New Strain of Cyphellophora olivacea Exhibits Striking Tolerance to Sodium Bicarbonate
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Culturing Media
2.2. Isolation of the Unknown Fungal Strain
2.3. Morphological Observations and Microscopy
2.4. Tolerance Tests
2.5. Interaction Tests
2.6. DNA Analysis Methods
2.7. Bioinformatics
2.8. Statistical Analyses
2.9. Data Availability
3. Results
3.1. Taxonomic Note on the SAG 88.79 Strain
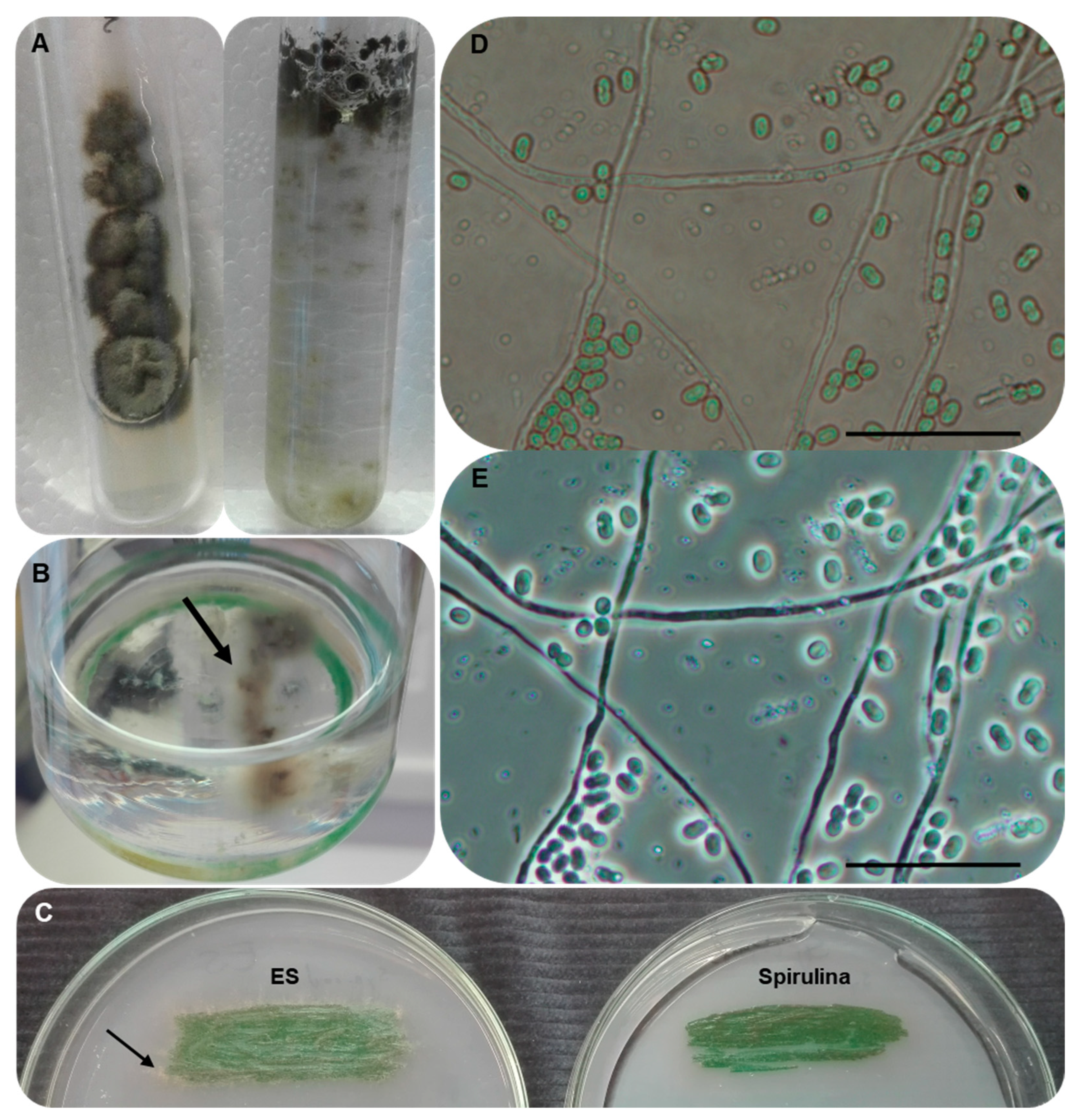
