Abstract
Diversity in Pistacia has been evaluated at all molecular levels using the internal transcribed spacer 1 (ITS1) marker in three species (Pistacia atlantica subsp. atlantica; Pistacia vera and Pistacia terebinthus), and compared with other Pistacia species. Results showed that the ITS amplification and sequencing, followed by phylogenetic analyses, identify the species and confirm their classification, which revealed that it can be used as a marker. Our results suggest that ITS1 analyses might provide a simple and inexpensive approach to validate the species of samples collected from the natural population, where species identification can be difficult, especially if hybrids are present or if the season is not optimal for identifying differences in morphological traits.
1. Introduction
Species of the genus Pistacia are dioecious trees which belong to the family Anacardiaceae and are widely distributed in the Mediterranean region, Middle Eastern areas and Canary Islands [,]. Pistacia genus is believed to have originated in Central Asia 80 million years ago [].
Although the most economically important species is P. vera L., due to nut production, other species such as P. atlantica Desf., P. terebinthus L. and P. integerrima L. are of interest, as they are used as rootstocks for pistachio cultivation []. In addition, the cultivation of pistachio and other Pistacia species is also particularly important for some Mediterranean countries, as they are drought-resistant, and are used as windbreaks and for soil conservation against erosion [].
Due to its resistance to drought, the Atlas pistachio (Pistacia atlantica Desf. subsp. atlantica) has been proposed for use in reforestation programs and to help prevent advance of the desert []. Furthermore, this species has traditionally been used for many medical and pharmaceutical applications [].
Unfortunately, this species is affected by climatic conditions, but above all by anthropogenic activity, which leads to a degradation of the stands. Therefore, it is important to preserve its genetic variability, but it is equally important to distinguish this species from others of the genus Pistacia which occur in the same area of dispersal.
For a long time, diversity among and within the Pistacia species has been mainly based on morphological and phenotypic characteristics. Nowadays, molecular markers are a useful complement to these characteristics, because they are used for the detection of genetic variability. They present a high degree of polymorphism and they are abundant in the genome, independent of tissue or environmental effects and allow cultivar identification in the early stages of development [,].
Taxonomic classification and identification of mastic trees is extremely limited and not yet fully carried out. The use of molecular data has significantly increased the understanding of their systematic at different taxonomic levels []. Molecular classification of 10 Pistacia species was first reported by Parfitt and Badenes []. Nevertheless, it is not easy to distinguish these different genotypes. In Turkey, Kafkas and Perl-Treves [] have reported the taxonomic relationships and genetic variations of wild Pistacia germplasm, including P. atlantica, P. terebinthus and P. eurycarpa using morphological data and random amplified polymorphic DNA (RAPD). In Syria, a molecular evaluation was carried out using the Amplified Fragment Length Polymorphism (AFLP) technique on P. vera varieties [].
Commonly used molecular markers for lower-level phylogenetic analysis in plants are the internal transcribed spacer (ITS) regions of nuclear ribosomal DNA [,]. This makes the ITS region an interesting subject for evolutionary investigations [], as well as biogeographic investigations [,]. In fact, the phylogeny of Pistacia has previously been reconstructed with ITS sequences [,], and ITS1 has also been studied previously to assess genetic diversity in some Pistacia species [,,]. Recently, ITS, along with Inter Simple Sequence Repeat (ISSR), Simple Sequence Repeat (SSR) and Random Amplification of Polymorphic DNA (RAPD) markers, were used to evaluate patterns of genetic variation and phylogenetic relationships in 24 wild-type mastic trees of P. lenticus L. in Turkey [].
The main objective of this study is the validation of ITS1 sequences for the assignment of species in the genus Pistacia and the reconstruction of their phylogenetic relationships. For that reason, we confirm the identification of Algerian samples of Atlas pistachio trees (Pistacia atlantica subsp atlantica) and Italian samples of P. vera and P. terebinthus. This was carried out based on the comparison of their ITS1 sequences, characterized in this study with those available in GenBank for the genus Pistacia (https://www.ncbi.nlm.nih.gov/taxonomy/?term=Pistacia; accessed on 27 July 2021). Afterward, intra- and inter- specific phylogenetic relationships were studied for these sequences.
