Effects of David Deer Grazing on Soil Bacterial and Fungal Communities in an Eastern Coastal Wetland of China
Abstract
1. Introduction
2. Materials and Methods
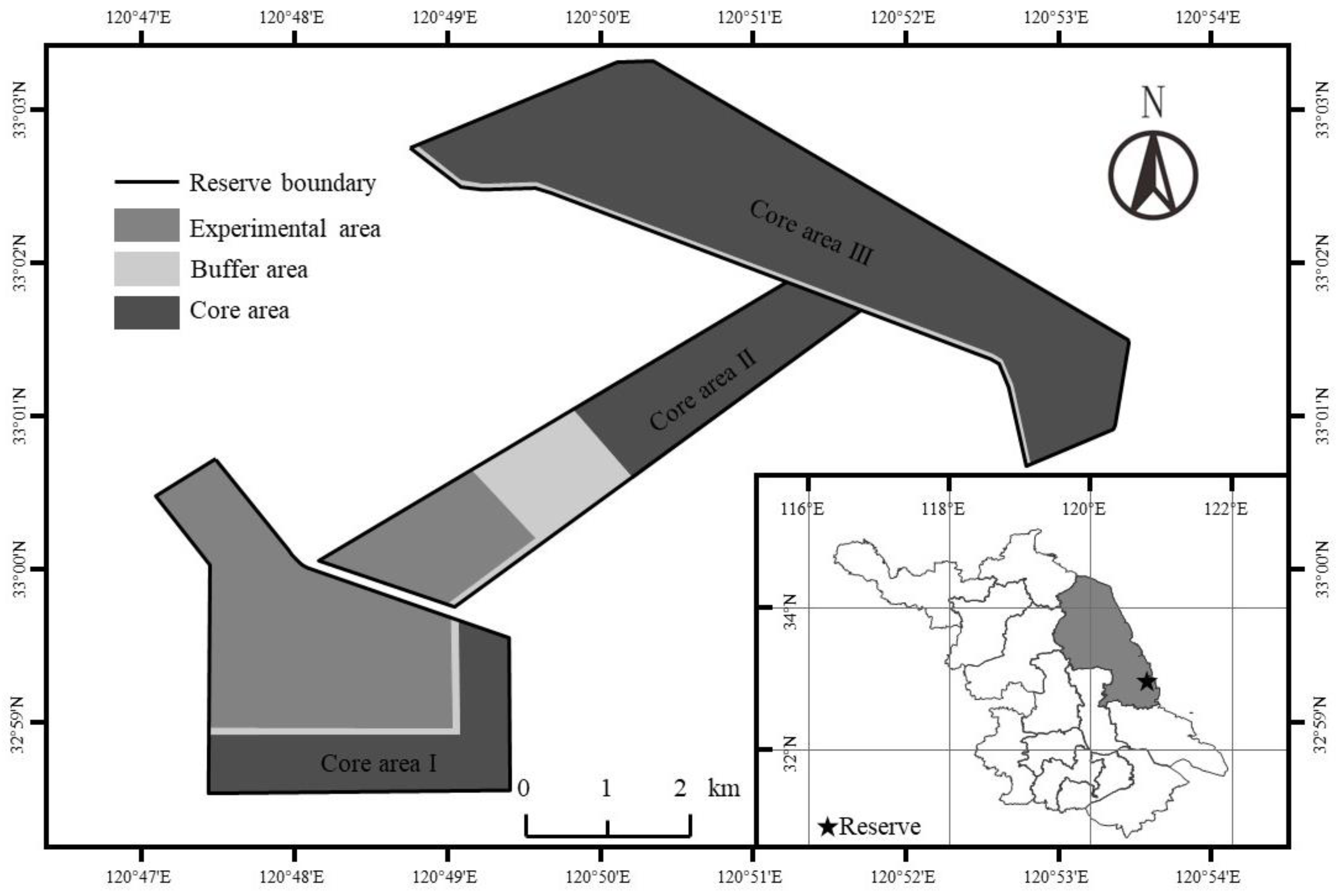
2.1. Study Area
2.2. Experimental Design
2.3. Determination of Soil Environmental Factors
2.4. Soil Microbial Community DNA Extraction and Amplification and Sequencing
2.5. Data Processing and Analysis
3. Results
3.1. Effect of Grazing on Soil Properties
3.2. Diversity of Soil Bacterial and Fungal Communities
3.2.1. Soil Bacterial and Fungal Communities Alpha Diversity
3.2.2. Soil Bacterial and Fungal Communities Beta Diversity
3.3. Soil Microbial Community Composition
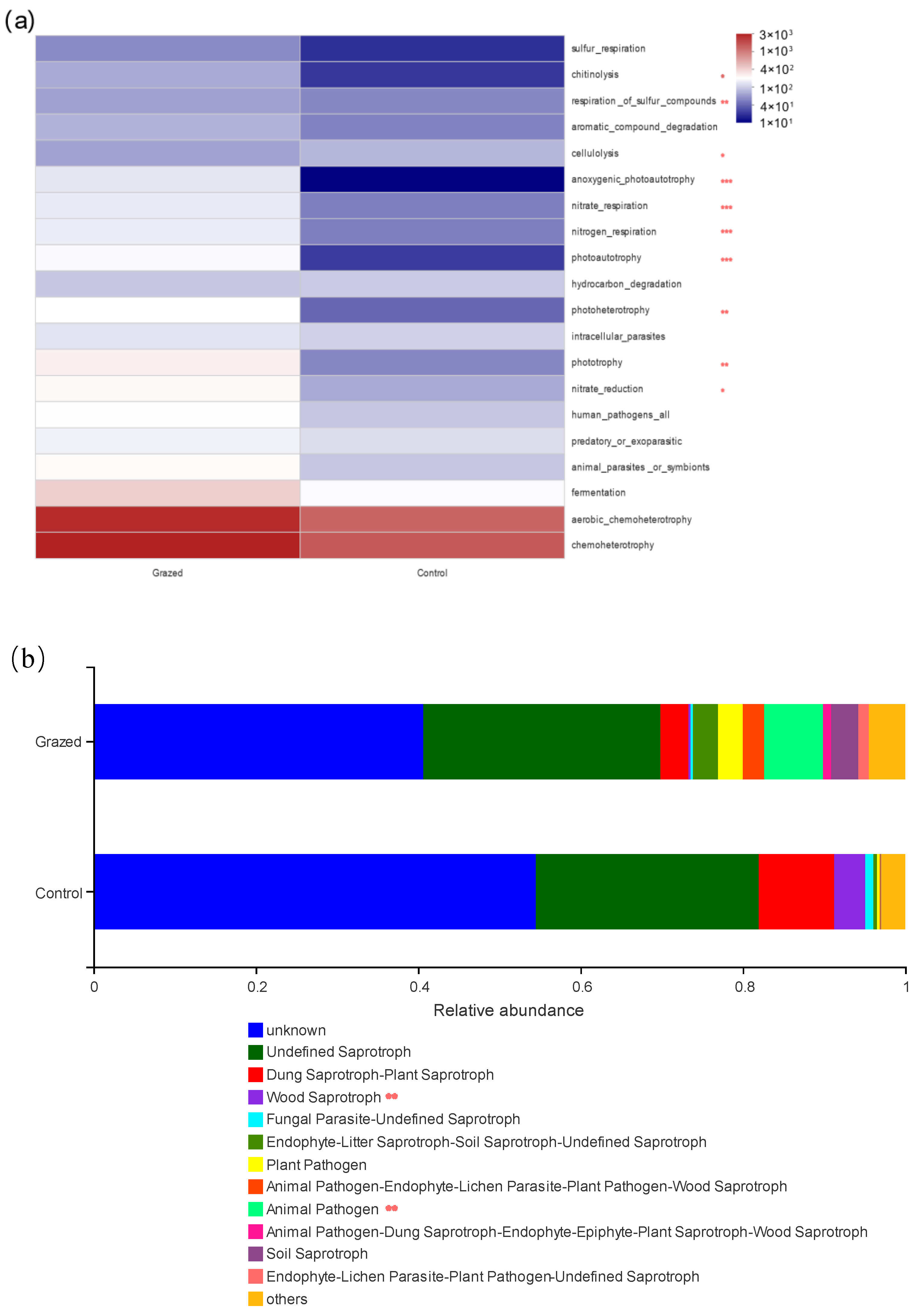
3.4. Predicting the Function of Soil Bacteria and Fungi
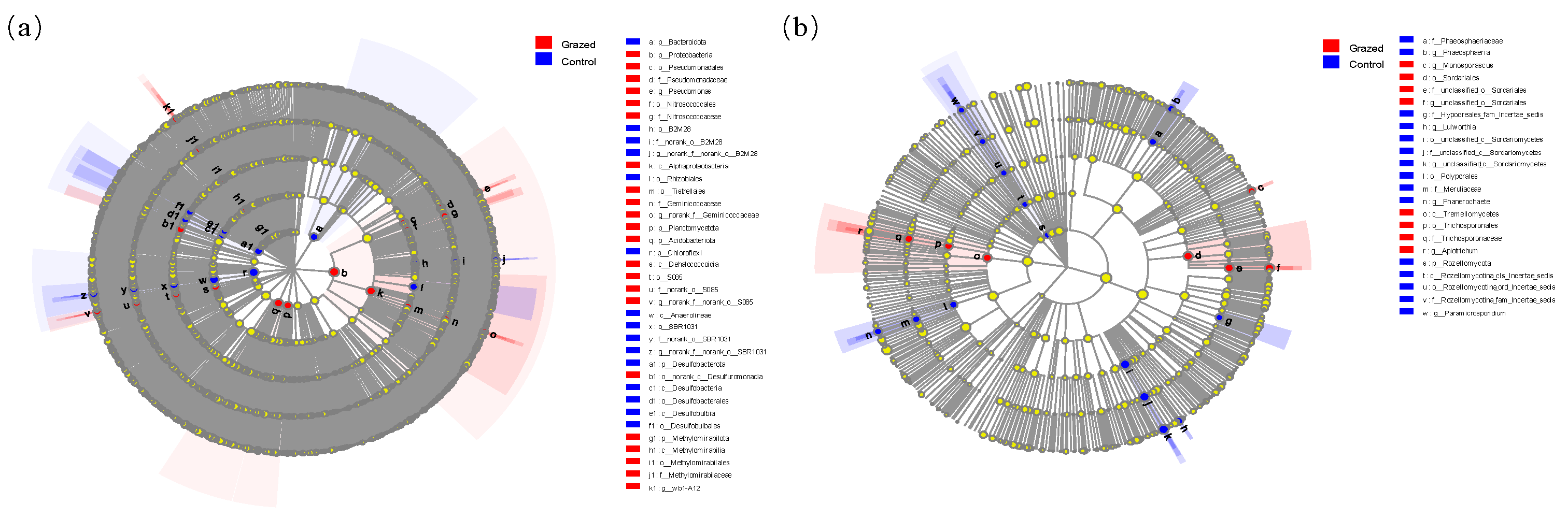
3.5. Soil Microbial Community LEfSe Analysis
3.6. RDA Analysis of Soil Microbial Community and Soil Environmental Factors
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khan, R.W.A.; Shaheen, H.; Mehmood, A.; Awan, S.N. Grazing intensity impacts on soil carbon stocks of Western Himalayan Alpine paddocks. Carbon Manag. 2019, 10, 533–540. [Google Scholar] [CrossRef]
- Fenetahun, Y.; Yuan, Y.; Xu, X.W.; Fentahun, T.; Nzabarinda, V.; Wang, Y.D. Impact of Grazing Intensity on Soil Properties in Teltele Rangeland, Ethiopia. Front. Environ. 2021, 9, 664104. [Google Scholar] [CrossRef]
- Doran, J.W.; Zeiss, M.R. Soil health and sustainability: Managing the biotic component of soil quality. Appl. Soil Ecol. 2000, 15, 3–11. [Google Scholar] [CrossRef]
- Chen, L.L.; Saixi, Y.L.; Yi, R.; Baoyin, T. Characterization of soil microbes associated with a grazing-tolerant grass species, Stipa breviflora, in the Inner Mongolian desert steppe. Ecol. Evol. 2020, 10, 10607–10618. [Google Scholar] [CrossRef]
- Lienhard, P.; Terrat, S.; Prevost-Boure, N.C.; Nowak, V.; Regnier, T.; Sayphoummie, S.; Panyasiri, K.; Tivet, F.; Mathieu, O.; Leveque, J.; et al. Pyrosequencing evidences the impact of cropping on soil bacterial and fungal diversity in Laos tropical grassland. Agron. Sustain. Dev. 2014, 34, 525–533. [Google Scholar] [CrossRef]