3.2. Isolation of the Unknown Fungal Strain
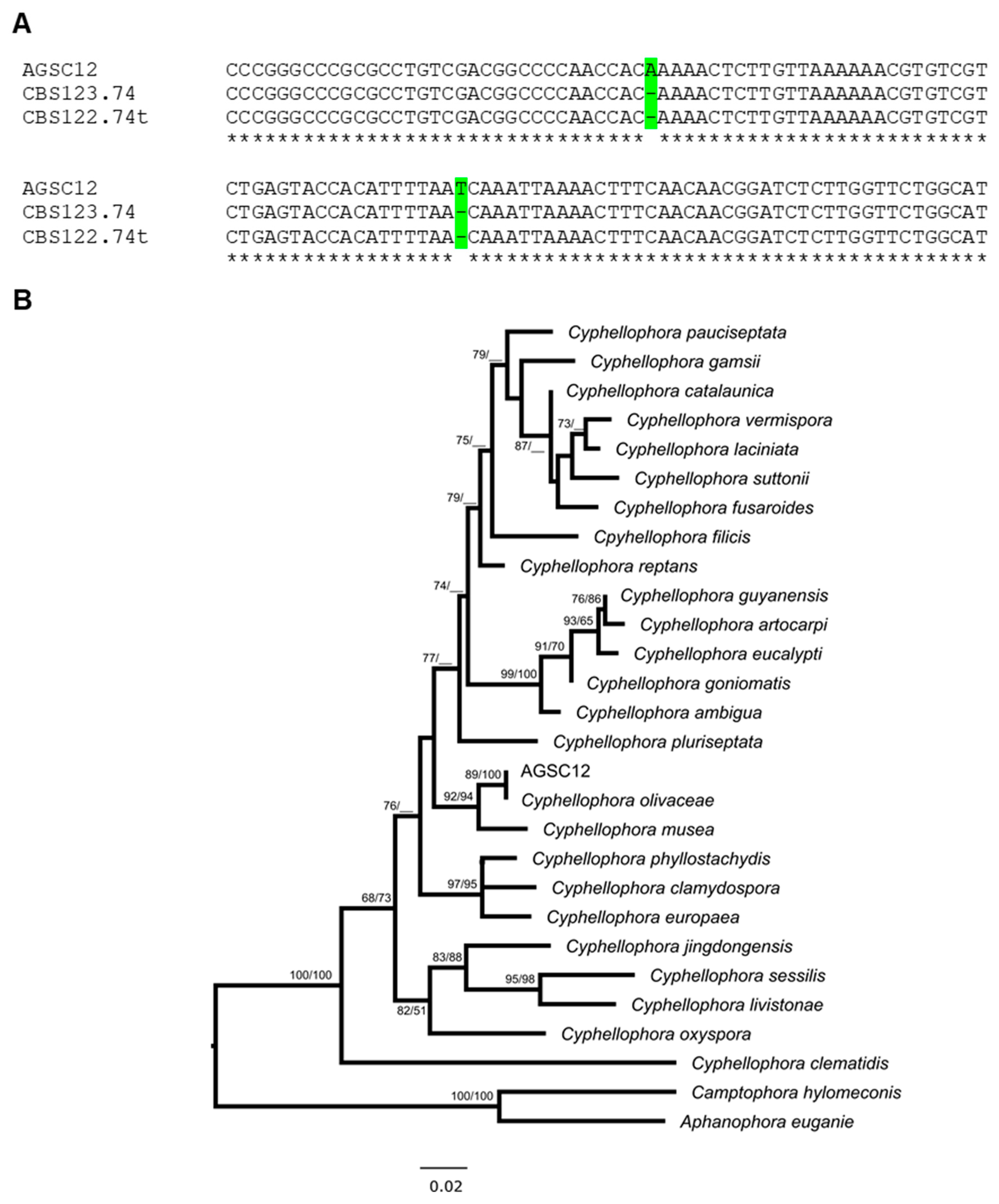
3.3. Molecular Identification of the AGSC12 Strain
3.4. Phylogenetic Analyses of the AGSC12 Strain and Its Relatives
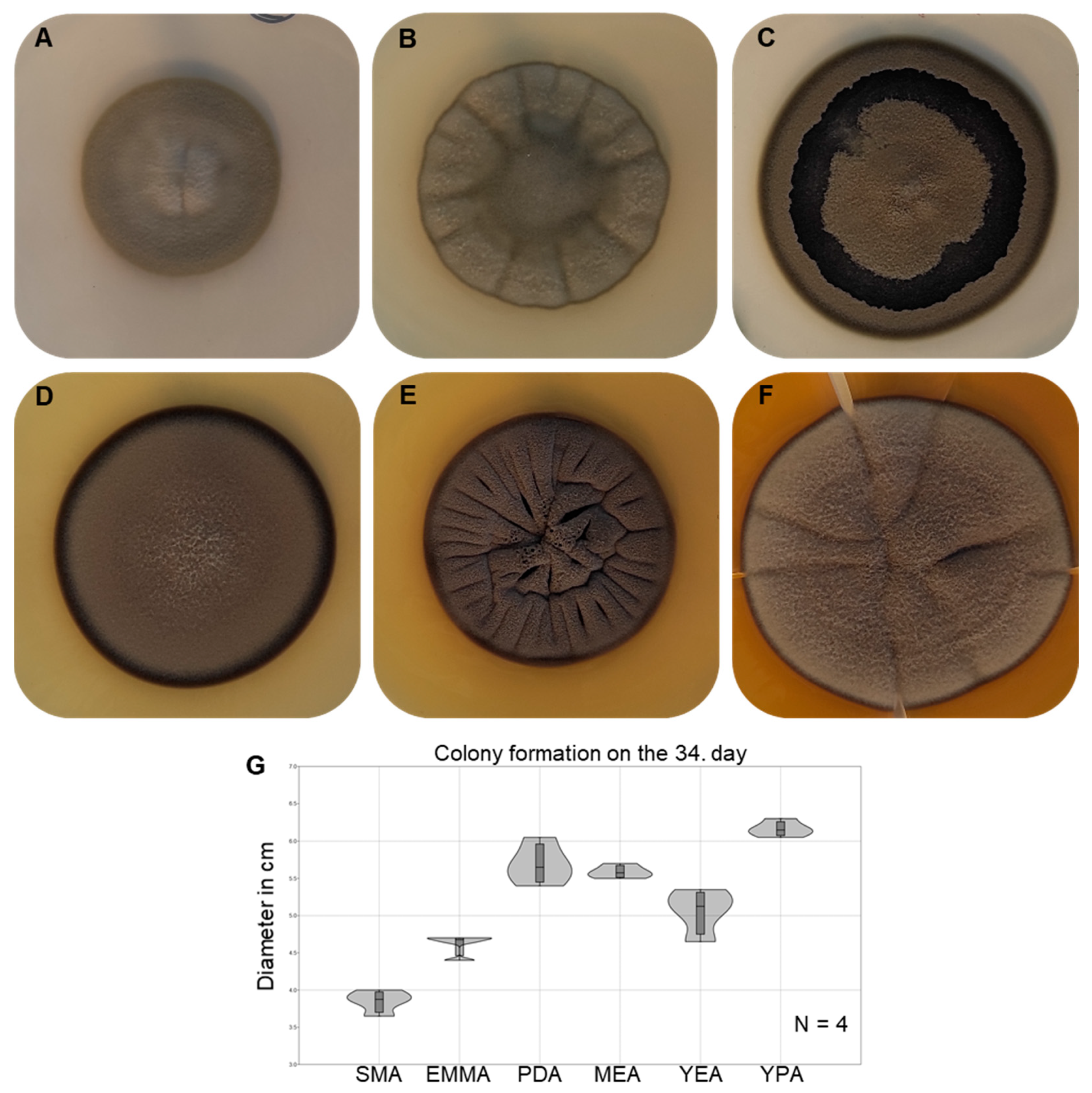
3.5. Morphological Characterisation and Growth Ability of the Strain AGSC12
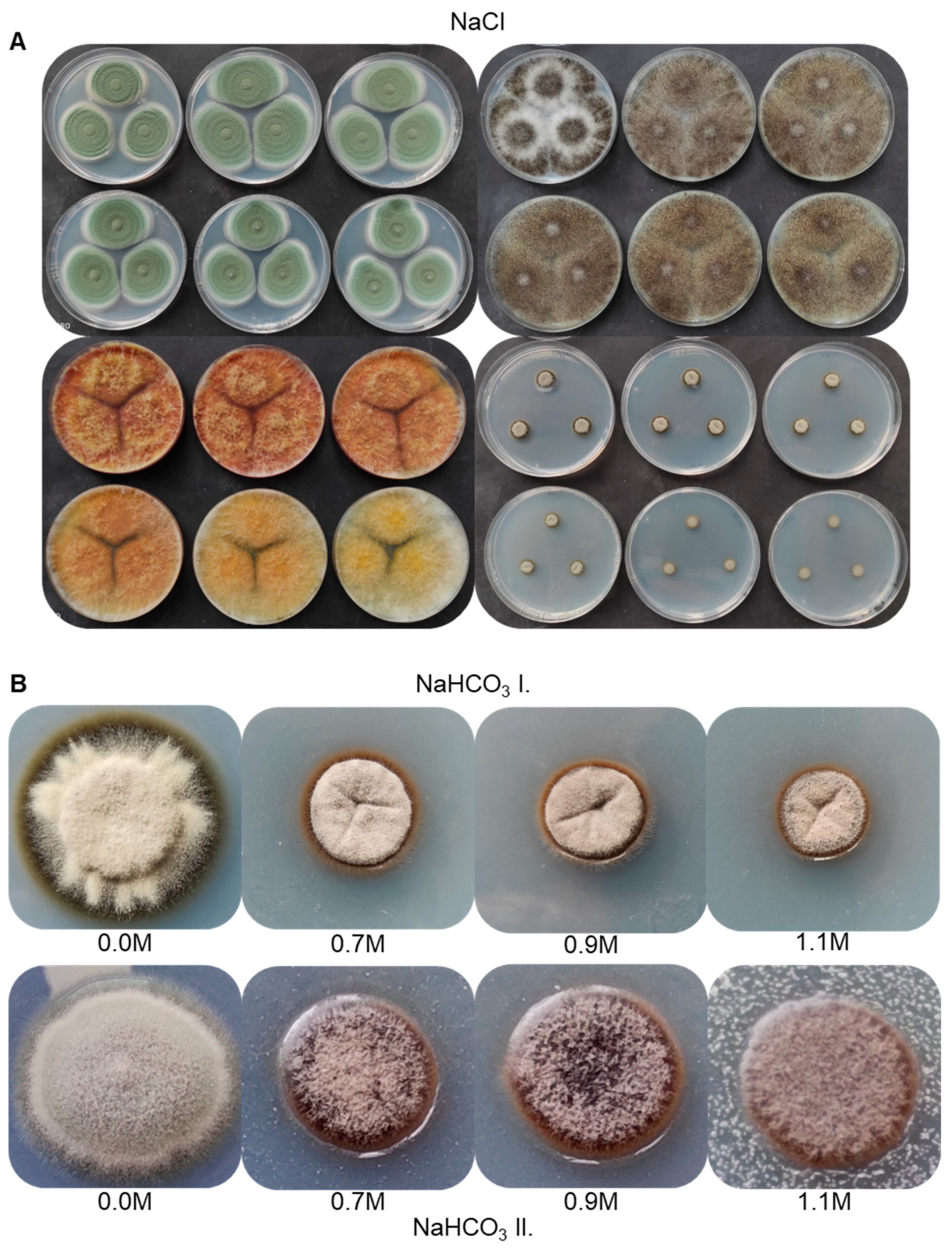
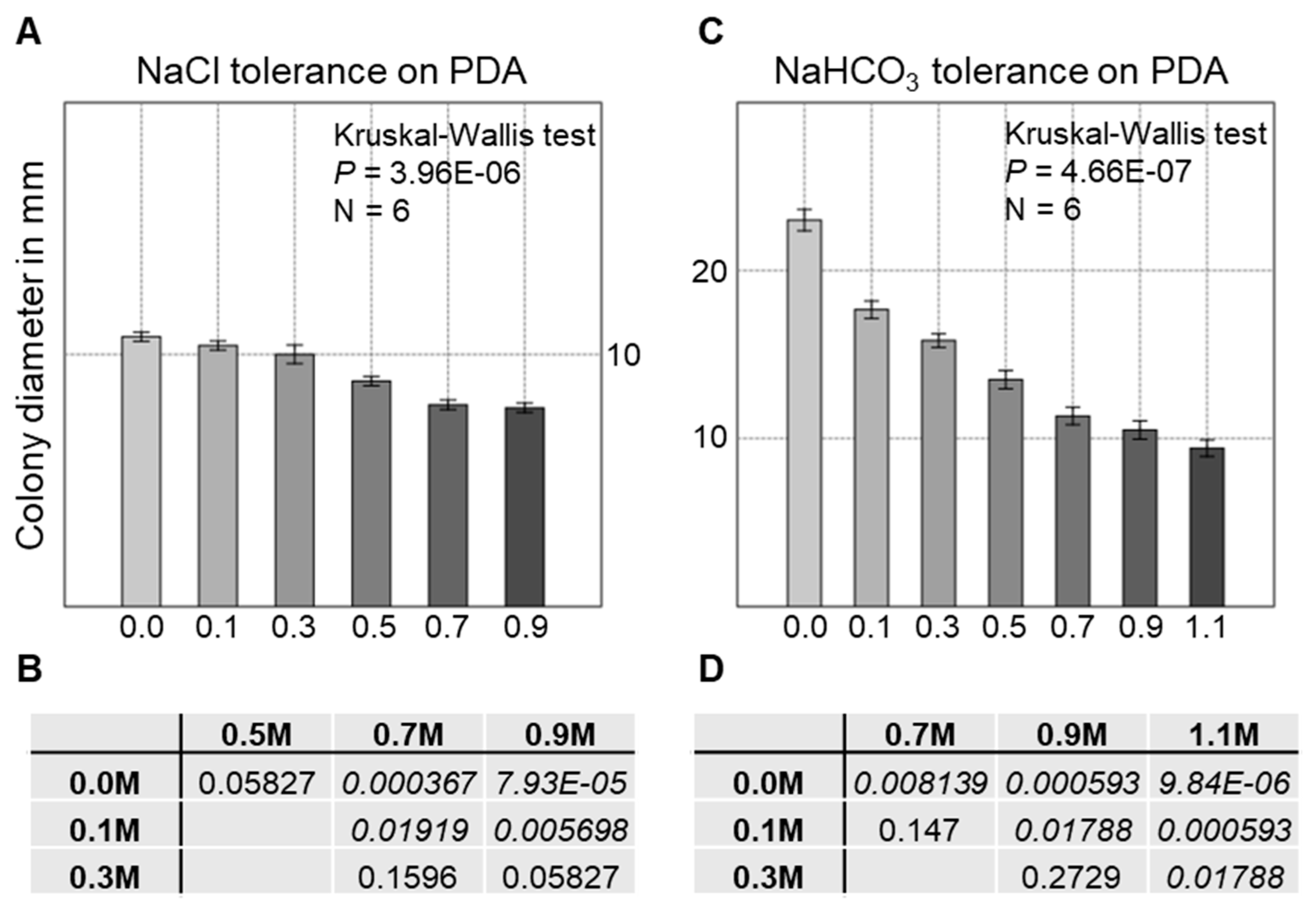
3.6. Salt Tolerance of the Strain AGSC12 Compared to Other Fungal Strains
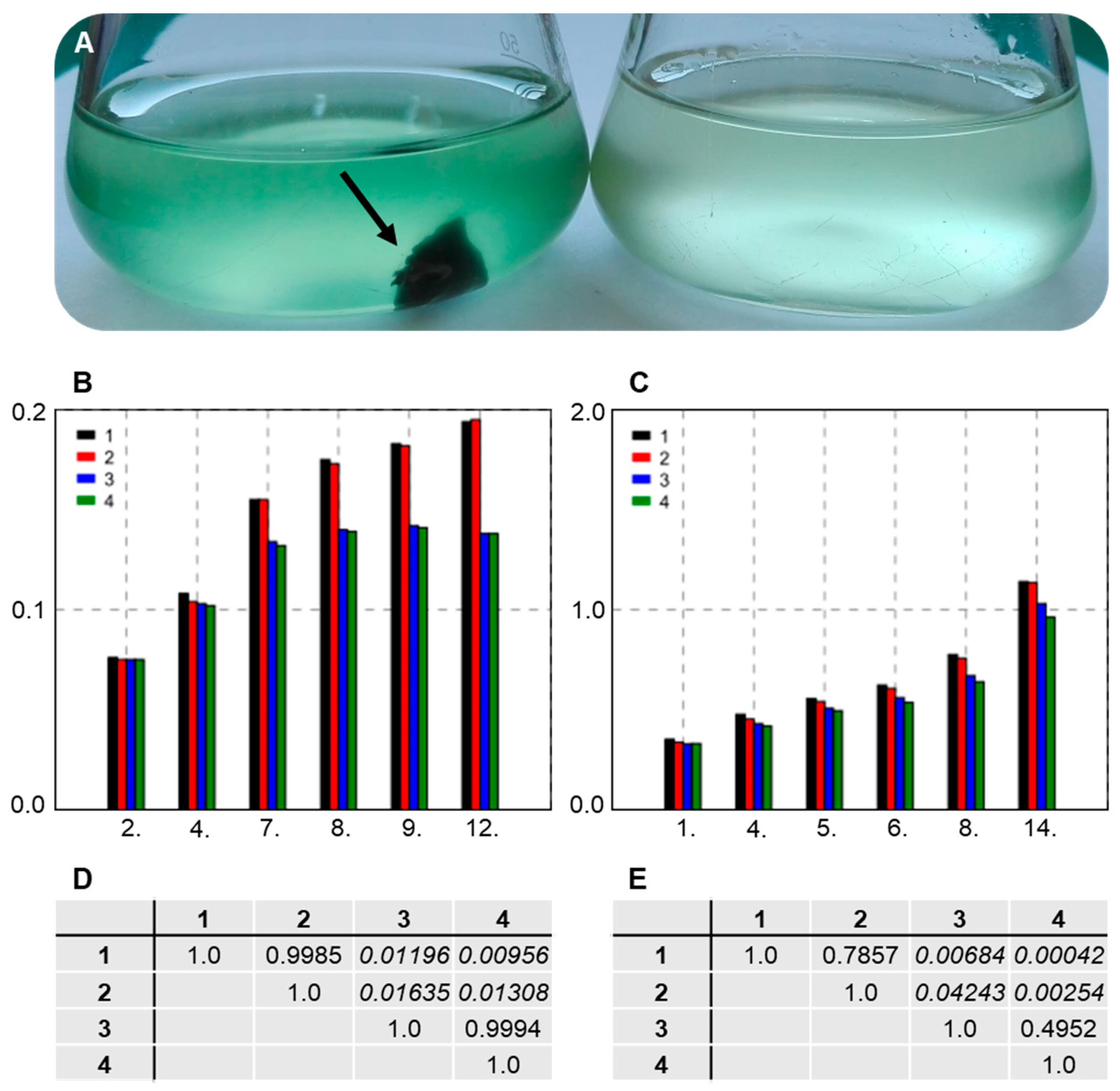
