Abstract
Striking geographic variation in male advertisement calls was observed in frogs formerly referred to as Limnonectes doriae and L. limborgi, respectively. Subsequent analyses of mtDNA and external morphological data brought supporting evidence for the recognition of these populations as distinct species. We describe two new frog species of the genus Limnonectes (i.e., L. bagoensis sp. nov. and L. bagoyoma sp. nov.) from Myanmar. Limnonectes bagoensis sp. nov. is closely related to L. doriae whereas L. bagoyoma sp. nov. is closely related to L. limborgi. Results of this integrative study provide evidence for the presence of additional undescribed species in these species complexes but due to the lack of bioacoustical data, we consider these additional diverging populations as candidate species that need further study to resolve their respective taxonomic status. Both new species are distributed in Lower Myanmar. Limnonectes doriae is restricted to southern Myanmar along the Malayan Peninsula whereas L. limborgi is known to occur in eastern Myanmar and northwestern Thailand. The remaining populations formerly referred to as either L. doriae or L. limborgi are considered representatives of various candidate species that await further study. We further provide a de novo draft genome of the respective holotypes of L. bagoensis sp. nov. and L. bagoyoma sp. nov. based on short-read sequencing technology to 25-fold coverage.
1. Introduction
The anuran family Dicroglossidae has its center of distribution and greatest species diversity in South, Southeast, and East Asia as well as on the islands of the Sunda Shelf and the Philippines but is also known from northwestern and sub-Saharan Africa across the southern Arabian Peninsula to Pakistan [1]. The great number of new species described every year (e.g., [2,3,4]) shows that this group of frogs is understudied. Obviously, much more field and lab work is needed to evaluate the actual diversity and better understand their phylogenetic relationships. Additionally, many of the supposedly widespread species actually consist of several species. This further drives the discovery of new species with intensified study (e.g., [5,6,7,8]).
During fieldwork in Myanmar, we came across variations in the male advertisement call of two frog species that belong to the genus Limnonectes, L. doriae (Boulenger, 1887) and L. limborgi (Sclater, 1892), respectively. In fact, the calls were so different that we immediately suspected multiple species and initiated a taxonomic study of these two species complexes to evaluate if the bioacoustically distinct populations were also differentiated in morphology and molecular genetics and warrant describing them as distinct species.
The species currently recognized as L. limborgi was originally described by Sclater (1892) as Rana limborgi. The holotype, however, is lost and a neotype (specimen MSNG 29422) collected by Leonard Fea at “Karin Bia-po”, Myanmar, was designated [9]. In the descriptions of his travel activities in Myanmar, Fea [10] reported that some of his zoological collections were made at “Leitò, villaggio di Carin Chebà o Bia-Po, posto a 900 metri sul livello del mare” (=Leitò, village of Carin Chebà or Bia-Po, located at 900 m above sea level). Today this village is called Leiktho and is located at 19.2212°, 96.5818°, 865–885 m above sea level. Other authors published erroneous GPS coordinates for Lea’s “Bia-Po” such as “19°03′ N/96°30′ E” (=19.05°, 96.50°) [9] or “19°15′ N/97°30′ E” (=19.25°, 97.50°) [11]. The names Carin or Karin refer to the Independent State of Carin that included “Leitò” at the time of Lea’s travels in Myanmar. Today, this village is in Kayin State.
Dubois [12] established a new genus, Taylorana, to accommodate the species of Limnonectes with direct development, comprising L. hascheanus and L. limborgi. However, phylogenetic analyses show (e.g., [13,14]) that these two species are nested within the remaining species currently assigned to Limnonectes. Thus, the placement of these two species in Taylorana was never widely accepted, though some authors recognized Taylorana as a subgenus within Limnonectes (e.g., [9,15]). Smith [16] considered L. hascheanus and L. limborgi to be synonyms until L. limborgi was resurrected from synonymy with L. hascheanus by Dubois [12], and Inger and Stuart [9] demonstrated that these two nominal species differ in body size, degree of toe webbing, development of odontoids in adult males as well as in molecular genetics.
In 1887, Boulenger [16] introduced his new species Rana doriae based on material he had obtained from Leonardo Fea. The geographic origin of the type series is given as “Thagatà Juwa; Kaw-ka-riet” (now called Kawkareik, about 66 km east of the city of Mawlamyine, northern Tanintharyi, Myanmar; see also [1]). The GPS coordinates for this locality were given as 16°33′ N, 98°14′ E [11]. Ohler and Dubois [17] restricted the geographic distribution of L. doriae to the Tenasserim region in Myanmar and adjacent Thailand on the Malayan Peninsula. However, Inger and Stuart [9] identified a specimen from Bago Yoma, Bago Region, Myanmar, as L. doriae.
Smith [18] associated Rana doriae with a group of species that all tend to develop a swelling or flap-like structure in the occipital region on top of the head as well as the presence of well-developed odontoids on the lower jaw on either side of the symphysis. Later, the species was transferred to the genus Limnonectes [12]. Ohler and Dubois [17] placed Limnonectes doriae in the subgenus Elachyglossa along with L. dabanus, L. gyldenstolpei (=the type species of this subgenus), L. kohchangae, L. macrognathus, L. maw-phlangensis, L. plicatellus, and L. toumano. Lambertz et al. [19] restricted the subgenus Elachyglossa to the four caruncle-bearing species of Limnonectes that were recognized at the time (i.e., L. dabanus, L. gyldenstolpei, L. macrognathus, and L. plicatellus). However, the phylogeny published by Phimmachak et al. [20] indicated L. kohchangae, L. macrognathus, and L. plicatellus to be more closely related to the non-caruncle-bearing species L. doriae, L. hascheanus, and L. limborgi than to the other caruncle-bearing species (i.e., L. dabanus, L. gyldenstolpei, L. lauhachindai, L. savan). Most recently, Yodthong et al. [21] described a new species related to L. doriae. Obviously, additional phylogenetic studies are needed including also nuclear DNA loci in order to resolve the relationships of the species of Limnonectes.
2. Materials and Methods
The methods and format of the presentation follow previous works coauthored by the senior author [2,5,22]. Specimens examined for this study were collected by GK, NLT, and PT (see Appendix A for specimens examined). Specimens labeled with GK field numbers were deposited in the collections of Senckenberg Forschungsmuseum Frankfurt (SMF) or at East Yangon University (EYU), Thanlyin, Myanmar, and those with PT field numbers were deposited in the collection of the Chulalongkorn University Museum of Natural History (CUMZ), Bangkok.
Prior to the preservation of collected specimens in the field, we took color photographs of each individual in life. We euthanized the frogs with a pericardial injection of T61 or MS 222. We cut tissue samples from one forelimb or from the tongue and preserved these in 98% non-denatured ethanol for DNA extraction. The tissue samples were deposited in the collection of SMF and CUMZ. Specimens were then preserved by injecting a solution of 5 mL absolute (i.e., 36%) formalin in 1 L of 96% ethanol into the body cavity, and stored in 70% ethanol. Coordinates and elevation were recorded using Garmin GPS receivers with built-in altimeters. All coordinates were recorded in decimal degrees, WGS 1984 datum. Capitalized colors and color codes (in parentheses) followed [23].
In evaluating species’ boundaries among populations, we followed the evolutionary species concept [24,25]. As lines of evidence for species delimitation, we applied a phenotypic criterion (external morphology), the genetic distinctness of a mitochondrial genetic marker as well as a criterion for reproductive isolation (bioacoustic data).
Abbreviations used are EYD (eye diameter); FL (foot length); HL (head length); HW (head width); IND (internasal diameter); IOD (interorbital diameter); LAL (lower arm length; measured from elbow to base of palmar tubercle); NED (nostril–eye distance); HNL (hand length), measured from base of palmar tubercle to tip of third finger); SHL (shank length); SL (snout length); SVL (snout–vent length); TED (tympanum–eye distance); THL (thigh length); TYD (longitudinal tympanum diameter); UEW (upper eyelid width). Webbing formulae follow [26]. Terminology of snout shape follows [27]. Morphological data for Limnonectes alpinus, L. liui, and L. medogensis were taken from [15].
We recorded anuran vocalizations using a digital audio recorder (Olympus LS-12) with a Sennheiser ME 66 shotgun microphone capsule and a Sennheiser K6 powering module. The microphone was positioned between 0.5 and 1.5 m from the calling frog. Aside from the GPS coordinates and elevation above sea level of the locality we also noted ambient air temperature and determined the SVL of the recorded individual. Files were recorded as uncompressed 24-bit WAV files at a sampling frequency of 96 kHz. Audio files were deposited in the Fonoteca Zoológica, Museo Nacional de Ciencias Naturales, Madrid, Spain.
The spectral and temporal parameters were analyzed and the power spectra were calculated in RAVEN PRO 1.4. (Bioacoustics Research Program 2011). Spectrograms were obtained using the Blackman window function at 3db Filter Bandwidth of 158 Hz; grid spacing of 21.5 Hz; overlap 90%. Frequency information was generated through Fast Fourier Transformation (FFT, width 2048 samples). Temporal measurements of calls such as repetition rates, duration of notes, and the number of pulses, were measured on the waveforms. Terminology in call descriptions follows [28]. The map was created using ArcMap 10.4.