2. Materials and Methods
2.1. Plant Materials
Extraction and molecular assessments were carried out from samples of young leaves from two Atlas pistachio trees originating from the Batna forest (395 km East of Algiers; 35°37′10″ N, 6°22′13″ E; altitude 1027 m), and from four trees from the Botanical Garden of Padova (492 km north of Rome, Italy, 45°23′34″ N, 11°54′10″ E, altitude 25 m) relating to the species P. vera and P. terebinthus (Figure 1).

Figure 1.
Pistachio Terebinth tree of Botanical Garden in Padova (Italy) (A); Atlas pistachio tree in region of Batna (Algeria) (B). Photos: Labdelli A.
2.2. DNA Extraction
Total DNA was extracted from fresh-leaf material using the modified CTAB method of Doyle and Doyle []. A 100 mg sample of young leaf tissue were manually ground into a smooth powder in liquid nitrogen using a plastic micro-pestle, added to 2 mL Eppendorf tube and mixed with 900 µL of CTAB extraction buffer, containing 1 M TRIS-HCl (pH 8), 5 M NaCl, 0.5 M EDTA, 2% CTAB, 2% polyvinylpyrrolidone PVP, 0.2% b-mercaptoethanol and 0.1 % NaHSO3. Samples were incubated at 65 °C for 1 h, mixed with an equal volume of chloroform-isoamyl alcohol (24:1) and centrifuged for 5 min at 20,784× g. The supernatant was transferred into a new 2 mL Eppendorf tube and mixed with an equal volume of ice-cold isopropanol to precipitate the DNA and left at −80 °C for 1 h [,]. The tubes were gently inverted several times. The nucleic acid pellet was recovered by centrifugation (20,784× g for 5 min), washed with 1 mL of 10 mM ammonium acetate in 76% ethanol for a few minutes, dried for 15 min at room temperature and re-suspended in 100 µL modified TE buffer (pH 8); (10 mM TRIS-HCI, 0.1 mM EDTA) with RNAase (5 µg/mL) []. DNA integrity was ascertained by DNA staining after agarose electrophoresis (EuroSafe Nucleic Acid Stain, Euroclone, Italy). Extracted DNA was diluted with milli-Q purified water to 20 ng/µL and used for polymerase chain reaction (PCR) amplification.
2.3. Amplification and Purification of ITS1 Region of rDNA
This study has only targeted the ITS1 region located between the regions encoding the large subunits (18 S and 5.8 S) of ribosomal RNA. This region is among the most conserved in plants [,], and has already been used in the genus Pistacia []. The PCR amplification reaction volume is 20 μL, consisting of 1.7 µL of DNA, 0.4 µL of each primer (Forward: GCGAACCTGTCTCATCATCATCATCATCATCG and Reverse: CACCAAGTATCGCATTTCTCGCGCGCGCGCGCGC), 4 µL reaction buffer (5X Green GoTaq® Reaction Buffer, Milano, Italy), 0.8 µL MgCl2 (50 mM), 0.8 µL dNTPs, 0.2 µL pfu (Promega, PFU DNA Polymerase, 5 U/µL) and 11.7 µL ultrapure water. The reactions were carried out according to the following procedure: the first phase of initial denaturation of the DNA (94 °C for 10 min) was followed by 35 cycles of amplification of the target region (each cycle comprising: 45 s denaturation at 94 °C, 45 sechybridization at 54 °C and 2 min extension at 72 °C), and a final step of extension at 72 °C for 10 min []. The primers used for the amplification of the ITS1 region of interest were synthesized by BMR Genomics (http://www.bmr-genomics.it/, accessed on 11 April 2016).
The amplified products were detected by electrophoresis on a 1% agarose gel containing a Eurosafe nucleic acid staining solution (Nucleic acid staining solution 20,000×). The PCR products were purified prior to sequencing using the PureLink® PCR purification kit (Invitrogen by Life Technologies, Wiesbaden-Nordenstadt, Germany).
2.4. Sequencing and Alignment of ITS Sequences
Sequencing was performed with PCR-amplified ITS1 region rDNA (400 ng) by BMR GENOMICS SRL, Padova, Italy.