- Zeilinger, S.; Gupta, V.K.; Dahms, T.E.S.; Silva, R.N.; Singh, H.B.; Upadhyay, R.S.; Gomes, E.V.; Tsui, C.K.M.; Nayak, S.C.; van der Meer, J.R. Friends or foes? Emerging insights from fungal interactions with plants. FEMS Microbiol. Rev. 2016, 40, 182–207. [Google Scholar] [CrossRef]
- Zhao, F.Z.; Ren, C.J.; Shelton, S.; Wang, Z.T.; Pang, G.W.; Chen, J.; Wang, J. Grazing intensity influence soil microbial communities and their implications for soil respiration. Agric. Ecosyst. Environ. 2017, 249, 50–56. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, Q.; Staley, C.; Gao, H.L.; Ishii, S.; Wei, X.R.; Liu, J.; Cheng, J.M.; Hao, M.D.; Sadowsky, M.J. Impact of long-term grazing exclusion on soil microbial community composition and nutrient availability. Biol. Fert. Soils 2019, 55, 121–134. [Google Scholar] [CrossRef]
- Zhang, H.R.; Fu, G. Responses of plant, soil bacterial and fungal communities to grazing vary with pasture seasons and grassland types, northern Tibet. Land Degrad. Dev. 2021, 32, 1821–1832. [Google Scholar] [CrossRef]
- Yang, F.; Niu, K.C.; Collins, C.G.; Yan, X.B.; Ji, Y.G.; Ling, N.; Zhou, X.H.; Du, G.Z.; Guo, H.; Hu, S.J. Grazing practices affect the soil microbial community composition in a Tibetan alpine meadow. Land Degrad. Dev. 2019, 30, 49–59. [Google Scholar] [CrossRef]
- Ma, X.Y.; Zhang, Q.T.; Zheng, M.M.; Gao, Y.; Yuan, T.; Hale, L.; Van Nostrand, J.D.; Zhou, J.Z.; Wan, S.Q.; Yang, Y.F. Microbial functional traits are sensitive indicators of mild disturbance by lamb grazing. ISME J. 2019, 13, 1370–1373. [Google Scholar] [CrossRef]
- Hijuelos, A.C.; Dijkstra, J.T.; Carruthers, T.J.B.; Heynert, K.; Reed, D.J.; van Wesenbeeck, B.K. Linking management planning for coastal wetlands to potential future wave attenuation under a range of relative sea-level rise scenarios. PLoS ONE 2019, 14, e0216695. [Google Scholar] [CrossRef] [PubMed]