Marker-based analysis: We extracted DNA following the protocol of [29]. To eliminate potential PCR-inhibiting contaminants, the tissue samples were incubated for 14 h at 4 °C in 200 µL low PBS buffer (20 µL PBS in 180 µL of water) before overnight digestion with the vertebrate lysis buffer at 56 °C. After extraction, DNA was eluted in 50 µL TE buffer. Fragments of the mitochondrial 16S rRNA (16S) were amplified in an Eppendorf Mastercycler® Pro using the following protocol: initial denaturation for 2 min at 94 °C; followed by 40 cycles with denaturation for 35 s at 94 °C, hybridization for 35 s at 48.5 °C, and elongation for 60 s at 72 °C; final elongation for 10 min at 72 °C. The reaction mix for each sample contained 1 µL DNA template, 14 µL water, 2.5 µL PCR-buffer, 1 µL 25 mM MgCl2, 4 µL 2.5 mM dNTPs (Invitrogen), 0.5 µL (containing 2.5 units) Taq Polymerase (PeqLab), and 1 µL of the primer (16S: forward: L2510, 5′–CGCCTGTTTATCAAAAACAT–3′; reverse: H3056, 5′–CCGGTCTGAACTCAGATCACGT–3′; from Eurofins MWG Operon). DNA extraction, PCR, and sequencing were performed at SMF for samples from Myanmar and at CUMZ for samples from Thailand. We generated 43 new sequences for this study (see Appendix B). Additionally, we downloaded relevant 16S sequences from GenBank (Appendix B). Our dataset contains topotypic specimens of Limnonectes limborgi and near-topotypic specimens of L. doriae.
We aligned the sequences with MUSCLE [30] using the default settings in Geneious 6.1.2. [31]. For software applications, sequence data formatting was converted using the online server Alter [32]. The best substitution model for each gene was identified using PartitionFinder2 [33], with linked branch lengths via PhyML 3.0 [34]. Model selection used the corrected (for finite sample size) Akaike Information Criterion (AICc) [35]. Fejervarya limnocharis (GenBank number AF206466) was used as an outgroup [14].
Bayesian Inference analyses (BI) used MrBayes 3.2 [36] with five runs with eight chains. The first 25% of trees were discarded as burn-in. MCMC runs used an initial set of 1,000,000 generations with sampling every 500 generations and adding 500,000 generations until chains reached convergence. Convergence was considered achieved when the standard deviation of split frequencies was 0.015 or less. Additionally, convergence was diagnosed by PSRF (Potential Scale Reduction Factor), which should approach 1.0 as runs converge [37]. We used the IQTree webserver [38] to run a Maximum Likelihood (ML) analysis using 10,000 ultrafast Bootstrap approximation (UFBoot) replicates with 10,000 maximum iterations and minimum correlation coefficient of 0.99 [39], plus 10,000 replicates of Shimodaira–Hasegawa approximate likelihood ratio (SH-aLRT), which proved to be accurate with a high statistical power [34]. We used FigTree 1.3.1 (http://tree.bio.ed.ac.uk/software/figtree/, accessed on 2 April 2021) for viewing the trees. We estimated evolutionary genetic divergence, computing uncorrected pairwise distances with MEGA 7.0.26 [40] to assess the degree of intra- and interspecific differences, using a Bootstrap estimation method of 10,000 replications. We built a species tree using BEAST 2.4.7 [41,42] with 1,000,000 generations for the MCMC model. The resulting tree was visualized using DensiTree 2.2.6 [43].
Whole-genome sequencing and assembly: Genomic DNA was extracted from muscle tissue of the respective holotypes (SMF 104090 and SMF 106034) of the two new species of Limnonectes described herein according to standard phenol/chloroform procedures [44]. The resulting DNA was then resuspended in TE buffer (10 mM Tris-Cl, 0.1 mM EDTA) and stored at −20 °C. Quality checks for high molecular weight DNA were performed by agarose gel electrophoresis [44]. The DNA samples were shipped on dry ice to Novogene (UK) for short-read Illumina genome sequencing. One genomic library for each of the holotypes (insert size: 350 bp) was prepared and 150 bp paired-end reads were sequenced on an Illumina NovaSeq 6000 platform (San Diego, CA, USA). A long-read genomic sequencing was performed for the holotype SMF 106034 using PacBio Sequel sequencing platform.
A k-mer profile analysis was performed from the raw reads with the help of Jellyfish 2.3.0 software [45] and GenomeScope webserver [46]. The quality checks of the raw reads were performed by trimming the low-quality regions and adapter sequences and filtered for possible contamination using Trimmomatic 0.39 [47] and Kraken 2.0.9 [48], respectively. Nuclear genome assembly of the holotype SMF 106034 was conducted in the SPAdes assembler [49]. Scaffolds smaller than 500 in length were discarded. To assemble the nuclear genome of the holotype SMF 104090, the short reads were mapped over the genome of SMF 106034 with the help of Bowtie2 software [50]. The resulting SAM file was then converted to a sorted bam file by using Samtools [51]. The consensus sequence was then generated with the help of Samtools and BCFtools [52] software. Scaffolds smaller than 500 in length were discarded.
The mitochondrial genome of SMF 104090 was assembled from the genomic data separately using NOVOplasty 4.2 [53]. For the holotype, SMF 106034, mitochondrial genome of the holotype SMF 104090 was taken as reference and Pacbio reads which had homology to the reference were fetched using NCBI Blast 2.11 [54]. These reads were then used in Geneious Prime software 2020.2.3 (www.geneious.com) to generate the consensus sequence. The consensus sequence was then polished by Illumina short reads using Pilon v1.23 software [55]. Annotation of the mitochondrial genome was performed by MITOS2 webserver [56] and visualization pictures were generated using Geneious Prime software 2020.2.3.
3. Results and Conclusions
The alignment of the 16S sequences used in this study had a length of 583 bp. General Time Reversible using a discrete Gamma distribution with five rate categories and by assuming that a certain fraction of sites are evolutionarily invariable (GTR + I + G) was determined to be the best fitting model of sequence evolution. The trees generated by BI, ML, and BEAST analyses agree well at most well-supported nodes, but show differences in branch arrangement at weakly supported nodes (Figure 1 and Figure 2).
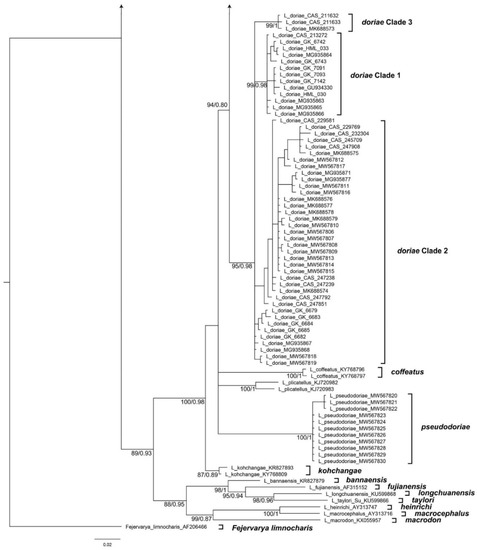
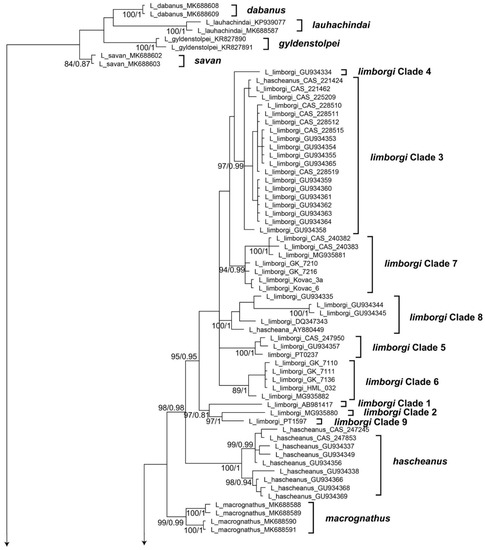
Figure 1.
Phylogenetic relationships among frogs of the genus Limnonectes from a BI analysis inferred from the mtDNA marker 16S. The numbers at nodes are bootstrap values (left) and Bayesian posterior probabilities (right), but only for nodes with bootstrap values higher than 75. A scale bar of genetic distance is indicated. The tree is rooted using Fejervarya limnocharis.
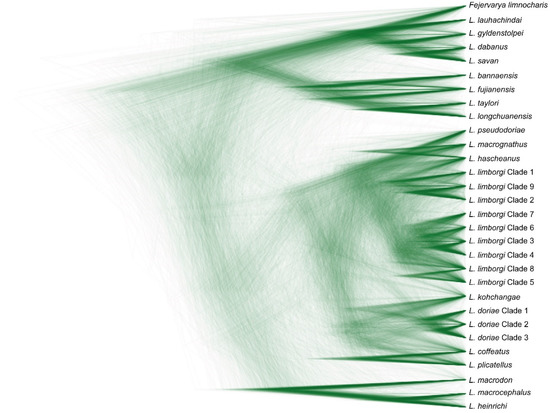
Figure 2.
Species tree indicating phylogenetic relationships among frogs of the genus Limnonectes from a BEAST analysis showing density of trees proportional to frequency of occurrence drawn in DensiTree.
Limnonectes doriae species complex. Among the specimens formerly referred to as Limnonectes doriae, three major clades are recognizable (Figure 1). Clade 1 contains specimens from lower Myanmar in Bago and Yangon Regions (Figure 3). Clade 2 includes samples from southern Myanmar including the Malayan Peninsula and adjacent regions in extreme western Thailand; only in the ML analysis, these specimens formed a weakly supported clade, whereas, in the Bayes analysis, these are recovered as a polytomy. Finally, Clade 3 contains specimens from western Myanmar in Rakhine state. The genetic distances among these clades vary from 1.9% to 2.4% (Table 1). We consider these three clades as candidate species, that should be assessed by including additional data types. Clade 3 includes specimens from near the type locality of L. doriae and is therefore considered to represent the “true” L. doriae.