The obtained DNA sequences were compared with homologous sequences available in the NCBI (National Centre for Biotechnology Information) GenBank database using the BLAST program (Basic Local Alignment Search Tool) []. Our sequences were then entered into a matrix and aligned with homologous sequences of pistachio trees recovered from GenBank using Fast Fourier Transform (MAFFT) program (version 7) []. The resulting aligned sequences file in FASTA format [] was viewed and verified using the sequence editor BioEdit software (Version 7.0) [].
Thus, the first matrix of 107 aligned ITS sequences was constructed, comprising our six sequences (P. atlantica Desf. subsp atlantica, P. vera L., P. terebinthus L.) and a set of ITS sequences (obtained by Blast) of the genus Pistacia. Following a preliminary phylogenetic analysis of this total matrix, only 44 sequences were retained, including six sequences of interest, thirty-six representatives of the phylogenetic diversity of Pistacia and two outgroups (Cotinus coggygria, AY510157; Rhus chinensis, KP093194), to simplify the analysis and visualisation of the results (Table 1).

Table 1.
List of GenBank accession numbers of the ITS sequences used in this study.
2.5. Phylogenetic Analysis
Phylogenetic analyses of the aligned ITS sequence matrices were performed using MEGA-7 software version 7.0 [], using the maximum parsimony (MP) method. The robustness of the clades in percent confidence was evaluated by a bootstrap test with 1000 replicates [].
3. Results
ITS Amplification and Sequence Comparison
The amplified band size of the ITS1 region of all samples (P. vera L. (Pver5; Pver7), P. terebinthus L. (Pter44; Pter45) and P. atlantica Desf. subsp atlantica (PatlB4; PatlB7) was approximately 300 bp (Figure 2).
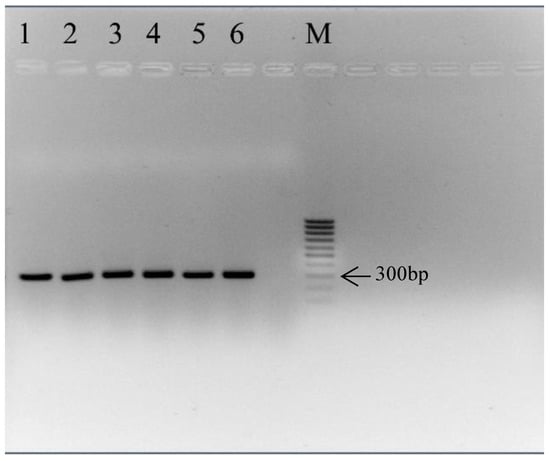
Figure 2.
Agarose gel electrophoresis of the ITS1 region of: P. vera L., line 1 (Pver5) and line 2 (Pver7); P. terebinthus L., line 3 (Pter44) and line 4 (Pter45); and P. atlantica Desf. subsp atlantica, line 5 (PatlB4) and line 6 (PatlB7); M: Marker 100 bp ladder.
Multiple sequence alignment analysis (Table 2; Figure S1) showed a length of 248 nucleotide sites, of which 61 sites were variable (V) (24.6%), 185 were conserved (C) (74.60%), 40 were informational sites (Pi) (Parsimony-informative: 16.13%) and 21 were automorphic sites or Singletons (S) (8.41%).

Table 2.
Site characterization of the sequences obtained.