- An, Y.T.; Liu, B.; Wang, L.B.; Xie, S.B.; Xue, D.D.; Wu, Y.B. Effects of Père David deer (Elaphurus davidianus) grazing on soil physicochemical properties. Acta Ecol. Sin. 2020, 40, 3571–3578. [Google Scholar]
- Liu, J.G.; Xue, J.H.; Wang, L.; Ding, J.J.; Ma, W.L.; Liu, C.G.; Rong, Y. Habitat degradation features of Père David’s Deer Natural Reserve in Dafeng of Jiangsu Province, East China. Chin. J. Ecol. 2011, 30, 1793–1798. [Google Scholar]
- Nunan, N.; Singh, B.; Reid, E.; Ord, B.; Papert, A.; Squires, J.; Prosser, J.I.; Wheatley, R.E.; McNicol, J.; Millard, P. Sheep-urine-induced changes in soil microbial community structure. FEMS Microbiol. Ecol. 2006, 56, 310–320. [Google Scholar] [CrossRef]
- Siciliano, S.D.; Palmer, A.S.; Winsley, T.; Lamb, E.; Bissett, A.; Brown, M.V.; van Dorst, J.; Ji, M.K.; Ferrari, B.C.; Grogan, P.; et al. Soil fertility is associated with fungal and bacterial richness, whereas pH is associated with community composition in polar soil microbial communities. Soil Biol. Biochem. 2014, 78, 10–20. [Google Scholar] [CrossRef]
- Ramirez, P.B.; Fuentes-Alburquenque, S.; Diez, B.; Vargas, I.; Bonilla, C.A. Soil microbial community responses to labile organic carbon fractions in relation to soil type and land use along a climate gradient. Soil Biol. Biochem. 2020, 141, 107692. [Google Scholar] [CrossRef]
- Zhang, G.L.; Bai, J.H.; Tebbe, C.C.; Zhao, Q.Q.; Jia, J.; Wang, W.; Wang, X.; Yu, L. Salinity controls soil microbial community structure and function in coastal estuarine wetlands. Environ. Microbiol. 2021, 23, 1020–1037. [Google Scholar] [CrossRef]
- Shezi, T.A.; O’Connor, T.G.; Witkowski, E.T.F. Impact of livestock grazing intensity on plant diversity of montane grassland in the northern Drakensberg, South Africa. Afr. J. Range For. Sci. 2021, 38, 67–79. [Google Scholar] [CrossRef]
- Xu, N.; Tan, G.C.; Wang, H.Y.; Gai, X.P. Effect of biochar additions to soil on nitrogen leaching, microbial biomass and bacterial community structure. Eur. J. Soil Biol. 2016, 74, 1–8. [Google Scholar] [CrossRef]