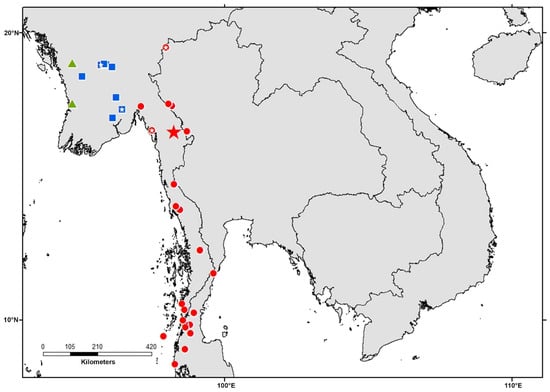
Figure 3.
Map showing collecting localities of the Limnonectes species related to L. doriae included in this study Each symbol can represent one or more adjacent localities. Blue squares = Clade 1; red circles = Clade 2; green triangles = Clade 3; red star = type locality of L. doriae. Small white stars within symbols indicate localities from which bioacoustics data have been used in this study.

Table 1.
Mean genetic distances (in %) of Clades 1–3 (see Figure 1) of frogs referred to as Limnonectes doriae based on the 16S dataset. See text for details.
Bioacoustical data are available for our Clades 1 and 2 (see also Table 2). The male advertisement calls of these two clades are strikingly different (Figure 4; Table 2): The call of SMF 104084 from Clade 2 from the region of the species’ type locality (Figure 4A) consists of a single series of 15–26 notes, each note varying from 51 to 103 ms (mean 65 ms) in duration with 11.6–12.1, mean 11.8, notes per second, their amplitude rising during the first 3–8 notes, then remaining about the same level. There is no offset single note anterior to the series of notes. The call has a duration of 1.25–2.24 s and a dominant frequency of 689–990 Hz, a mean of 947 Hz. The analyzed call of a Clade 2 male (ZMKU 1552) from a locality in Mae Hong Son Province, Thailand, about 320 airline km NNE of the SMF 104084 locality (see Figure 3), is similar in structure (i.e., a single series of notes without an off-set single note anterior to this series), dominant frequency (840–1055 Hz, mean 902 Hz), note duration (60–72 ms, mean 68 ms), and number of notes per second (11.7). It differs in the length of the call (6.75–7.71 s) and thus in the number of notes per call (79–90, mean 83.7). Whether this difference in call duration is of individual or geographical nature can only be clarified once the calls of additional individuals and localities can be included in the analyses.

Table 2.
Selected bioacoustic parameters of the species related to Limnonectes doriae. Range is followed by mean value and standard deviation in parentheses. Dom. Freq. = dominant frequency; Freg. = frequency.
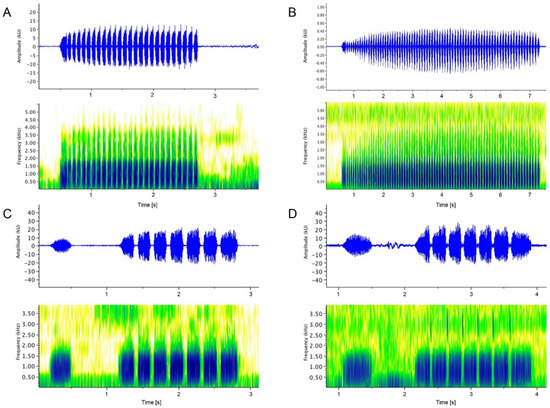
Figure 4.
Advertisement calls of male Limnonectes. (A) L. doriae, SMF 104084 (Mon State, Myanmar); (B) L. doriae, ZMKU 1552 (Mae Hong Son Province, Thailand); (C) L. bagoensis n. sp., SMF 104090 (Bago, Bago Region, Myanmar); (D) L. bagoensis n. sp., SMF 106039 (Bagoyoma, Bago Region, Myanmar). See text for details and Figure 4 for localities plotted on map.
The call of Clade 1 males (Figure 4C,D) consists of two portions that are separated by a break of 573–1043 ms (mean 792 ms). The first portion of the call is a single unpulsed sound emission (note duration 225–291 ms) and with a slightly weaker amplitude than the second portion which consists of 4–9 notes. Depending on the number of notes, the second portion of the call has a duration of 1202 (4 notes) to 1954 ms (9 notes), mostly around 1600 ms (7 notes); each note varying from 155 to 239 ms in duration with 4.2–4.6, mean 4.4, notes per second, their amplitude rising during the first note, then remaining about the same level. The dominant frequency is at 861–990 Hz (mean 945 Hz) for both call portions. We analyzed the advertisement calls of Clade 1 males (Figure 4C,D) from two localities that are about 190 air-line km apart from another (see Figure 3), and the calls from the different localities agree very well in all examined characters (see Table 2). The mean genetic distance (16S) between Clade 1 and 2 is 1.9% (Table 1).
In external morphology, specimens of these three clades are very similar but show differences in some morphometric values (Figure 5) as well as in the amount of toe webbing and in some details of coloration (see Table 3 and respective diagnosis sections below). Additionally, there is sexual dimorphism evident in several morphometric characters such as SVL, IOD, TYD, and HL. In particular, individuals of Clades 1 and 2 differ in the pattern on the posterior surface of the thigh (distinct and contrasting in Clade 1 versus weak and indistinct in Clade 2), development of a dermal fringe along the outer edge of Toe 5 (absent or weak in Clade 1 versus distinct in Clade 2), and the gular coloration in adult males (pale in Clade 1 versus dark gray in Clade 2). However, since our sample sizes are small for characters of external morphology, these putative differences between the clades need to be evaluated with more specimens in order to test their actual diagnostic value.
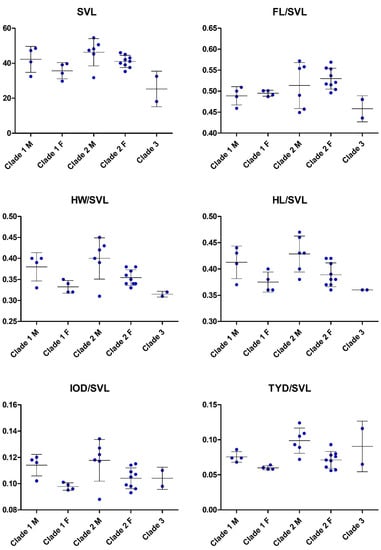
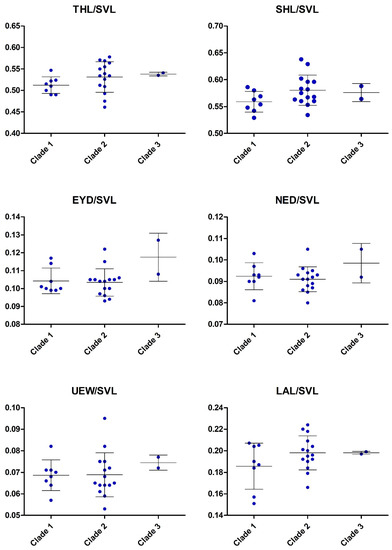
Figure 5.
Scatter plots illustrating morphological variation in the clades frogs related to Limnonectes doriae. M = males; F = females. For definitions of Clades and abbreviations of morphological characters see text.

Table 3.
Selected measurements and proportions of the two new species of Limnonectes described herein and the species they are most closely related to. Range is followed by mean value and standard deviation in parentheses. For abbreviations see text.
Thus, based on the results of the integrative analyses of mtDNA data, morphology, and bioacoustics, we recognize Clades 1 and 2 as species-level units. Clade 3 remains a candidate species awaiting further study, especially based on nuclear DNA data and bioacoustics. Based on the respective type locality, our Clade 2 is assigned to Limnonectes doriae whereas our Clade 1 represents an undescribed species for which no name is available. Thus, we describe the Limnonectes related to L. doriae from lower Myanmar (Bago Region and Yangon Region, our Clade 1) as a new species below.
Limnonectes limborgi species complex. Among the specimens formerly referred to as Limnonectes limborgi, we found nine major clades that we recognize as candidate species (Figure 1 and Figure 2). The genetic distances among these clades vary from 1.8% to 4.9%. (Table 4). Of these clades, five (i.e., Clades 1, 2, 4, 8, 9) are represented only by sequences (mostly from GenBank) and we are unable to study these in any detail due to the lack of morphological or bioacoustical data. Therefore, these clades were not included in the integrative taxonomic analysis.

Table 4.
Mean genetic distances (in %) of Clades 1–12 (see Figure 1) of frogs referred to as Limnonectes limborgi as well as L. hascheanus based on the 16S dataset. Cl = Clade; hasch. = hascheanus. See text for details.
Of the remaining four clades (i.e., 3, 5–7), we also examined voucher specimens for characters of external morphology. Clade 3 contains specimens from upper Myanmar in Kachin State and Sagaing Region, Myanmar (Figure 6). Clade 5 includes samples from Tanintharyi Division, Malayan Peninsula, Myanmar. Clade 6 contains specimens from lower Myanmar (Yangon and Bago Regions). Clade 7 is distributed in the eastern portion of lower Myanmar (Kayin and Mon States) and northwestern Thailand (Mae Hong Son province).
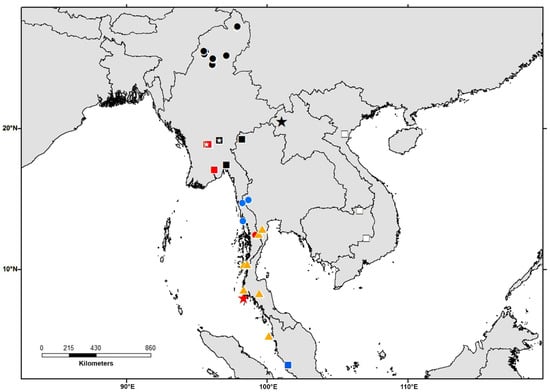
Figure 6.