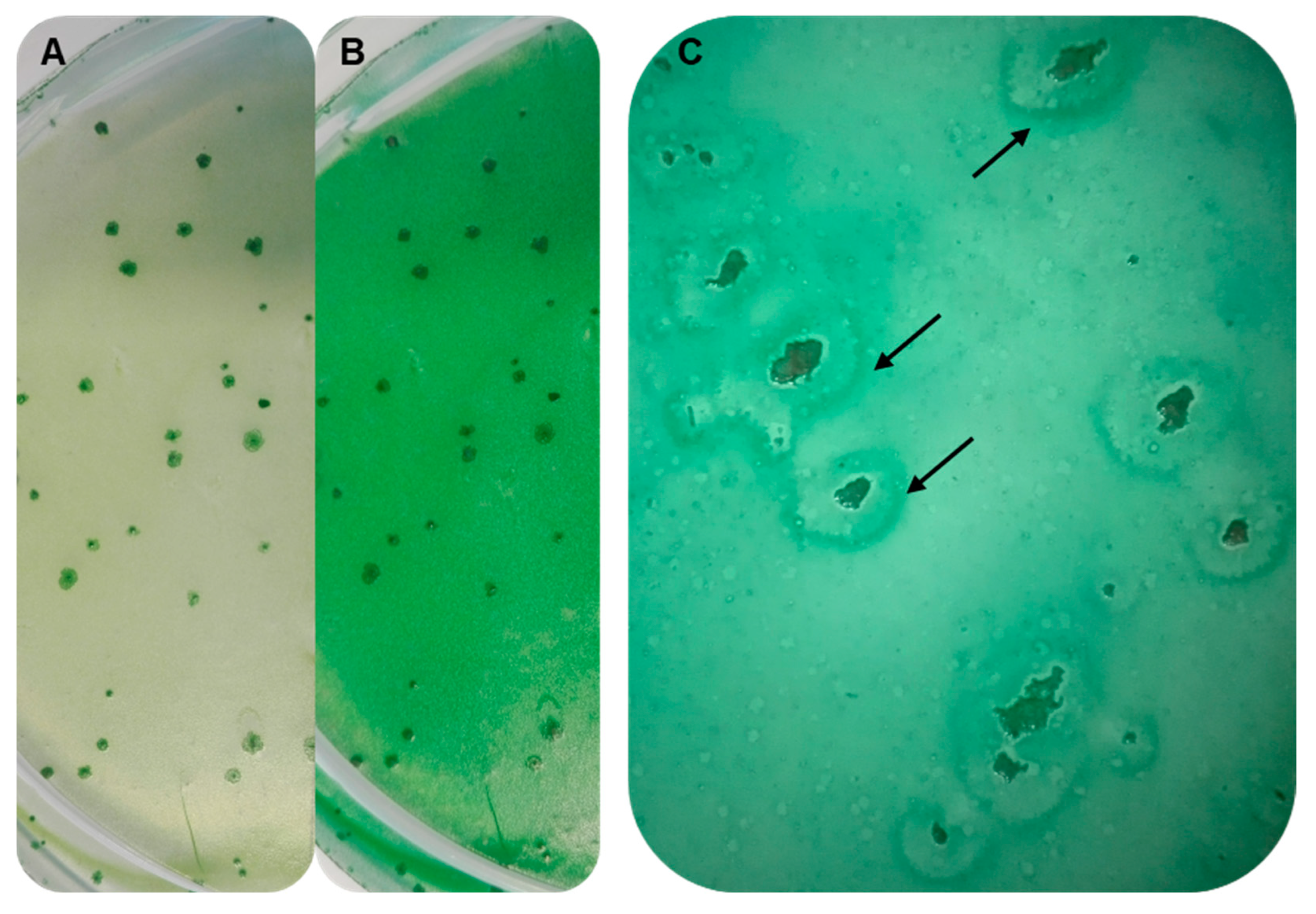
3.7. Establishing Interaction between the Strains SAG 88.79 and AGSC12
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rippka, R.; Cohen-Bazire, G. The Cyanobacteriales: A legitimate order based on the type strain Cyanobacterium stanieri? Ann. Microbiol. 1983, 134, 21–36. [Google Scholar] [CrossRef] [PubMed]
- Komárek, I.; Cepák, V.; Kaštovský, J.; Sulek, J. What are the cyanobacterial genera Cyanothece and Cyanobacterium? Contribution to the combined molecular and phenotype taxonomic evaluation of cyanobacterial diversity. Algol. Stud. 2004, 113, 1–36. [Google Scholar] [CrossRef]
- Moro, I.; Rascio, N.; La Rocca, N.; Di Bella, M.; Andreoli, C. Cyanobacterium aponinum, a new Cyanoprokaryote from the microbial mat of Euganean Thermal Springs (Padua, Italy). Algol. Stud. 2007, 123, 1–15. [Google Scholar] [CrossRef]
- Komárek, J.; Kaštovský, J.; Mareš, J.; Johansen, J.R. Taxonomic classification of cyanoprokaryotes (cyanobacterial genera) 2014, using a polyphasic approach. Preslia 2014, 86, 295–335. [Google Scholar]
- Sauvageau, C. Sur les algues d’eau douce recoltées en Algérie pendant le session de la Societé Botanique en 1892. Bulletin de la Société Botanique de France 1892, 39, civ–cxxviii. [Google Scholar] [CrossRef]
- NCBI Taxonomy Browser: Cyanobacterium stanieri PCC 7202. Available online: https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=292563&lvl=3&lin=f&keep=1&srchmode=1&unlock (accessed on 15 October 2022).