The phylogenetic tree resulting from the analysis of ITS sequences is shown in Figure 3. It can be observed that all of the samples relating to pistachio tree species are descended from a common ancestor, thus forming a monophyletic group distinct from the outgroup. In addition, P. mexicana presented as an ancient lineage in a sister position to all other pistachio trees. The latter form a large monophyletic group which is well-supported by ITS1, with 93% bootstrap. This large group diversified from a common ancestor into three divergent evolutionary lines equidistant from each other (polytomy) represented by clades A, B and C (Figure 3). Despite having a significant number of informative sites for the genus, these three clades did not present bootstraps values higher than 70, although the synapomorphies were significant in two of the three groups (with values of 3 and 10). All of our newly sequenced samples (coloured blue for P. vera L., green for P. terebintus L., and red for P. atlantica Desf.) were included in a monophyletic group, clade C, which is mainly composed of all samples of P. vera L., including cultivars, P. atlantica Desf., their hybrids and those of P. terebinthus L. This line C has itself diversified into three subclades, c1, c2 and c3, the latter showing the highest divergence (three synapomorphies and a bootstraps value greater than 70) from the other two. The two individuals of the local population of Atlas pistachio of Batna (PatlB4, PatlB7) position themselves unambiguously with the three reference sequences of P. atlantica Desf. Hence, they form a monophyletic subgroup (c2) of various origins, distributed from Asia to the Mediterranean region []. This confirms the taxonomic identification of our PatlB4 and PatlB7 samples. Moreover, it is highly interesting to note that this subgroup c2, although it is not robustly distinguishable not only from the samples of P. vera L. and P. terebinthus L. that we obtained from the Botanical Garden of Padova (Italy), but also from the few individuals of GenBank identified with hybrids of P. atlantica Desf. and P. vera L., which are positioned with P. vera L. samples in subgroups c1 and c3. Concerning the two P. vera L. samples from the Botanical Garden of Padova, it is conspicuous that they are clearly distinguished from each other; one relates to subgroup c1 (Pvera7) and the other to c3 (Pvera5). Finally, as demonstrated on the tree, the two samples, Pter44 and Pter45, belong to the same lineage (P. terebinthus L.) within subclade c1, where it is surprisingly positioned as a sister species to Pvera7, and not in clade A with other P. terebinthus L. specimens.
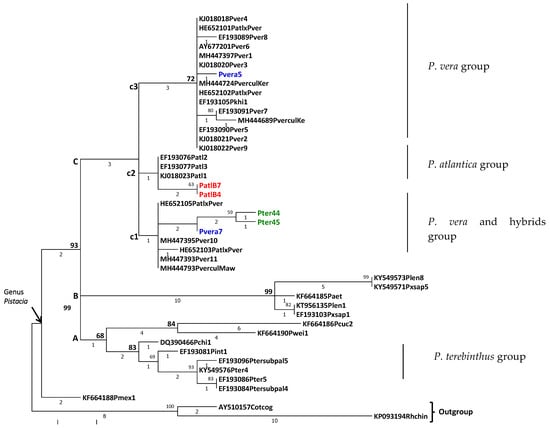
Figure 3.
Phylogenetic tree of Pistacia pecies based in Maximum Parsimony analysis of taxa.
The evolutionary history was inferred using the Maximum Parsimony method. The most parsimonious tree with length = 86 is shown (Figure 3). Bootstrap values (with 1000 replicates) are shown above the branches when they are greater than 70. Below the inner branches, the numbers of synapomorphy changes are given, and the terminal branches give the numbers of autapomorphy. Newly sequenced samples are coloured blue for P. vera, green for P. terebinthus and red for P. atlantica Desf. The consistency index is (0.725806), the retention index is (0.912371) and the composite index is 0.729897 (0.662205) for all sites and parsimony-informative sites (in parentheses). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (636 replicates) is shown next to the branches []. The MP tree was obtained using the Subtree-Pruning-Regrafting (SPR) algorithm [] (p.126) with search level 1, in which the initial trees were obtained by the random addition of sequences (10 replicates). The names of the samples are indicated as accession number, initials of the species name and clone number (see Table 1).
4. Discussion
Our results show a relatively high variability of ITS1 sequences within the genus Pistacia (about 16%) (Table 2), indicating that the ITS1 region appears to contain a good proportion of phylogenetically informative sites in these species. Pi sites are useful for determining phylogenetic relationships, and the higher their rate, the more they condition the most parsimonious phylogenetic tree and, thus, the most reliable hypothesis of kinship relationships.
In the literature, the genetic diversity found in ITS sequences of P. atlantica Desf. was relatively low in relation to other species of Pistacia. In our study, all five sequences of this species showed greater than 99% identity and a low value of diversity compared to the identity of P. vera, which was lower than 94% identity. Their sequences appear in two different subclades. Esmailpour [] has explained that this may be due to a significant decline in its habitat, in addition to over-harvesting, resulting in genetic erosion. In this sense, Pazouki et al. [] have suggested that the low genetic diversity of P. atlantica subsp. kurdica could be sensitive to environmental changes and disease attacks, and that protection against loss of genetic diversity is an urgent need.