Map showing collecting localities of the Limnonectes species related to L. limborgi. Each symbol can represent one or more adjacent localities. Orange triangles = L. hascheanus; blue square = L. limborgi Clade 1; red circle = L. limborgi Clade 2; black circles = L. limborgi Clade 3; black star = L. limborgi Clade 4; blue circles = L. limborgi Clade 5; red squares = L. limborgi Clade 6; black squares = L. limborgi Clade 7; white squares = L. limborgi Clade 8; red star = L. limborgi Clade 9. Small white stars within symbols indicate localities from which bioacoustics data have been used in this study.
Clade 7 includes topotypic specimens from the neotype locality of L. limborgi and are therefore considered to represent the “true” L. limborgi. Bioacoustical data are available for our Clades 6 and 7 (see also Table 5). The male advertisement calls of these two clades differ clearly (Figure 7): the call of males of “true” L. limborgi (Clade 7) consists of a series of three to five notes whereas males from Clade 6 emit a single “quaaak”. The two calls differ also in the length of the notes (0.218–0.252 s, usually <0.250 s, in males of Clade 6 versus 0.241–0.419 s, usually >0.260 s, in males of Clade 7). The genetic distance (16S) between these two clades is 2.8% (Table 5).

Table 5.
Selected bioacoustic parameters of the species related to Limnonectes limborgi. Range is followed by mean value and standard deviation in parentheses. Dom. Freq. = dominant frequency; Freg. = frequency.
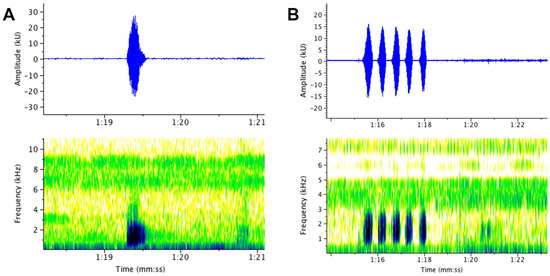
Figure 7.
Advertisement calls of male Limnonectes. (A) P. bagoyoma n. sp., SMF 106035 (B) L. limborgi, GK-7209. See text for details and Figure 6 for localities plotted on map.
In external morphology, specimens of these five clades differ in details such as in the shape and orientation of the supratympanic fold, the development of the tarsal fold as well as in the amount of toe webbing, and in some details of coloration (see Table 3 and respective diagnosis sections below). Additionally, they show differences in some morphometric values (Figure 8), Finally, there is sexual dimorphism evident in several morphometric characters, most evident in relative head width.
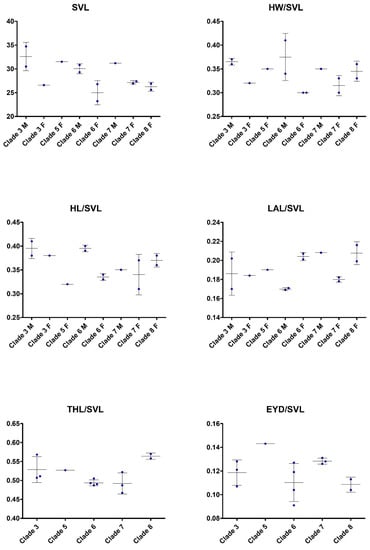
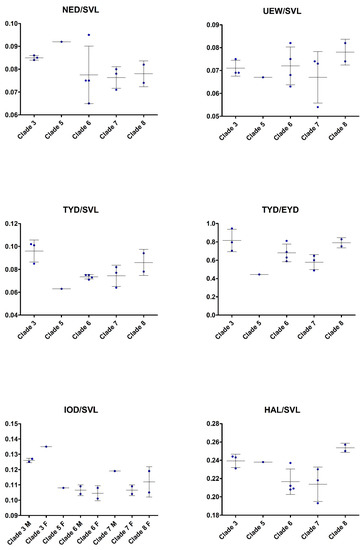
Figure 8.
Scatter plots illustrating morphological variation in the clades frogs related to Limnonectes limborgi. M = males; F = females. For definitions of Clades and abbreviations of morphological characters see text.
Thus, based on the results of the integrative analyses of mtDNA data, morphology, and bioacoustics, we recognize Clades 6 and 7 as species-level units whereas the other clades remain candidate species awaiting further study, especially based on nuclear DNA data and bioacoustics. Based on the respective type locality, our Clade 7 is assigned to Limnonectes limborgi whereas our Clade 6 represents an undescribed species for which no name is available. Thus, we describe the Limnonectes related to L. limborgi from lower Myanmar (Yangon and Bago Regions) as a new species below.
This published work and the nomenclatural acts it contains have been registered in ZooBank, the online registration system for the International Commission on Zoological Nomenclature (ICZN). The ZooBank LSIDs (Life Science Identifiers) can be resolved and the associated information can be viewed through any standard web browser by appending the LSID to the prefix http://zoobank.org. The LSID for this publication is as follows: urn:lsid:zoobank.org:pub:4C463126-CD59-4935-96E3-11AF4131144C. The LSID registration and any associated information can be viewed in a web browser by adding the LSID to the prefix “http://zoobank.org/”.
Limnonectes bagoensis sp. nov.
Gunther Köhler, Britta Zwitzers, Ni Lar Than and Panupong Thammachoti
Holotype. SMF 104090, an adult male collected in Bago, near the airport (17.32922, 96.44025; 35 m a.s.l.), Bago Region, Myanmar, collected 7 July 2017 by Gunther Köhler and Ni Lar Than. Field tag number GK-6742.
Paratypes. All Bago or Yangon Region, Myanmar. SMF 104091 (field tag number GK-6743), adult female, same collecting data as holotype. SMF 106038 (field tag number GK-7091), adult male, Bago Yoma (18.89255, 95.69519; 135 m a.s.l.), collected 16 June 2019 by Gunther Köhler and Ni Lar Than. 106039 (field tag number GK-7093), adult male, Bago Yoma (18.89854, 95.87848; 420 m a.s.l.), collected 16 June 2019 by Gunther Köhler and Ni Lar Than. SMF 106040-41 (field tag numbers GK-7142, 7147), adult females, Bago Yoma (18.89410, 95.88054; 405 m a.s.l.), collected 20 June 2019 by Gunther Köhler and Ni Lar Than. SMF 106042-43 (field tag numbers GK-7147, 7148), adult females, Bago Yoma (18.90105, 95.87756; 440 m a.s.l.), collected 20 June 2019 by Gunther Köhler and Ni Lar Than. CAS 213272, adult male, Mingalardon Township, Hlawga Wildlife Park (17.04656, 96.11078), Yangon Region, Myanmar, collected 17 December 1999 by T. Thin, S. W. Kyi and Y. M. Win.
Referred specimens. Myanmar:Bago: Bago Yoma (18.910667, 95.801350; 250 m a.s.l.): HML-30, 33.
Diagnosis. A species of the genus Limnonectes to which it is assigned because of its inferred phylogenetic position, males with hypertrophied heads, and the presence of odontoids on the lower jaw in adult males [19,57]. Limnonectes bagoensis sp. nov. is assigned to the subgenus Elachyglossa [17] because of its close phylogenetic position to the type species of this subgenus (i.e., L. gyldenstolpei). Limnonectes bagoensis sp. nov. differs from all congeners by having (1) a medium body size (males 32–49 mm; females 30–47 mm); (2) slightly enlarged toe disks; (3) adult males with small odontoids in the lower jaw; (4) adult males without a knob-like or flap-like structure (caruncle) in the interorbital and parietal region; also lacking swelling in the occipital region; (5) in adult males head not conspicuously enlarged; (6) webbing formula I 1–2 II 1–2.4 III 1.2–3 IV 3.3–2 V to I 1.8–2.2 II 1.5–2.8 III 2.2–3.2 IV 3.5–2 V; (7) a feeble dermal fringe along the outer edge of Toe 5; (8) male advertisement call consists of two portions that are separated by a break of slightly less than 1 s. The first portion of the call is a single note with a duration of 225–291 ms whereas the second portion consists of 4–9 notes with a duration of 1200 (4 notes) to about 2000 ms (9 notes) with about 4.5 notes per second; the dominant frequency is at 861–990 Hz (mean 945 Hz). Limnonectes bagoensis differs from all other species currently assigned to the subgenus Elachyglossa except L. doriae by lacking a knob-like or flap-like structure (caruncle) in the interorbital and parietal region in adult males (versus such structure present in adult males), and also lacking swelling in the occipital region (versus such swelling present in adult males). Limnonectes bagoensis differs from L. doriae in having a male advertisement call that consists of two portions, separated by a break of slightly less than 1s, the second portion with a series of 4–9 notes at about 4.5 notes per second (versus a single series of notes in L. doriae with about 12 notes per second); gular coloration in adult males not dark (versus gular region dark gray in adult males of L. doriae).