- Chen, B.S.; Du, K.Q.; Sun, C.; Vimalanathan, A.; Liang, X.L.; Li, Y.; Wang, B.H.; Lu, X.M.; Li, L.J.; Shao, Y.Q. Gut bacterial and fungal communities of the domesticated silkworm (Bombyx mori) and wild mulberry-feeding relatives. ISME J. 2018, 12, 2252–2262. [Google Scholar] [CrossRef]
- Rodrigues, J.L.M.; Pellizari, V.H.; Mueller, R.; Baek, K.; Jesus, E.D.; Paula, F.S.; Mirza, B.; Hamaoui, G.S.; Tsai, S.M.; Feigl, B.; et al. Conversion of the Amazon rainforest to agriculture results in biotic homogenization of soil bacterial communities. Proc. Natl. Acad. Sci. USA 2013, 110, 988–993. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.J.; Yin, Y.; Zhu, W.X.; Zhou, Y.B. Variations in Soil Bacterial Community Diversity and Structures among Different Revegetation Types in the Baishilazi Nature Reserve. Front. Microbiol. 2018, 9, 2874. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.C.; Dong, Y.H.; An, S.S. Bacterial Community Responses to Soils along a Latitudinal and Vegetation Gradient on the Loess Plateau, China. PLoS ONE 2016, 11, e0152894. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an ecological classification of soil bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef]
- Lesschen, J.P.; van den Berg, M.; Westhoek, H.J.; Witzke, H.P.; Oenema, O. Greenhouse gas emission profiles of European livestock sectors. Anim. Feed Sci. Technol. 2011, 166–167, 16–28. [Google Scholar] [CrossRef]
- Yoshitake, S.; Soutome, H.; Koizumi, H. Deposition and decomposition of cattle dung and its impact on soil properties and plant growth in a cool-temperate pasture. Ecol. Res. 2014, 29, 673–684. [Google Scholar] [CrossRef]
- Liao, C.Z.; Luo, Y.Q.; Jiang, L.F.; Zhou, X.H.; Wu, X.W.; Fang, C.M.; Chen, J.K.; Li, B. Invasion of Spartina alterniflora enhanced ecosystem carbon and nitrogen stocks in the Yangtze Estuary, China. Ecosystems 2007, 10, 1351–1361. [Google Scholar] [CrossRef]
- Hug, L.A.; Castelle, C.J.; Wrighton, K.C.; Thomas, B.C.; Sharon, I.; Frischkorn, K.R.; Williams, K.H.; Tringe, S.G.; Banfield, J.F. Community genomic analyses constrain the distribution of metabolic traits across the Chloroflexi phylum and indicate roles in sediment carbon cycling. Microbiome 2013, 1, 22. [Google Scholar] [CrossRef]