The results of Al Sousli et al. [], based on molecular data, have shown that P. vera L. was closer to P. atlantica Desf. than P. terebinthus L. These results are consistent with Kafkas and Perl-Treves [], who found that P. terebinthus L. was the most diverged species based on their RAPD data. In relation with the two analysed samples of P. terebinthus L., Pter44 and Pter45 are positioned in clade C with P. vera L. specimens and not with other P. terebinthus L. samples (clade A). This result would suggest an identification problem for these samples, or that we may be dealing with P. vera × P. terebinthus hybrids. This will need to be examined in more detail in the future by other molecular markers to clarify the nature of the relationships between these samples with such little divergence.
Despite the near-universal usage of ITS sequence data in plant phylogenetic studies, its complex and unpredictable evolutionary behaviour reduces its application for phylogenetic analysis. In fact, in the tree obtained in this study, some clades were not well resolved. It has been suggested that more reliable results are likely to emerge from the use of single-copy or low-copy nuclear genes [].
ITS sequence data have provided, and may continue to provide, insights into phylogenetic history, polyploid ancestry, genome relationships, historical introgression and other evolutionary questions [], but the data generated will only have the most lasting value if compared with data gained with other sequences.
The data on ITS sequence length and diversity may be a useful parameter for the assessment of genetic diversity at the intra-specific level, although the level of diversity is much higher at the inter-specific level in different groups of plants [].
The sequencing of ribosomal DNA (rDNA) has become a commonly used procedure and one of the most widely applied molecular markers for reconstructing plant phylogenies at the intra-family level (intra- and inter-generic and inter-specific levels) [], as demonstrated by the increasing number of publications discussing polymorphic rDNA [,,].
Perhaps even more striking is the fact that 34% of all published phylogenetic hypotheses are based exclusively on ITS sequences []. Optimally, phylogeographic analyses should be based on more than one genome, and nuclear phylogeographic based on ribosomal ITS, introns or exons of genes will become increasingly important as a result of decreasing costs of DNA sequencing [].
5. Conclusions
Our results are in agreement with previous findings, and suggest that ITS1 analyses could provide a simple and inexpensive approach to validate the assignment of some species where identification is not always easy, especially if hybrids are present or if the season is not optimal to highlight differences in morphological traits. This study has shown that the analysed samples of pistachio species from Algeria and Italy come from a common ancestor. In addition, it has verified the low variability among samples of the Atlantic pistachio compared to other species. The pistachio tree is of great ecological and economic interest, so it is advisable to preserve it from various natural and anthropogenic threats. Studies, such as those carried out in this work, help to identify genetic diversity between and within species.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/d14121051/s1, Figure S1: Multiple alignment of nucleotide sequences of the ITS1 region of the DNAnr (5.8 S) performed by Geneiuos Prime software on 44 samples of the genus Pistacia.