Description of the holotype (Figure 9). Adult male, as indicated by the presence of odontoids, broad head, and folds along mandibular arch; SVL 40.8 mm; habitus robust; head broad, about as wide as long, ratio HL/HW 1.07; snout nearly rounded in dorsal view, projecting beyond lower jaw, rounded in profile; nostril dorsolateral, closer to tip of snout than eye; canthus rounded; ratio EYD/SVL 0.10; IOD (4.8 mm) greater than width of upper eyelid (3.1 mm); tympanum distinct, slightly recessed relative to skin of temporal region, tympanic rim distinctly elevated relative to tympanum; ratio TYD/EYD 0.85; four to five vomerine teeth on slightly raised oblique ridges between choanae, separated from choanae by about 1/4 length of one group, gap between groups about 1/2 length of one group; tips of all four fingers rounded, not expanded into discs; relative finger lengths III > I > IV > II; no webbing; distinct subarticular tubercles; supernumerary tubercles absent; palmar tubercle distinct, flat, bifid; thenar tubercle large, elongate, raised, about twice the size of palmar tubercle; tips of toes rounded, slightly expanded into discs; relative toe lengths IV > III > V > II > I; feet almost fully webbed, webbing formula I 1–1.5 II 0.5–2 III 1–2.5 IV 2.5–1 V; a well-developed dermal ridge along lateral side of Toe V from level of outer metatarsal tubercle to distal subarticular tubercle; large, flap-like inner metatarsal tubercle, >0.5 length of first toe; distinct fold on distal one-half of tarsus beginning at inner metatarsal tubercle; outer metatarsal tubercle tiny; skin on dorsal and lateral surfaces of head, body, and limbs smooth except for a few scattered tubercles in sacral region and on dorsal surface of shank; skin on throat and venter smooth; distinct supratympanic ridge from posterior edge of upper eyelid along upper margin of tympanum and then obliquely down to shoulder; no dorsolateral fold. Measurements (mm) of holotype: SVL 40.8; HL 17.4; HW 16.3; SL 6.6; EYD 4.1; IOD 4.8; TYD 3.5; TED 1.7; SHL 22.1; THL 20.0; HNL 10.2; FL 20.4; NED 4.2; IND 4.6.
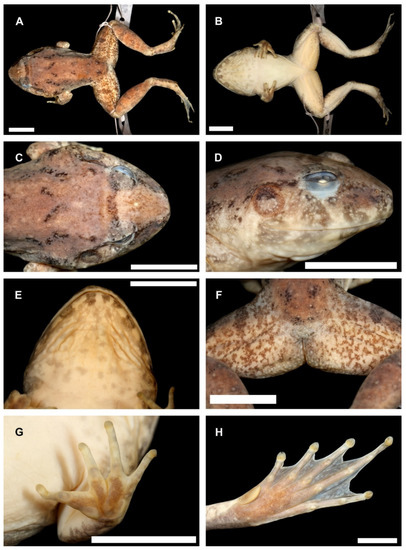
Figure 9.
Holotype of Limnonectes bagoensis n. sp. (SMF 104090). (A). dorsal view; (B). ventral view; (C). dorsal view of head; (D). lateral view of head; (E). ventral view of head; (F). cloacal and thigh region; (G). palmar surface of hand; (H). plantar surface of foot. Scale bars equal 5.0 mm. Photos by G.K.
Coloration in life was recorded as follows (Figure 10A): Dorsal surfaces of head and body Olive Horn Color (16) with a suffusion of Smoke Gray (267) in the occipital region and with a Dark Drab (45) interorbital bar; dorsum with a few Mikado Brown (42) and Grayish Olive (273) splotches; labial region Light Buff (2) with prominent broad Dark Drab (45) vertical bars; dorsal surfaces of hind limbs Pale Cinnamon (55) with Olive-Brown (278) transverse bars; iris Buff (5) above and whitish below and with a Sepia (279) streak in upper and lower edge as well as a suffusion of Grayish Olive (274) in lateral portions.

Figure 10.
Limnonectes bagoensis n. sp. in life. (A) Male holotype, SMF 104090; (B) female paratype, SMF 104091; (C–F) male paratype, SMF 106038. Photos by G.K.
Coloration after about three and half years preservation in 70% ethanol was recorded as follows: Dorsal and lateral ground color of head and body Burnt Umber (48) with broken Sepia (286) transverse zigzag line across anterior dorsum and with a Sepia (286) interorbital bar; labial region Light Buff (2) with prominent broad Sepia (279) vertical bars; dorsal surfaces of hind limbs Grayish Horn Color (268) with Olive-Brown (278) transverse bars; ventral surfaces of head, body, and limbs Light Buff (2) with indistinct Smoke Gray (266) mottling in the gular region; palmar surfaces Ground Cinnamon (270); plantar surfaces Fawn Color (258).
Variation. The paratype and the referred specimens agree well with the holotype in general appearance, morphometrics, and coloration. The coloration of an adult male (SMF 106038; Figure 10C–F) from Bago Yoma was noted as follows: Dorsal background color of head and body Olive-Brown (278) with Sepia (286) splotches on dorsum and with a Raw Umber (280) interorbital bar, edged anteriorly by a Clay Color (20) bar; labial region Clay Color (18) with broad Sepia (279) vertical bars; tympanum Clay Color (20); occipital region suffused with Hair Brown (277); flank region Chamois (84); dorsal surfaces of hind limbs Olive-Brown (278) with Sepia (286) transverse bars; tips of fingers and toes Salmon Color (83); posterior surface of thigh Tawny Olive (17) with Olive-Brown (278) mottling; ventral surfaces of head and body Whitish Lime Green (111) with a suffusion of Sulphur White (96) on posterior body; area adjacent to mandibular arch Medium Neutral Gray (298) with Whitish Lime Green (111) speckles; ventral surfaces of limbs Hair Brown (277) with Sulphur White (96) splotches; palmar and plantar surfaces suffused with Sepia (279); iris Drab (19) with a suffusion of iris Tawny Olive (17) in upper portion.
Etymology. The species name “bagoensis” refers to the city of Bago where the holotype of this species was collected, and ensis denoting place.
Natural history notes. At the type locality at night time, adult males were heard calling on the surface hidden under flat rocks. Figure 11 shows the holotype (SMF 104090) that continued calling even after the covering rocks had been removed. Additional individuals were encountered sitting on the forest floor at night without cover.

Figure 11.
Holotype of Limnonectes bagoensis n. sp. (SMF 104090) in life, at its calling site after the covering rocks had been removed.
Geographic Distribution and Conservation. As currently known, Limnonectes bagoensis sp. nov. is known from several localities in Bago Region, Myanmar. Given how little we know about this species, we classify Limnonectes bagoensis sp. nov. as Data Deficient based on the IUCN Red List Categories and Criteria [58].
Genomic characterization. Whole-genome sequencing and assembly (Table 6): Illumina sequencing produced 302,829,670 short-read pairs with a total sequence amount of 90 Gb. K-mer analysis estimated the genome size of 2.16 Gb with 0.78% heterozygosity. (Figure 14) The mitochondrial genome was assembled into one 17,480 bp long circular contig. A total of 13 protein-coding genes, two rRNAs, 22 tRNAs were annotated in the mitochondrial genome (Figure 12). A D-loop region could not be annotated in the mitochondrial genome sequence.

Table 6.
Genomic details of the holotypes of the two new species are described herein.
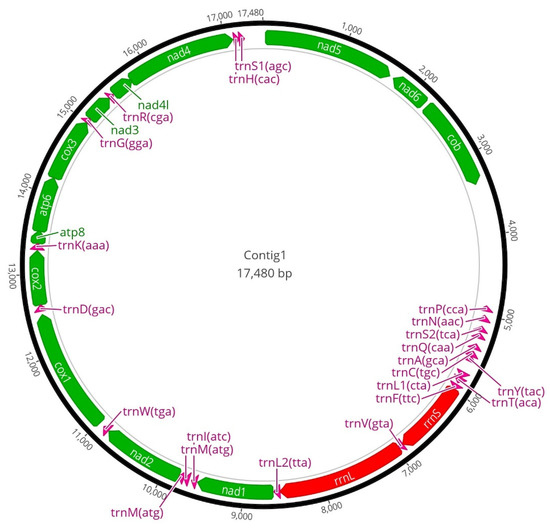
Figure 12.
Annotated mitochondrial genome of the holotype of Limnonectes bagoensis n. sp. (SMF 104090) see text for details.
The nuclear genome obtained by reference-based assembly consisted of 1,941,736,683 bases fragmented in 647,264 scaffolds. The longest scaffold was 42,771 bases and the N50 scaffold was 4270 bases whereas GC content was only 34.74%. Except for the high-quality mitochondrial genomes, the simple assembly from Illumina short-reads was predicted to be 26.2% complete as per the BUSCO gene space [59] (C:12.4%(S:12.3%, D:0.1%), F:13.9%, M:73.7%, n:5310). Raw reads and the draft-genome assemblies can be found within the BioProject PRJNA745544.
Limnonectes bagoyoma sp. nov.
Gunther Köhler, Britta Zwitzers, Ni Lar Than and Panupong Thammachoti
Holotype. SMF 106034, an adult female collected in Bago Yoma (18.89255, 95.69519; 135 m a.s.l.), Bago Region, Myanmar, collected 16 June 2019 by Gunther Köhler and Ni Lar Than. Field tag number GK-7110.
Paratypes. All paratypes were collected in Bago Yoma, Bago Region, Myanmar. SMF 106035, adult male, same collecting data as holotype; SMF 106036, adult female, collected 20 June 2019 by Gunther Köhler and Ni Lar Than at 18.89876, 95.87889; 435 m a.s.l. SMF 106037, adult male, collected 20 June 2019 by Gunther Köhler and Ni Lar Than at 18.90128, 95.87737; 440 m a.s.l.
Referred specimens. Myanmar:Bago: Bago Yoma (18.910667, 95.801350; 250 m a.s.l.): HML-32.