- Xiong, J.B.; Liu, Y.Q.; Lin, X.G.; Zhang, H.Y.; Zeng, J.; Hou, J.Z.; Yang, Y.P.; Yao, T.D.; Knight, R.; Chu, H.Y. Geographic distance and pH drive bacterial distribution in alkaline lake sediments across Tibetan Plateau. Environ. Microbiol. 2012, 14, 2457–2466. [Google Scholar] [CrossRef]
- Eichorst, S.A.; Kuske, C.R.; Schmidt, T.M. Influence of Plant Polymers on the Distribution and Cultivation of Bacteria in the Phylum Acidobacteria. Appl. Environ. Microb. 2011, 77, 586–596. [Google Scholar] [CrossRef]
- Davis, K.E.R.; Sangwan, P.; Janssen, P.H. Acidobacteria, Rubrobacteridae and Chloroflexi are abundant among very slow-growing and mini-colony-forming soil bacteria. Environ. Microbiol. 2011, 13, 798–805. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Y.; Hu, B.X.; Ren, H.J.; Zhang, J. Composition and functional diversity of microbial community across a mangrove-inhabited mudflat as revealed by 16S rDNA gene sequences. Sci. Total Environ. 2018, 633, 518–528. [Google Scholar] [CrossRef] [PubMed]
- Rooney, D.; Kennedy, N.; Deering, L.; Gleeson, D.; Clipson, N. Effect of sheep urine deposition on the bacterial community structure in an acidic upland grassland soil. Appl. Environ. Microb. 2006, 72, 7231–7237. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.Y.; Zhou, X.; Guo, D.; Zhao, J.H.; Yan, L.; Feng, G.Z.; Gao, Q.; Yu, H.; Zhao, L.P. Soil pH is the primary factor driving the distribution and function of microorganisms in farmland soils in northeastern China. Ann. Microbiol. 2019, 69, 1461–1473. [Google Scholar] [CrossRef]
- Huang, L.; Gao, X.; Liu, M.; Du, G.; Guo, J.S.; Ntakirutimana, T. Correlation among soil microorganisms, soil enzyme activities, and removal rates of pollutants in three constructed wetlands purifying micro-polluted river water. Ecol. Eng. 2012, 46, 98–106. [Google Scholar] [CrossRef]