Author Contributions
A.L., F.R. and L.T. designed the methodology; R.D.L.H. and A.L. performed the data analysis; A.L., F.R., R.D.L.H., M.T. and O.M., software; A.L., R.D.L.H., M.T., F.R. and O.M. participated in writing and original draft preparation; and all authors reviewed the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
Abdelkader Aïnouche (Université de Rennes 1, UMR-CNRS 6553, Ecobio, Campus de Beaulieu, Bat. 14, 35042 Rennes Cedex, France) is gratefully acknowledged for his revisions on the phylogenetic tree of this manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ak, B.E.; Parlakçı, H. Pistacia lentiscus in the Mediterranean Region in Turkey. Acta Hortic. 2009, 818, 77–82. [Google Scholar] [CrossRef]
- Ahmed, Z.B.; Yousfi, M.; Viaene, J.; Dejaegher, B.; Demeyer, K.; Heyden, Y.V. Four Pistacia atlantica subspecies (atlantica, cabulica, kurdica and mutica): A review of their botany, ethnobotany, phytochemistry and pharmacology. J. Ethnopharmacol. 2021, 265, 113329. [Google Scholar] [CrossRef] [PubMed]
- Al-Saghir, M.G. Phylogenetic Analysis of the Genus Pistacia L. (Anacardiaceae) Based on Morphological Data. Asian J. Plant Sci. 2010, 9, 28–35. [Google Scholar] [CrossRef][Green Version]
- El Zerey-Belaskri, A. Taxonomic and botanical retrospective review of Pistacia atlantica Desf. (Anacardiaceae). Arab. J. Med. Aromat. Plants 2019, 5, 47–77. [Google Scholar] [CrossRef]
- Belhadj, S.; Derridj, A.; Auda, Y.; Gers, C.; Gauquelin, T. Analyse de la variabilité morphologique chez huit populations spontanées de Pistacia atlantica en Algérie. Can. J. Bot. 2008, 86, 520–532. [Google Scholar] [CrossRef]
- El Zerey-Belaskria, A.; Ribeirob, T.; Alcarazc, M.L.; El Zereyd, W.; Castroe, S.; Loureiroe, J.; Benhassainia, H.; Hormaza, J.I. Molecular characterization of Pistacia atlantica Desf. subsp. atlantica (Anacardiaceae) in Algeria: Genome size determination, chromosome count and genetic diversity analysis using SSR markers. Sci. Hortic. 2018, 227, 278–287. [Google Scholar] [CrossRef]
- Labdelli, A.; De La Herrán, R.; Arafeh, R.; Resentini, F.; Trainotti, L.; Halis, Y.; Adda, A.; Merah, O. Genetic Variation in Damaged Populations of Pistacia atlantica Desf. Plants 2020, 9, 1541. [Google Scholar] [CrossRef]
- Soltis, P.S.; Soltis, D.E. Multiple origins of the Allotetraploid Tragopogon mirus (Compositae): rDNA evidence. Syst. Bot. 1991, 16, 407–413. [Google Scholar] [CrossRef]
- Parfitt, D.E.; Badenes, M.L. Phylogeny of the genus Pistacia as determined from analysis of the chloroplast genome. Proc. Natl. Acad. Sci. USA 1997, 94, 7987–7992. [Google Scholar] [CrossRef]
- Kafkas, S.; Perl-Treves, R. Inter-specific relationships in the genus Pistacia L. (Anacardiaceae) based on RAPD fingerprints. Hortic. Sci. 2002, 37, 168–171. [Google Scholar] [CrossRef]
- Basha, A.I.; Padulosi, S.; Chabane, K.; Hadj-Hassan, A.; Dulloo, E.; Pagnotta, M.A.; Porceddu, E. Genetic diversity of Syrian pistachio varieties evaluated by AFLP markers. Genet. Resour. Crop Evol. 2007, 54, 1807–1816. [Google Scholar] [CrossRef]
- Baldwin, B.G.; Sanderson, M.J.; Porter, J.M.; Wojciechowski, M.F.; Campbell, C.S.; Donoghue, M.J. The ITS region of nuclear ribosomal DNA: A valuable source of evidence on angiosperm phylogeny. Ann. Mo. Bot. Gard. 1995, 82, 247–277. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods Applications; Innis, M.A., Gelfand, D.H., Shinsky, J.J., White, T.J., Eds.; Academic Press: Cambridge, MA, USA, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- Baldwin, B.G. Phylogenetic utility of the internal transcribed spacer of nuclear ribosomal DNA in plants: An example from the Compositae. Mol. Phylogenet. Evol. 1992, 1, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Suh, Y.; Thien, L.B.; Reeve, H.E.; Zimmer, E.A. Molecular evolution and phylogenetic implications of ribosomal DNA in Winteraceae. Am. J. Bot. 1993, 80, 1042–1055. [Google Scholar] [CrossRef]