Diagnosis. A species of the genus Limnonectes to which it is assigned because of its inferred phylogenetic position (Figure 1), males with hypertrophied heads, and the presence of odontoids on the lower jaw in adult males [19,57]. Limnonectes bagoyoma sp. nov. is assigned to the subgenus Taylorana because of its close phylogenetic position to the type species of this subgenus (i.e., L. limborgi; Figure 1). Limnonectes bagoyoma sp. nov. differs from all congeners by having (1) a small body size (males 29.3–30.7 mm; females 23.1–26.6 mm); (2) males with slightly enlarged odontoids; (3) inner metatarsal tubercle large, raised, usually >0.5 length of the first toe; (4) webbing formula I 1–2 II 1–2.4 III 1.2–3 IV 3.3–2 V to I 1.8–2.2 II 1.5–2.8 III 2.2–3.2 IV 3.5–2 V; (5) females with clutches of enlarged, non-pigmented eggs; (6) male advertisement call consisting of a single note with a duration of 220–250 ms and a dominant frequency mostly in the 1300–1400 Hz range. Limnonectes bagoyoma differs from the remaining species of the subgenus Taylorana (see also [15]) as follows: from L. limborgi by having a male advertisement call consisting of a single note with a duration of 220–250 ms (versus call consisting of a series of 3–5 notes, each note with a duration of usually >260 ms in L. limborgi), and no dark lateral face mask (versus dark face mask present). Limnonectes bagoyoma differs from L. hascheanus by its larger body size, reaching 30 mm SVL in adult males (versus SVL <26 mm in both sexes in L. hascheanus), and by having more toe webbing, usually three phalanges free of webbing on the medial side of Toe 4 (versus more than three phalanges free of webbing on the medial side of Toe 4). Limnonectes bagoyoma differs from L. liui by its smaller body size, SVL not exceeding 31 mm in both sexes (versus SVL 32.0–38.5 mm in males of L. liui, only female in type series measuring 32.7 mm), toe webbing well-developed (versus rudimentary), and inner metatarsal tubercle usually >0.5 length of the first toe (versus “just half” of the first toe in L. liui according to [15]). Limnonectes bagoyoma differs from L. medogensis by having the nostril positioned closer to the snout than to the eye (versus closer to eye in L. medogensis), vomerine teeth present (versus absent), venter patternless (versus marbled markings present), toe webbing well-developed (versus rudimentary), inner metatarsal tubercle distinctly shorter than the first finger (versus about length of Toe 1), and tarsal fold indistinct but present (versus absent). Limnonectes bagoyoma differs from L. xizangensis by having the toe tips enlarged into small disks (versus toe tips pointed), vomerine teeth present (versus absent), toe webbing well-developed (versus absent), and the presence of tiny tubercles on the dorsal surface of the shank (versus smooth), venter patternless (versus black reticulated markings present), and smooth skin on venter (granular skin on venter). Limnonectes bagoyoma differs from L. alpinus by having the toe webbing well-developed (versus toe webbing absent in L. alpinus), vomerine teeth present (versus absent), and tympanum distinct (versus hidden).
Description of the holotype (Figure 13). Adult female, as indicated by the rather slender head, absence of odontoids, and absence of folds along mandibular arch; SVL 26.8 mm; habitus robust; head narrow, slightly longer than wide, ratio HL/HW 1.14; snout subovoid in dorsal view, projecting beyond lower jaw, rounded in profile; nostril dorsolateral, closer to tip of snout than eye; canthus rounded; ratio EYD/SVL 0.13; IOD (2.7) greater than width of upper eyelid (1.8 mm); tympanum distinct, slightly recessed relative to skin of temporal region, tympanic rim distinctly elevated relative to tympanum; ratio TYD/EYD 0.59; four to five vomerine teeth on slightly raised oblique ridges between choanae, separated from choanae by about 1/2 length of one group, gap between groups about 1/2 length of one group; tips of all four fingers rounded, not expanded into discs; relative finger lengths III>I>II>VI; no webbing; distinct subarticular tubercles; supernumerary tubercles absent; palmar tubercle distinct, flat, bifid; thenar tubercle large, elongate, raised, about twice the size of palmar tubercle; tips of toes rounded, slightly expanded into discs; relative toe lengths IV>III>V>II>I; feet almost fully webbed, webbing formula I 1.5–2 II 1.5–2.5 III 2.2–3 IV 3.5–2 V; a weakly developed dermal ridge along lateral side of Toe V from level of outer metatarsal tubercle to distal subarticular tubercle; large, flap-like inner metatarsal tubercle, >0.5 length of first toe; outer metatarsal tubercle tiny; tarsal fold indistinct; skin on dorsal and lateral surfaces of head, body, and limbs smooth except for a few scattered tiny tubercles in sacral region and on dorsal surface of shank; skin on throat and venter smooth; indistinct supratympanic ridge from posterior edge of upper eyelid along upper margin of tympanum and then obliquely down to shoulder; no dorsolateral fold. Measurements (mm) of holotype: SVL 26.8; HL 9.2; HW 8.1; SL 3.8; EYD 3.4; IOD 2.7; TYD 2.0; TED 1.3; SHL 14.0; THL 13.2; HNL 9.2; FL 12.7; NED 2.0; IND 2.5.
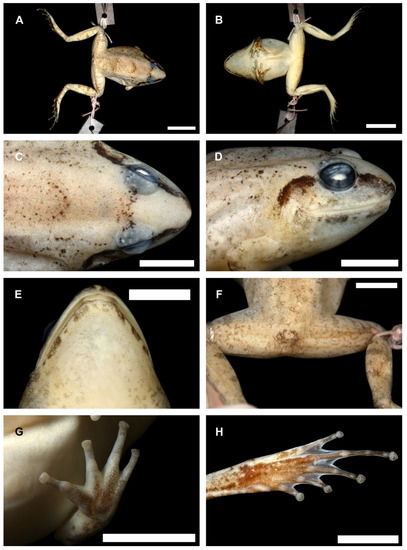
Figure 13.
Holotype of Limnonectes bagoyoma n. sp. (SMF 106034). (A). dorsal view; (B). ventral view; (C). dorsal view of head; (D). lateral view of head; (E). ventral view of head; (F). cloacal and thigh region; (G). palmar surface of hand; (H). plantar surface of foot. Scale bars equal 5.0 mm. Photos by G.K.
Coloration in life was recorded as follows (Figure 14A): Dorsal ground color of the head and body Dark Salmon Color (59) with a suffusion of Light Orange Yellow (7) on the dorsal head in front of broken Burnt Umber (48) interorbital bar; a Raw Umber (280) band below canthus rostralis and along the supratympanic ridge; flank region suffused with Buff-Yellow (6); dorsal surfaces of limbs Orange-Rufous (56) with Brownish-Olive (276) transverse bars; iris Cream Color (12) in the upper portion, Grayish Horn Color (268) below and with a suffusion of Warm Sepia (40) in anterior and posterior corners.
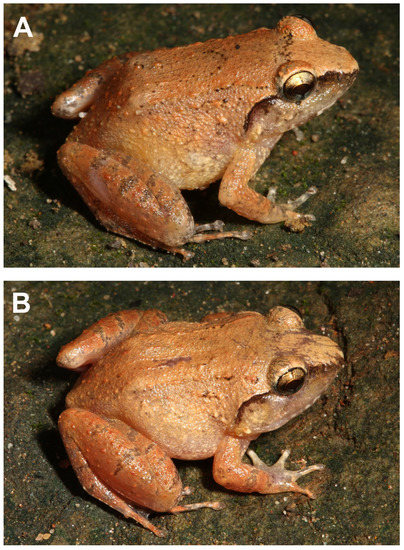
Figure 14.
Limnonectes bagoyoma n. sp. in life. (A) Female holotype, SMF 106034; (B) Male paratype, SMF 106035. Photos by G.K.
Coloration after about one and half years preservation in 70% ethanol was recorded as follows: Dorsal and lateral ground color of the head and body Drab (19), but paler, with a suffusion of Smoke Gray (266) on the dorsal head and with indistinct Vandyke Brown (281) stipples on dorsum and as a broken interorbital bar as well as a band below canthus rostralis and along the supratympanic ridge; Glaucous (289) vertical bars on the lower lip; dorsal surfaces of the thigh and shank Drab (19) with indistinct Glaucous (289) transverse bars; ventral surfaces of head, body, and limbs Cream White (52); palmar surfaces Ground Cinnamon (270); plantar surfaces Natal Brown (49).
Variation. The paratypes agree well with the holotype in general appearance; morphometrics and coloration (see Table 3). The two adult paratype males both have broad heads, folds along the mandibular arch, distinct odontoids, and four to five vomerine teeth. The coloration of a topotypic adult male (SMF 106035; Figure 14B) was noted as follows: Dorsal surface of head Pale Pinkish Buff (3) with Natal Brown (49) streaks; loreal region Cinnamon-Drab (50); a Raw Umber (280) band below canthus rostralis and along the supratympanic ridge; body Salmon Color (83) above and grading into Chamois (84) on the flank; dorsal surfaces of limbs Olive-Brown (278) with Hair Brown (277) transverse bars; iris Tawny Olive (17) in the upper portion, Grayish Horn Color (268) below and with a suffusion of Warm Sepia (40) in anterior and posterior corners.
Etymology. The species name “bagoyoma” refers to the type locality Bago Yoma, a large but relatively low mountain range that runs in a north-south direction between the Irrawaddy (=Ayeyarwady) and the Sittaung River in Myanmar. It is likely that Limnonectes bagoyoma is geographically restricted to this mountain range.
Natural history notes. At the type locality, the specimens were collected during the daytime in leaf litter in the rainforest. The two males we collected were heard calling before we caught them. Each male was sitting in a shallow depression, covered by leaf litter. Other frog species collected at the type locality include Duttaphrynus melanostictus, Fejervarya orissaensis, Ingerana tenasserimensis, Limnonectes bagoensis, and Leptobrachium smithi.
Geographic Distribution and Conservation. As currently known, Limnonectes bagoyoma sp. nov. is only known from the vicinity of its type locality. Given the little we know about this species, we classify Limnonectes bagoyoma sp. nov. as Data Deficient based on the IUCN Red List Categories and Criteria [58].
Genomic characterization. Whole-genome sequencing and assembly (Table 6): Pacbio sequencing resulted in 16,749,860 reads of size 90 Gb whereas Illumina sequencing produced 278,081,641 short read pairs with a total sequence amount of 83.4 Gb. K-mer analysis estimated the genome size of 2.4 Gb with 0.78% heterozygosity (Figure 15). A mitochondrial genome of 17,406 bases was assembled using Pacbio reads which matched to the complete annotated mito-genome of the holotype SMF 104090. The annotation resulted in 13 protein-coding genes, three rRNAs, 22 tRNAs and three replication origin sites. A D-loop region was also annotated in the mitochondrial genome sequence. (Figure 16).