- Wang, H.Y.; Guo, S.Y.; Huang, M.R.; Thorsten, L.H.; Wei, J.C. Ascomycota has a faster evolutionary rate and higher species diversity than Basidiomycota. Sci. China Life Sci. 2010, 53, 1163–1169. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.L.; Xu, T.L.; Veresoglou, S.D.; Hu, H.W.; Hao, Z.P.; Hu, Y.J.; Liu, L.; Deng, Y.; Rillig, M.C.; Chen, B.D. Plant diversity represents the prevalent determinant of soil fungal community structure across temperate grasslands in northern China. Soil Biol. Biochem. 2017, 110, 12–21. [Google Scholar] [CrossRef]
- Treseder, K.K.; Maltz, M.R.; Hawkins, B.A.; Fierer, N.; Stajich, J.E.; McGuire, K.L. Evolutionary histories of soil fungi are reflected in their large-scale biogeography. Ecol. Lett. 2014, 17, 1086–1093. [Google Scholar] [CrossRef]
- Yelle, D.J.; Ralph, J.; Lu, F.C.; Hammel, K.E. Evidence for cleavage of lignin by a brownrot basidiomycete. Environ. Microbiol. 2008, 10, 1844–1849. [Google Scholar] [CrossRef]



| Areas | Samples | Hoofprint Density/pcs/m2 | Fecal Residue/pcs/m2 | Vegetation Coverage/% | Dominant Plant |
|---|---|---|---|---|---|
| Grazed | G1 | 67 | 89 | 0 | -- |
| G2 | 52 | 73 | 0 | -- | |
| G3 | 71 | 98 | 0 | -- | |
| Control | C1 | 0 | 0 | 80 | S. alterniflora |
| C2 | 0 | 0 | 75 | S. alterniflora | |
| C3 | 0 | 0 | 79 | S. alterniflora |
| Properties | Treats | |
|---|---|---|
| Grazed | Control | |
| BD/(g/cm3) | 1.7034 ± 0.10165 | 1.6421 ± 0.14977 |
| SWC/(%) | 13.6527 ± 3.27637 | 15.3944 ± 5.1625 |
| pH | 9.2907 ± 0.12256 A | 8.8247 ± 0.11122 B |
| Na+/(g/kg) | 0.2677 ± 0.14046 | 0.2901 ± 0.03025 |
| SOC/(g/kg) | 2.2074 ± 0.76054 B | 7.1627 ± 2.72498 A |
| TN/(g/kg) | 0.2063 ± 0.03146 B | 0.6987 ± 0.25665 A |
| TP/(g/kg) | 1.2186 ± 0.4882 B | 1.8456 ± 0.2672 A |
| TK/(g/kg) | 7.3589 ± 0.61162 b | 7.8083 ± 0.28175 a |
| CAT/(mg/g) | 8.1981 ± 3.29221 | 8.077 ± 2.06997 |
| URE/(mg/g) | 36.5701 ± 10.35941 B | 82.3233 ± 13.19902 A |
| SUC/(mg/g) | 0.1181 ± 0.03546 B | 0.4468 ± 0.26713 A |
| Samples | Shannon | Simpson | Ace | Chao | |
|---|---|---|---|---|---|
| Bacteria | Grazed | 6.12 ± 0.36 b | 0.01 ± 0.006 A | 3145.96 ± 632.27 | 3122.43 ± 625.61 |
| Control | 6.38 ± 0.22 a | 0.005 ± 0.001 B | 3286.51 ± 432.99 | 3270.72 ± 431.40 | |
