. Evol. Ecol. 2012, 26, 825–845. [Google Scholar] [CrossRef]
- Husak, J.F.; Fox, S.F. Field use of maximal sprint speed by collared lizards (Crotaphytus collaris): Compensation and sexual selection. Evolution 2006, 60, 1888–1895. [Google Scholar]
- Lappin, A.K.; Husak, J.F. Weapon performance, not size, determines mating success and potential reproductive output in the collared lizard (Crotaphytus collaris). Am. Nat. 2005, 166, 426–436. [Google Scholar] [CrossRef] [PubMed]
- Herrel, A.; Aerts, P.; Fret, J.; De Vree, F. Morphology of the feeding system in agamid lizards: Ecological correlates. Anat. Rec. Off. Publ. Am. Assoc. Anat. 1999, 254, 496–507. [Google Scholar] [CrossRef]
- Herrel, A.; Spithoven, L.; Van Damme, R.; De Vree, F. Sexual dimorphism of head size in Gallotia galloti: Testing the niche divergence hypothesis by functional analyses. Funct. Ecol. 1999, 13, 289–297. [Google Scholar] [CrossRef]
- Husak, J.F.; Lappin, A.K.; Van Den Bussche, R.A. The fitness advantage of a high-performance weapon. Biol. J. Linn. Soc. 2009, 96, 840–845. [Google Scholar] [CrossRef]
- Jardim Botânico UTAD. Espécie Bituminaria bituminosa. Available online: https://jb.utad.pt (accessed on 17 November 2022).
- Flora-On|Flora de Portugal. Bituminaria bituminosa. Available online: https://flora-on.pt (accessed on 17 November 2022).
- Pérez-Mellado, V.; Romero-Beviá, M.; Ortega, F.; Martín-García, S.; Perera, A.; López-Vicente, M.; Galache, C. El uso de los recursos troficos en Gallotia simonyi (Sauria, Lacertidae) de la isla de El Hierro (Islas Canarias). Monogr. Herpetológicas 1999, 4, 63–83. [Google Scholar]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rato, C.; Dellinger, T.; Carretero, M.A. Dietary Variation Is Driven by Landscape Heterogeneity in an Insular Omnivorous Endemic Lizard, Revealed by DNA Metabarcoding. Diversity 2022, 14, 1078. https://doi.org/10.3390/d14121078
Rato C, Dellinger T, Carretero MA. Dietary Variation Is Driven by Landscape Heterogeneity in an Insular Omnivorous Endemic Lizard, Revealed by DNA Metabarcoding. Diversity. 2022; 14(12):1078. https://doi.org/10.3390/d14121078
Chicago/Turabian StyleRato, Catarina, Thomas Dellinger, and Miguel A. Carretero. 2022. "Dietary Variation Is Driven by Landscape Heterogeneity in an Insular Omnivorous Endemic Lizard, Revealed by DNA Metabarcoding" Diversity 14, no. 12: 1078. https://doi.org/10.3390/d14121078
APA StyleRato, C., Dellinger, T., & Carretero, M. A. (2022). Dietary Variation Is Driven by Landscape Heterogeneity in an Insular Omnivorous Endemic Lizard, Revealed by DNA Metabarcoding. Diversity, 14(12), 1078. https://doi.org/10.3390/d14121078