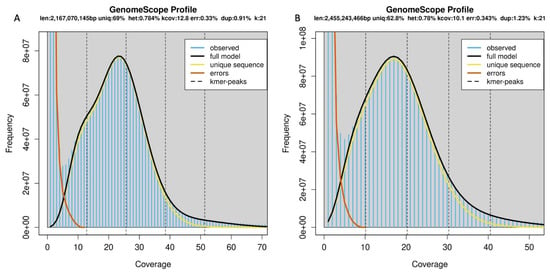
Figure 15.
Results of K-mer analysis. See text for details.
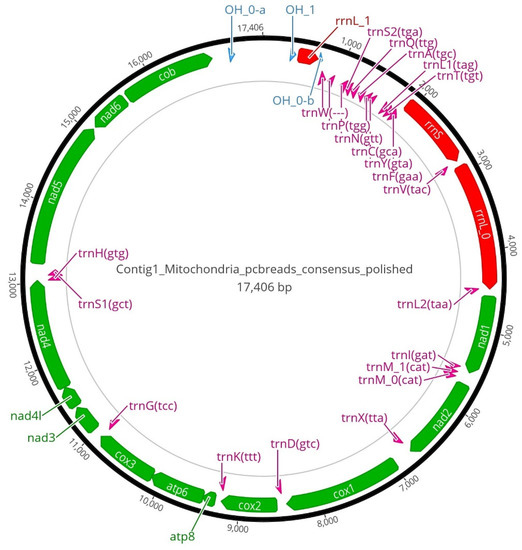
Figure 16.
Annotated mitochondrial genome of the holotype of Limnonectes bagoyoma n. sp. (SMF 106034) see text for details.
The draft nuclear genome assembly contains 691,036 scaffolds with a total length of 2,099,953,831 bps, an N50 of 4,236 bps and a GC content of 42.54%. The BUSCO [57] search allowed to identify 31.1% of the BUSCO genes [59] (C:13.2% [S:13.1%, D:0.1%], F:17.9%, M:68.9%, n:5310). Raw reads and the draft-genome assemblies can be found within the BioProject PRJNA745544.
4. Discussion
The present study presents further examples for putative geographically wide-spread species that actually represent species complexes of several taxa that are similar in external morphology but distinct in bioacoustics and genetics ([5,21,60]). To resolve the diversity in these complexes, additional field research is needed to obtain fresh voucher specimens from across their range for genetic and bioacoustics characterization of the geographically distinct populations. The case of Limnonectes bagoensis reported here, shows that in some cases the genetic differentiation in mtDNA can be rather low (i.e., just 2% in 16S between L. bagoensis and its putatively most closely related sister species L. doriae). However, given their rather drastically different male advertisement calls, there is no doubt that these two forms are distinct species. This hypothesis is reinforced by the observation that the male advertisement call of L. bagoensis does not vary across a large geographic distance (i.e., almost 200 air-line km). In the case of the species pair L. limborgi and L. bagoyoma, the genetic differentiation in mtDNA data (16S) is close to 3%, and the species are distinct in bioacoustics, too. In the course of this study, we identified additional candidate species in either species complex. Due to the lack of additional evidence lines we refrained from naming any of these groups defined only by mtDNA data. For “L. limborgi” from northern Vietnam (which corresponds with our Clade 10) the male advertisement call has been described as “single notes, repeated in very large intervals from 30 s to several minutes”, an observation confirmed by [61]. The latter author reports the call to have a dominant frequency of 1000–1100 Hz and a duration of 278–331 ms. Both accounts describe the male sitting in a hole with just the head visible after the covering leaves had been removed. Except for some details, this call description agrees very well with our analyses of male advertisement calls of L. bagoyoma and is unlike the call of the geographically closer L. limborgi from the neotype locality of the latter species. Both the Vietnamese “L. limborgi” and L. bagoyoma have a male advertisement call consisting of a single note as opposed to a series of 3–5 notes in “true” L. limborgi. The calls of males from Vietnam have a dominant frequency of 1000–1100 Hz and a duration of 278–331 ms versus a dominant frequency mostly in the 1300–1400 Hz range and a duration of 220–250 ms in L. bagoyoma. The genetic distance in 16S between these two clades with similar calls (i.e., our Clades 6 and 10) is 3.2% whereas it is 2.5% between the Vietnamese “L. limborgi” and “true” L. limborgi. In order to determine whether the two similar calls are cases of convergence or have both remained in the ancestral state, it would be essential to know which call type represents the plesiomorphic and which one the apomorphic character state. A more robust genetic phylogeny, including nuclear markers as well as bioacoustical data from additional populations of this species complex, is needed to gain a better understanding of call evolution in this group of frogs. In frogs, bioacoustic data and analyses are as useful as are molecular genetics and often yield initial information on the presence of cryptic taxonomic diversity [60,62,63]. The male advertisement call in frogs works as an effective isolating mechanism to avoid hybridization among similar species under natural conditions [28].
With this description of two new species, we provide the complete, annotated mitochondrial genome as well as a draft genome of short-read genome resources of the holotypes of both species described herein. This genomic characterization of the species based on the name-bearing specimen will be a genomic resource for any forthcoming studies.
Author Contributions
Conceptualization, G.K.; methodology, G.K., P.T. and D.K.G.; Marker-based analysis, G.K., B.Z. and P.T.; bioinformatics genomic data, D.K.G.; fieldwork and specimen acquisition, G.K., N.L.T. and P.T.; writing—original draft preparation, G.K.; writing—review and editing, all authors; funding acquisition, A.J., S.U.P. and P.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded partly by Chulalongkorn University: CU_GR_63_66_23_10 and also partly financially supported by the Sci-Super VI fund from Faculty of Science, Chulalongkorn University. The genomic work was funded by the Centre for Translational Biodiversity Genomics (LOEWE-TBG) through the program “LOEWE—Landes-Offensive zur Entwicklung wissenschaftlich-okonomischer Exzellenz” of Hesse’s Ministry of Higher Education, Research, and the Arts.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Chulalongkorn University Animal Care and Use Committee of Chulalongkorn University, Bangkok (protocol code 2123010; 1 January 2021).
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Acknowledgments
We would like to express our gratitude to Nyi Nyi Kyaw, Director General, Forest Department, Ministry of Natural Resources and Environmental Conservation, Myanmar, for issuing our collecting permits, U Hla Maung Thein, Director General, Environmental Conservation Department, Ministry of Natural Resources and Environmental Conservation, Myanmar, for issuing our permission to access the genetic resources as in compliance with the Nagoya Protocol, and to Naing Zaw Htun, Director, Nature and Wildlife Conservation Division, Ministry of Natural Resources and Environmental Conservation, Myanmar, for issuing the exportation permits. Our special thanks are due to Kyaw Kyaw Khaung, Rector, East Yangon University, Thanlyin, for supporting us to obtain all necessary permissions from the Ministry of Natural Resources and Environmental Conservation, Myanmar. We thank Linda Mogk, Senckenberg Research Institute, Frankfurt, for doing molecular lab work that resulted in some of the new DNA sequences included in this contribution. We are grateful to the following colleagues who provided audio files with recordings of Limnonectes: Anchalee Aowphol and Attapol Rujirawan, Bangkok, Thailand; Sandra Goutte, Taiwan; Thomas Ziegler, Köln. The audio files were made available to us via the Fonoteca Zoológica, Museo Nacional de Ciencias Naturales, Madrid, Spain.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A. Comparative Specimens Examined
Limnonectes bagoensis n. sp.—Myanmar: Bago: Bago, near airport: SMF 104090–91; Bago Yoma: HML-30, 33; SMF 106038-43.
Limnonectes bagoyoma n. sp.—Myanmar: Bago: Bago Yoma: HML-32, SMF 106034-37.
Limnonectes doriae—Myanmar:Mawlamyine: Paung Township: SMF 104084–89; Mon State: Kyaikto Township, along the trail from Kinmon to Kyaikto Pagoda: CAS 240316; Rakhine State: Rakhine Yoma Mountain Range: CAS 211632–33; Tanintharyi: Bokpyin Township, Pakchan Reserve Forest, Main Ma The camp: CAS 229581; Khamaukgyi Township: CAS 247792, 247851; Khamaukgyi Township, Kawthaung-Bokpyin road: CAS 247238–39; Thayetchaung Township, between Mal Ke Village and Ye Bya Village: CAS 229769; Thayetchaung Township, E Mal Ke Village, Nwalabo Reserve Forest: CAS 232304; Yebyu Township, Tanintharyi Nature Reserve: CAS 247908–09.
Limnonectes hascheanus—Myanmar: Tanintharyi: Khamaukgyi Township: CAS 247853; Khamaukgyi Township, Khamaukgyi Creek: CAS 247245.
Limnonectes limborgi—Myanmar: Kachin State: Machanbaw Township, the road between Ahtonga and Babaw: CAS 221462; Machanbaw Township, between Ahtonga and Aureinga: CAS 221424; Mohnyin Township, Indawgyi Wildlife Sanctuary, Nankan Forest Reserve, Mine Naung Village: CAS 228510–12, 228515, 228519; Nagmung Township, Au Yin Ga camp: CAS 225209; Kayin: Leiktho: GK-7209–10, 7216; Mon State: Kyaikto Township, Kinpon Chaung village, along waterfall stream: CAS 240382–83; Tanintharyi: Yebyu Township, Tanintharyi Nature Reserve: CAS 247950. Thailand: Kanchanaburi: 20 km N Tha Kah-nun: PT0236–37; Mae Hong Son: near Ban Nam Rin: Kovac_3a, Kovac_6; Phuket: near Kathu: PT1597.