| Fungi | Grazed | 1.83 ± 0.98 | 0.36 ± 0.28 | 52.01 ± 20.92 B | 48.36 ± 20.91 B |
| Control | 2.03 ± 0.81 | 0.31 ± 0.24 | 299.92 ± 122.27 A | 294.23 ± 124.18 A |
| Indexes | Bacteria | Fungi | ||||||
|---|---|---|---|---|---|---|---|---|
| RDA1 | RDA2 | r2 | p Values | RDA1 | RDA2 | r2 | p Values | |
| BD | −0.9877 | 0.1563 | 0.1019 | 0.251 | −0.9993 | 0.0381 | 0.0262 | 0.727 |
| SWC | 0.9478 | 0.3189 | 0.1473 | 0.139 | 0.9921 | 0.1254 | 0.0598 | 0.491 |
| pH | −0.9915 | 0.13 | 0.7076 | 0.001 | −0.3101 | 0.9507 | 0.1223 | 0.172 |
| Na+ | 0.9848 | 0.1735 | 0.01 | 0.883 | −0.9849 | 0.1732 | 0.0278 | 0.703 |
| SOC | 0.9771 | −0.2128 | 0.4081 | 0.001 | 0.8539 | −0.5205 | 0.1968 | 0.055 |
| TN | 0.9653 | −0.2613 | 0.4612 | 0.002 | 0.6885 | −0.7252 | 0.11 | 0.203 |
| TP | 0.9387 | −0.3448 | 0.4146 | 0.001 | 0.9307 | −0.3658 | 0.0794 | 0.318 |
| TK | 0.8061 | −0.5918 | 0.1981 | 0.05 | −0.0851 | −0.9964 | 0.2113 | 0.044 |
| CAT | 0.1707 | −0.9853 | 0.0873 | 0.27 | −0.9784 | 0.2065 | 0.0633 | 0.44 |
| URE | 0.9729 | −0.2312 | 0.6308 | 0.001 | 0.4821 | −0.8761 | 0.266 | 0.014 |
| SUC | 0.9882 | 0.153 | 0.625 | 0.001 | 0.4275 | −0.904 | 0.0475 | 0.541 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, Y.; Wu, Y.; An, Y. Effects of David Deer Grazing on Soil Bacterial and Fungal Communities in an Eastern Coastal Wetland of China. Diversity 2022, 14, 976. https://doi.org/10.3390/d14110976
Zhu Y, Wu Y, An Y. Effects of David Deer Grazing on Soil Bacterial and Fungal Communities in an Eastern Coastal Wetland of China. Diversity. 2022; 14(11):976. https://doi.org/10.3390/d14110976
Chicago/Turabian StyleZhu, Yi, Yongbo Wu, and Yuting An. 2022. "Effects of David Deer Grazing on Soil Bacterial and Fungal Communities in an Eastern Coastal Wetland of China" Diversity 14, no. 11: 976. https://doi.org/10.3390/d14110976
APA StyleZhu, Y., Wu, Y., & An, Y. (2022). Effects of David Deer Grazing on Soil Bacterial and Fungal Communities in an Eastern Coastal Wetland of China. Diversity, 14(11), 976. https://doi.org/10.3390/d14110976