- Komárek, J. Taxonomic review of the genera Synechocystis SAUV, 1892, Synechococcus NÄG, 1849 and Cyanothece gen. nov. (Cyanophyceae). Arch. Für Protistenkd. 1976, 118, 119–179. [Google Scholar]
- Komárek, J.; Kopecký, J.; Cepák, V. Generic characters of the simplest cyanoprokaryotes Cyanobium, Cyanobacterium and Synechococcus. Cryptogam. Algol. 1999, 20, 209–222. [Google Scholar] [CrossRef]
- Honegger, R. Functional aspects of the lichen symbiosis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1991, 42, 553–578. [Google Scholar] [CrossRef]
- Singh, G.; Dal Grande, F.; Divakar, P.K.; Otte, J.; Crespo, A.; Schmitt, I. Fungal-algal association patterns in lichen symbiosis linked to macroclimate. New Phytol. 2017, 214, 317–329. [Google Scholar] [CrossRef]
- Grimm, M.; Grube, M.; Schiefelbein, U.; Zühlke, D.; Bernhardt, J.; Riedel, K. The Lichens’ Microbiota, Still a Mystery? Front. Microbiol. 2021, 12, 623839. [Google Scholar] [CrossRef]
- Gimmler, H. Mutualistic Relationships Between Algae and Fungi (Excluding Lichens). In Progress in Botany; Esser, K., Lüttge, U., Kadereit, J.W., Beyschlag, W., Eds.; Springer: Berlin/Heidelberg, Germany, 2001; Volume 62, pp. 194–214. [Google Scholar] [CrossRef]
- Calcott, M.J.; Ackerley, D.F.; Knight, A.; Keyzers, R.A.; Owen, J.G. Secondary metabolism in the lichen symbiosis. Chem. Soc. Rev. 2018, 47, 1730–1760. [Google Scholar] [CrossRef]
- Corral, L.G.; Post, L.S.; Montville, T.J. Antimicrobial activity of sodium bicarbonate. A research note. J. Food Sci. 1988, 53, 981–982. [Google Scholar] [CrossRef]
- Smilanick, J.L.; Margosan, D.A.; Mlikota, F.; Usall, J.; Michael, I.F. Control of Citrus Green Mold by Carbonate and Bicarbonate Salts and the Influence of Commercial Postharvest Practices on Their Efficacy. Plant Dis. 1999, 83, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Droby, S.; Wisniewski, M.; Ghaouth, A.E.; Wilson, C. Influence of food additives on the control of postharvest rots of apple and peach and efficacy of the yeast-based biocontrol product aspire. Postharvest Biol. Technol. 2003, 27, 127–135. [Google Scholar] [CrossRef]
- Gamagae, S.U.; Sivakumar, D.; Wijeratnam, R.S.W.; Wijesundera, R.L.C. Use of sodium bicarbonate and Candida oleophila to control anthracnose in papaya during storage. Crop Prot. 2003, 22, 775–779. [Google Scholar] [CrossRef]
- Zamani, M.; Sharifi Tehrani, A.; Ali Abadi, A.A. Evaluation of antifungal activity of carbonate and bicarbonate salts alone or in combination with biocontrol agents in control of citrus green mold. Commun. Agric. Appl. Biol. Sci. 2007, 72, 773–777. [Google Scholar] [PubMed]
- Letscher-Bru, V.; Obszynski, C.M.; Samsoen, M.; Sabou, M.; Waller, J.; Candolfi, E. Antifungal activity of sodium bicarbonate against fungal agents causing superficial infections. Mycopathologia 2013, 175, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Türkkan, M.; Özcan, M.; Erper, I. Antifungal effect of carbonate and bicarbonate salts against Botrytis cinerea, the casual agent of grey mould of kiwifruit. Akademik Ziraat Dergisi 2017, 6, 107–114. [Google Scholar] [CrossRef]
- Aiba, S.; Ogawa, T. Assessment of growth yield of a blue-green alga: Spirulina platensis, in axenic and continuous culture. J. Gen. Microbiol. 1977, 102, 179–182. [Google Scholar] [CrossRef]
- Sipiczki, M.; Ferenczy, L. Protoplast fusion of Schizosaccharomyces pombe auxotrophic mutants of identical mating-type. Mol. Gen. Genet. 1977, 151, 77–81. [Google Scholar] [CrossRef]
- Mitchison, J.M. Physiological and cytological methods for Schizosaccharomyces pombe. Methods Cell Biol. 1970, 4, 131–165. [Google Scholar] [CrossRef]
- Beever, R.E.; Bollard, E.G. The Nature of the Stimulation of Fungal Growth by Potato Extract. J. Gen. Microbiol. 1970, 60, 273–279. [Google Scholar] [CrossRef]
- Thom, C.; Church, M.B. The Aspergilli; Williams & Wilkins, Co.: Baltimore, MD, USA, 1926; p. 272. [Google Scholar]
- Sipiczki, M.; Takeo, K.; Grallert, A. Growth polarity transitions in a dimorphic fission yeast. Microbiology 1998, 144, 3475–3485. [Google Scholar] [CrossRef] [PubMed]
- Balazs, A.; Batta, G.; Miklos, I.; Acs-Szabo, L.; Vazquez de Aldana, C.R.; Sipiczki, M. Conserved regulators of the cell separation process in Schizosaccharomyces. Fungal Genet. Biol. 2012, 49, 235–249. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, C.S.; Winston, F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 1987, 57, 267–272. [Google Scholar] [CrossRef] [PubMed]
- del Campo, E.M.; del Hoyo, A.; Royo, C.; Casano, L.M.; Alvarez, R.; Barreno, E. A single primer pair gives a specific ortholog amplicon in a wide range of Cyanobacteria and plastid-bearing organisms: Applicability in inventory of reference material from collections and phylogenetic analysis. Mol. Phylogenet. Evol. 2010, 57, 1323–1328. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. A Guide Methods Appl. 1990, 18, 315–322. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Standard ethanol precipitation of DNA in microcentrifuge tubes. CSH. Protoc. 2006, 2006, pdb.prot4456. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Larsson, K.H.; Taylor, A.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019, 47, D259–D264. [Google Scholar] [CrossRef]