- Yi, T.; Wen, J.; Golan-Glodhirsh, A.; Parfitt, D.E. Phylogenetics and reticulate evolution in Pistacia (Anacardiaceae). Am. J. Bot. 2008, 95, 241–251. [Google Scholar] [CrossRef]
- Aznarte-Mellado, C.; Sola-Campoy, P.J.; Robles, F.; Rejón, C.R.; Herrán, R.; Navajas-Pérez, R. Molecular characterization of the interspecific hybrid Pistacia vigros (P. vera L. × P. atlantica Desf.). Sci. Hort. 2014, 179, 180–183. [Google Scholar] [CrossRef]
- Alaei, H.; Mohammadi, A.H.; Dehghani, A. Molecular characterization of the rDNA-ITS sequence and a PCR diagnostic technique for Pileolaria terebinthi, the cause of pistachio rust. Phytopathol. Mediterr. 2012, 51, 488–495. [Google Scholar] [CrossRef]
- Abuduli, A.; Aydin, Y.; Sakiroglu, M.; Onay, A.; Ercisli, S.; Uncuoglu, A.A. Molecular Evaluation of Genetic Diversity in Wild-Type Mastic Tree (Pistacia lentiscus L.). Biochem. Genet. 2016, 54, 619–635. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bul. 1987, 19, 11–15. [Google Scholar]
- Kafkas, S. Phylogenetic analysis of the genus Pistacia by AFLP markers. Plant Syst. Evol. 2006, 262, 113–124. [Google Scholar] [CrossRef]
- Al-Sousli, M.; Faory, H.; Nakar, M.; Zaid, S.; Al-Safadi, B.; Al-Saghir, M. Genetic Relationships among Some Pistacia Species (Anacardiaceae) in Syria. Middle East J. Sci. Res. 2014, 21, 1487–1496. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software versions 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Pearson, W.R.; Lipman, D.J. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 1988, 85, 2444–2448. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: New York, NY, USA, 2000. [Google Scholar]
- Esmailpour, A. Distribution, use and conservation of pistachio in Iran. In Towards a Comprehensive Documentation and Use of Pistacia Genetic Diversity in Central and West Asia, North Africa and Europe. Report of the IPGRI Workshop; 14–17 December 1998, Ibrid, Jordan; Padulosi, S., Hadj-Hassan, A., Eds.; IPGRI: Rome, Italy, 2001; p. 16. [Google Scholar]
- Pazouki, L.; Mardi, M.; Shanjani, P.S.; Hagidimitriou, M.; Pirseyedi, S.M.; Naghavi, M.R.; Avanzato, D.; Vendramin, E.; Kafkas, S.; Ghareyazi, B.; et al. Genetic diversity and relationships among Pistacia species and cultivars. Conserv. Genet. 2010, 11, 311–318. [Google Scholar] [CrossRef]
- Kafkas, S.; Perl-Treves, R. Morphological and molecular phylogeny of Pistacia species in Turkey. Theor. Appl. Genet. 2001, 102, 908–915. [Google Scholar] [CrossRef]
- Álvarez, I.; Wendel, J.F. Ribosomal ITS sequences and plant phylogenetic inference. Mol. Phylogenet. Evol. 2003, 29, 417–434. [Google Scholar] [CrossRef]
- Wissemann, V. Hybridization and the evolution of the nrITS spacer region. In Plant Genome: Biodiversity and Evolution; Sharma, A.K., Sharma, A., Eds.; Science Publishers, Inc.: Enfield, NH, USA, 2003; pp. 57–66. [Google Scholar]
- Sharma, S.; Rustgi, S.; Balyan, H.S.; Gupta, P.K. Internal transcribed spacer (ITS) sequences of ribosomal DNA of wild barley and their comparison with ITS sequences in common wheat. Barley Genet. Newlet. 2002, 32, 38–45. [Google Scholar]
- Talebi, M.; Akbari, M.; Zamani, M.; Sayed-Tabatabaei, B.E. Molecular polymorphism in Pistacia vera L. using non-coding regions of chloroplast DNA. J. Genet. Engin. Biotechnol. 2016, 14, 31–37. [Google Scholar] [CrossRef]
- Muir, P.; Li, S.; Lou, S.; Wang, D.; Spakowicz, D.J.; Salichos, L.; Zhang, J.; Weinstock, G.M.; Isaacs, F.; Rozowsky, J.; et al. The real cost of sequencing: Scaling computation to keep pace with data generation. Genome Biol. 2016, 17, 53. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).