- Feng, P.; Lu, Q.; Najafzadeh, M.J.; Ende, A.H.; Sun, J.; Li, R.; Xi, L.; Vicente, V.; Lai, W.; Lu, C.; et al. Cyphellophora and its relatives in Phialophora: Biodiversity and possible role in human infection. Fungal Divers. 2012, 65, 17–45. [Google Scholar] [CrossRef]
- Réblová, M.; Untereiner, W.A.; Réblová, K. Novel evolutionary lineages revealed in the Chaetothyriales (Fungi) based on multigene phylogenetic analyses and comparison of ITS secondary structure. PLoS ONE 2013, 8, e63547. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Ma, Y.; Zhao, W.; Wei, Z.; Gleason, M.L.; Chen, H.; Hao, L.; Sun, G.; Zhang, R. Three New Species of Cyphellophora (Chaetothyriales) Associated with Sooty Blotch and Flyspeck. PLoS ONE 2015, 10, e0136857. [Google Scholar] [CrossRef] [PubMed]
- Crous, P.W.; Wingfield, M.J.; Guarro, J.; Cheewangkoon, R.; van der Bank, M.; Swart, W.J.; Stchigel, A.M.; Cano-Lira, J.F.; Roux, J.; Madrid, H.; et al. Fungal Planet description sheets: 154213. Persoonia 2013, 31, 188–296. [Google Scholar] [CrossRef] [PubMed]
- Madrid, H.; Hernández-Restrepo, M.; Gené, J.; Cano, J.; Guarro, J.; Silva, V. New and interesting chaetothyrialean fungi from Spain. Mycol. Prog. 2016, 15, 1179–1201. [Google Scholar] [CrossRef]
- Crous, P.W.; Schumacher, R.K.; Akulov, A.; Thangavel, R.; Hernández-Restrepo, M.; Carnegie, A.; Cheewangkoon, R.; Wingfield, M.; Summerell, B.; Quaedvlieg, W.; et al. New and Interesting Fungi. 2. Fungal Syst. Evol. 2019, 3, 57–134. [Google Scholar] [CrossRef]
- Cheewangkoon, R.; Groenewald, J.Z.; Summerell, B.A.; Hyde, K.D.; To-Anun, C.; Crous, P.W. Myrtaceae, a cache of fungal biodiversity. Persoonia 2009, 23, 55–85. [Google Scholar] [CrossRef]
- Crous, P.W.; Braun, U.; Wingfield, M.J.; Wood, A.R.; Shin, H.D.; Summerell, B.A.; Alfenas, A.C.; Cumagun, C.J.; Groenewald, J.Z. Phylogeny and taxonomy of obscure genera of microfungi. Persoonia 2009, 22, 139–161. [Google Scholar] [CrossRef] [PubMed]
- de Hoog, G.S.; Mayser, P.; Haase, G.; Horre, R.; Horrevorts, A.M. A new species, Phialophora europaea, causing superficial infections in humans. Mycoses 2000, 43, 409–416. [Google Scholar] [CrossRef]
- Phookamsak, R.; Hyde, K.; Jeewon, R.; Bhat, D.J.; Jones, E.; Maharachchikumbura, S.; Raspé, O.; Karunarathna, S.; Wanasinghe, D.N.; Hongsanan, S.; et al. Fungal diversity notes 929–1035: Taxonomic and phylogenetic contributions on genera and species of fungi. Fungal Divers. 2019, 95, 1–273. [Google Scholar] [CrossRef]
- Decock, C.; Delgado-Rodríguez, G.; Buchet, S.; Seng, J.M. A new species and three new combinations in Cyphellophora, with a note on the taxonomic affinities of the genus, and its relation to Kumbhamaya and Pseudomicrodochium. Antonie van Leeuwenhoek 2003, 84, 209–216. [Google Scholar] [CrossRef]
- Crous, P.W.; Wingfield, M.J.; Richardson, D.M.; Le Roux, J.J.; Strasberg, D.; Edwards, J.; Roets, F.; Hubka, V.; Taylor, P.W.; Heykoop, M.; et al. Fungal Planet description sheets: 400–468. Persoonia 2016, 36, 316–458. [Google Scholar] [CrossRef] [PubMed]
- Crous, P.W.; Wingfield, M.; Lombard, L.; Roets, F.; Swart, W.; Alvarado, P.; Carnegie, A.; Moreno, G.; Luangsa-Ard, J.; Thangavel, R.; et al. Fungal Planet description sheets: 951–1041. Pers.—Mol. Ph. Evol. Fungi 2019, 43, 223–425. [Google Scholar] [CrossRef] [PubMed]
- Crous, P.W.; Schubert, K.; Braun, U.; de Hoog, G.S.; Hocking, A.D.; Shin, H.D.; Groenewald, J.Z. Opportunistic, human-pathogenic species in the Herpotrichiellaceae are phenotypically similar to saprobic or phytopathogenic species in the Venturiaceae. Stud. Mycol. 2007, 58, 185–217. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Hyde, K.; Karunarathna, S.; Deng, C.; Gu, C.; Yang, S.; Zhang, Z. New species of Camptophora and Cyphellophora from China, and first report of sexual morphs for these genera. Phytotaxa 2018, 343, 149–159. [Google Scholar] [CrossRef]
- De Vries, G.A. Cyphellophora laciniata nov. gen., nov. sp. and Dactylium fusarioides Fragoso et Ciferri. Mycopathol. Mycol. Appl. 1962, 16, 47–54. [Google Scholar] [CrossRef]
- Gams, W.; Holubová-Jechová, V. Chloridium and some other dematiaceous hyphomycetes growing on decaying wood. Stud. Mycol. 1976, 13, 99. [Google Scholar]
- de Vries, G.A.; Elders, M.C.; Luykx, M.H. Description of Cyphellophora pluriseptata sp. nov. Antonie van Leeuwenhoek 1986, 52, 141–143. [Google Scholar] [CrossRef]
- Matsushima, T. Matsushima Mycological Memoirs; Matsushima Fungus Collection: Kobe, Japan, 1987; Volume 5, pp. 1–100. [Google Scholar]
- Walz, A.; de Hoog, G.S. A new species of Cyphellophora. Antonie van Leeuwenhoek 1987, 53, 143–146. [Google Scholar] [CrossRef]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008, 36, W465–W469. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Gascuel, O. BIONJ: An Improved Version of the NJ Algorithm Based on a Simple Model of Sequence Data. Mol. Biol. Evol. 1997, 14, 685–695. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar]
- Crous, P.W.; Braun, U.; McDonald, B.A.; Lennox, C.L.; Edwards, J.; Mann, R.C.; Zaveri, A.; Linde, C.C.; Dyer, P.S.; Groenewald, J.Z. Redefining genera of cereal pathogens: Oculimacula, Rhynchosporium and Spermospora. Fungal Syst. Evol. 2021, 7, 67–98. [Google Scholar] [CrossRef] [PubMed]
- de Hoog, G.S.; Weenink, X.O.; Ende, A.H. Taxonomy of the Phialophora verrucosa complex with the description of two new species. Stud. Mycol. 1999, 43, 107–121. [Google Scholar]
- Crous, P.W.; Cowan, D.A.; Maggs-Kölling, G.; Yilmaz, N.; Larsson, E.; Angelini, C.; Brandrud, T.E.; Dearnaley, J.D.W.; Dima, B.; Dovana, F.; et al. Fungal Planet description sheets: 1112–1181. Persoonia 2020, 45, 251–409. [Google Scholar] [CrossRef]
- Crous, P.W.; Boers, J.; Holdom, D.G.; Osieck, E.R.; Steinrucken, T.V.; Tan, Y.P.; Vitelli, J.S.; Shivas, R.G.; Barrett, M.; Boxshall, A.G.; et al. Fungal planet description sheets: 1383–1435. Pers.—Mol. Phylogeny Evol. Fungi 2022, 4, 261–371. [Google Scholar] [CrossRef]
- Zabel, R.A.; Lombard, F.F.; Wang, C.J.K.; Terracina, F. Fungi associated with decay in treated southern pine utility poles in the eastern United States. Wood Fiber Sci. 1985, 17, 75–91. [Google Scholar]
- Isola, D.; Selbmann, L.; de Hoog, G.S.; Fenice, M.; Onofri, S.; Prenafeta-Boldú, F.X.; Zucconi, L. Isolation and screening of black fungi as degraders of volatile aromatic hydrocarbons. Mycopathologia 2013, 175, 369–379. [Google Scholar] [CrossRef]
- Harper, D.D.; Farag, A.M.; Skaar, D. Acute toxicity of sodium bicarbonate, a major component of coal bed natural gas produced waters, to 13 aquatic species as defined in the laboratory. Environ. Toxicol. Chem. 2014, 33, 525–531. [Google Scholar] [CrossRef] [PubMed]
- Göttlich, E.; Van Der Lubbe, W.; Lange, B.; Fiedler, S.; Melchert, I.; Reifenrath, M.; Flemming, H.C.; De Hoog, S. Fungal flora in groundwater-derived public drinking water. Int. J. Hyg. Environ. Health 2002, 205, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.Y.; Zienkiewicz, K.; Vande Pol, N.; Ostrom, N.E.; Benning, C.; Bonito, G.M. Algal-fungal symbiosis leads to photosynthetic mycelium. eLife 2019, 8, e47815. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Li, T.; Jenkins, J.; Hu, Y.; Brueck, C.L.; Pei, H.; Betenbaugh, M.J. Evidence for a mutualistic relationship between the cyanobacteria Nostoc and fungi Aspergilli in different environments. Appl. Microbiol. Biotechnol. 2020, 104, 6413–6426. [Google Scholar] [CrossRef]
| Species | Strain | Origin | Maintaining Medium |
|---|---|---|---|
| Cyanobacterium stanierei | SAG 88.79 | SAG | Spirulina |
| Aspergillus niger | CBS 554.65 | CBS-KNAW | PDA |
| Penicillium expansum | SZMC 2175 | SZMC | PDA |
| Fusarium graminearum | FGSC 9075 | ATCC | PDA |
| unkown fungus | AGSC12 | This study | PDA |
| Species | NaCl Tolerance on PDA | ||||||
|---|---|---|---|---|---|---|---|
| 0.0 M | 0.1 M | 0.3 M | 0.5 M | 0.7 M | 0.9 M | ||
| A. niger | 3 | 3 | 3 | 3 | 3 | 3 | |
| P. expansum | 3 | 3 | 3 | 3 | 3 | 3 | |
| F. graminearum | 3 | 3 | 3 | 3 | 3 | 3 | |
| C. olivacea | 3 | 3 | 3 | 3 | 2 | 2 | |
| NaHCO3 I. Tolerance on PDA | |||||||
| 0.0 M | 0.1 M | 0.3 M | 0.5 M | 0.7 M | 0.9 M | 1.1 M | |
| A. niger | 3 | 0 | 0 | 0 | 0 | 0 | ND |
| P. expansum | 3 | 2 | 0 | 0 | 0 | 0 | ND |
| F. graminearum | 3 | 3 | 1 | 1 | 1 | 0 | 0 |
| C. olivacea | 3 | 3 | 3 | 3 | 2 | 2 | 2 |
| NaHCO3 II. Tolerance on PDA | |||||||
| 0.0 M | 0.1 M | 0.3 M | 0.5 M | 0.7 M | 0.9 M | 1.1 M | |
| A. niger | 3 | 0 | 0 | 0 | 0 | 0 | ND |
| P. expansum | 3 | 2 | 0 | 0 | 0 | 0 | ND |
| F. graminearum | 3 | 3 | 1 | 1 | 1 | 0 | ND |
| C. olivacea | 3 | 3 | 3 | 3 | 2 | 2 | 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Acs-Szabo, L.; Papp, L.A.; Csoma, H.; Miklos, I.; Sipiczki, M. New Strain of Cyphellophora olivacea Exhibits Striking Tolerance to Sodium Bicarbonate. Diversity 2022, 14, 1023. https://doi.org/10.3390/d14121023
Acs-Szabo L, Papp LA, Csoma H, Miklos I, Sipiczki M. New Strain of Cyphellophora olivacea Exhibits Striking Tolerance to Sodium Bicarbonate. Diversity. 2022; 14(12):1023. https://doi.org/10.3390/d14121023
Chicago/Turabian StyleAcs-Szabo, Lajos, Laszlo Attila Papp, Hajnalka Csoma, Ida Miklos, and Matthias Sipiczki. 2022. "New Strain of Cyphellophora olivacea Exhibits Striking Tolerance to Sodium Bicarbonate" Diversity 14, no. 12: 1023. https://doi.org/10.3390/d14121023
APA StyleAcs-Szabo, L., Papp, L. A., Csoma, H., Miklos, I., & Sipiczki, M. (2022). New Strain of Cyphellophora olivacea Exhibits Striking Tolerance to Sodium Bicarbonate. Diversity, 14(12), 1023. https://doi.org/10.3390/d14121023