Appendix B

Table A1.
GenBank Accession Numbers of Genetic Sequences (16S) Used.
Table A1.
GenBank Accession Numbers of Genetic Sequences (16S) Used.
| Species | GenBank Accession No. | Voucher |
|---|---|---|
| Fejervarya limnocharis | AF206466 | USNM 520407 |
| Limnonectes bagoensis | MZ578030 | HML-030 |
| Limnonectes bagoensis | MZ578031 | HML-033 |
| Limnonectes bagoensis | MZ578010 | CAS 213272 |
| Limnonectes bagoensis | MZ578025 | GK-6742/SMF 104090 |
| Limnonectes bagoensis | MZ578026 | GK-6743/SMF 104091 |
| Limnonectes bagoensis | MZ578027 | GK-7091/SMF 106038 |
| Limnonectes bagoensis | MZ578028 | GK-7093/SMF 106039 |
| Limnonectes bagoensis | MZ578029 | GK-7142/SMF 106040 |
| Limnonectes bagoyoma | MZ578045 | GK-7110/SMF 106034 |
| Limnonectes bagoyoma | MZ578046 | GK-7111/SMF 106035 |
| Limnonectes bagoyoma | MZ578047 | GK-7136/SMF 106036 |
| Limnonectes bagoyoma | MZ578050 | HML-032 |
| Limnonectes bannaensis | KR827879 | #2004.0342 |
| Limnonectes coffeatus | KY768796 | NCSM 77785 |
| Limnonectes coffeatus | KY768797 | NCSM 77786 |
| Limnonectes dabanus | MK688608 | MVZ 258240 |
| Limnonectes dabanus | MK688609 | MVZ 258244 |
| Limnonectes doriae | GU934330 | CAS 208425 |
| Limnonectes doriae | MG935863 | USNM 587327 |
| Limnonectes doriae | MG935864 | USNM 587096 |
| Limnonectes doriae | MG935865 | CAS 248173 |
| Limnonectes doriae | MG935866 | USNM 587326 |
| Limnonectes doriae | MG935867 | USNM 587306 |
| Limnonectes doriae | MG935868 | USNM 587303 |
| Limnonectes doriae | MG935871 | USNM 586919 |
| Limnonectes doriae | MG935877 | USNM 586913 |
| Limnonectes doriae | MK688573 | CAS 211632 |
| Limnonectes doriae | MK688574 | CAS 229581 |
| Limnonectes doriae | MK688575 | CAS 229714 |
| Limnonectes doriae | MK688576 | FMNH 268506 |
| Limnonectes doriae | MK688577 | FMNH 268509 |
| Limnonectes doriae | MK688578 | FMNH 270111 |
| Limnonectes doriae | MK688579 | FMNH 270113 |
| Limnonectes doriae | MW567806 | ZMKU 01521 |
| Limnonectes doriae | MW567807 | ZMKU 01522 |
| Limnonectes doriae | MW567808 | ZMKU AM 01523 |
| Limnonectes doriae | MW567809 | ZMKU AM 01524 |
| Limnonectes doriae | MW567810 | ZMKU AM 01525 |
| Limnonectes doriae | MW567811 | ZMKU AM 01526 |
| Limnonectes doriae | MW567812 | ZMKU AM 01527 |
| Limnonectes doriae | MW567813 | ZMKU AM 01528 |
| Limnonectes doriae | MW567814 | ZMKU AM 01530 |
| Limnonectes doriae | MW567815 | ZMKU AM 01534 |
| Limnonectes doriae | MW567816 | ZMKU AM 01543 |
| Limnonectes doriae | MW567817 | ZMKU AM 01545 |
| Limnonectes doriae | MW567818 | ZMKU AM 01547 |
| Limnonectes doriae | MW567819 | ZMKU AM 01552 |
| Limnonectes doriae | MZ578008 | CAS 211632 |
| Limnonectes doriae | MZ578009 | CAS 211633 |
| Limnonectes doriae | MZ578011 | CAS 229581 |
| Limnonectes doriae | MZ578012 | CAS 229769 |
| Limnonectes doriae | MZ578013 | CAS 232304 |
| Limnonectes doriae | MZ578014 | CAS 245709 |
| Limnonectes doriae | MZ578015 | CAS 247238 |
| Limnonectes doriae | MZ578016 | CAS 247239 |
| Limnonectes doriae | MZ578017 | CAS 247792 |
| Limnonectes doriae | MZ578018 | CAS 247851 |
| Limnonectes doriae | MZ578019 | CAS 247908 |
| Limnonectes doriae | MZ578020 | GK-6679 |
| Limnonectes doriae | MZ578021 | GK-6682 |
| Limnonectes doriae | MZ578022 | GK-6683 |
| Limnonectes doriae | MZ578023 | GK-6684 |
| Limnonectes doriae | MZ578024 | GK-6685 |
| Limnonectes fujianensis | AF315152 | not given |
| Limnonectes gyldenstolpei | KR827890 | #0155Y |
| Limnonectes gyldenstolpei | KR827891 | #0175Y |
| Limnonectes hascheana | AY880449 | MNHN 1997.5355 |
| Limnonectes hascheanus | GU934337 | FMNH 270118 |
| Limnonectes hascheanus | GU934338 | FMNH 270119 |
| Limnonectes hascheanus | GU934349 | LSUHC 6777 |
| Limnonectes hascheanus | GU934356 | CAS 229588 |
| Limnonectes hascheanus | GU934366 | THNHM 4488 |
| Limnonectes hascheanus | GU934368 | THNHM 4490 |
| Limnonectes hascheanus | GU934369 | THNHM 4491 |
| Limnonectes hascheanus | MZ578033 | CAS 247245 |
| Limnonectes hascheanus | MZ578034 | CAS 247853 |
| Limnonectes heinrichi | AY313747 | A167137 |
| Limnonectes kohchangae | KR827893 | #2003.0317 |
| Limnonectes kohchangae | KY768809 | NCSM 79546 |
| Limnonectes lauhachindai | KP939077 | ZMKU AM 01109 |
| Limnonectes lauhachindai | MK688587 | FMNH 266153 |
| Limnonectes limborgi | AB981417 | KUHE 15614 |
| Limnonectes limborgi | DQ347343 | isolate 1218 |
| Limnonectes limborgi | GU934334 | FMNH 271338 |
| Limnonectes limborgi | GU934335 | MVZ 258223 |
| Limnonectes limborgi | GU934344 | FMNH 262817 |
| Limnonectes limborgi | GU934345 | FMNH 262818 |
| Limnonectes limborgi | GU934353 | CAS 228513 |
| Limnonectes limborgi | GU934354 | CAS 228514 |
| Limnonectes limborgi | GU934355 | CAS 228516 |
| Limnonectes limborgi | GU934357 | CAS 229833 |
| Limnonectes limborgi | GU934358 | CAS 230378 |
| Limnonectes limborgi | GU934359 | CAS 232167 |
| Limnonectes limborgi | GU934360 | CAS 232168 |
| Limnonectes limborgi | GU934361 | CAS 232174 |
| Limnonectes limborgi | GU934362 | CAS 232189 |
| Limnonectes limborgi | GU934363 | CAS 232267 |
| Limnonectes limborgi | GU934364 | CAS 232274 |
| Limnonectes limborgi | GU934365 | CAS 232725 |
| Limnonectes limborgi | MG935880 | USNM 586920 |
| Limnonectes limborgi | MG935881 | USNM 587305 |
| Limnonectes limborgi | MG935882 | USNM 587100 |
| Limnonectes limborgi | MZ578032 | CAS 221424 |
| Limnonectes limborgi | MZ578035 | CAS 221462 |
| Limnonectes limborgi | MZ578036 | CAS 225209 |
| Limnonectes limborgi | MZ578037 | CAS 228510 |
| Limnonectes limborgi | MZ578038 | CAS 228511 |
| Limnonectes limborgi | MZ578039 | CAS 228512 |
| Limnonectes limborgi | MZ578040 | CAS 228515 |
| Limnonectes limborgi | MZ578041 | CAS 228519 |
| Limnonectes limborgi | MZ578042 | CAS 240382 |
| Limnonectes limborgi | MZ578043 | CAS 240383 |
| Limnonectes limborgi | MZ578044 | CAS 247950 |
| Limnonectes limborgi | MZ578048 | GK-7210 |
| Limnonectes limborgi | MZ578049 | GK-7216 |
| Limnonectes limborgi | MZ578051 | Kovac-3a |
| Limnonectes limborgi | MZ578052 | Kovac-6 |
| Limnonectes limborgi | MZ578053 | PT-1597 |
| Limnonectes limborgi | MZ578054 | PT-0237 |
| Limnonectes longchuanensis | KU599868 | KIZYPX 33448 |
| Limnonectes macrocephalus | AY313716 | TNHC 61913 |
| Limnonectes macrodon | KX055957 | not given |
| Limnonectes macrognathus | MK688588 | FMNH 268503 |
| Limnonectes macrognathus | MK688589 | FMNH 268505 |
| Limnonectes macrognathus | MK688590 | FMNH 270114 |
| Limnonectes macrognathus | MK688591 | FMNH 270115 |
| Limnonectes plicatellus | KJ720982 | LSUHC 6582 |
| Limnonectes plicatellus | KJ720983 | LSUHC 4001 |
| Limnonectes pseudodoriae | MW567820 | ZMKU AM 01553 |
| Limnonectes pseudodoriae | MW567821 | ZMKU AM 01555 |
| Limnonectes pseudodoriae | MW567822 | ZMKU AM 01563 |
| Limnonectes pseudodoriae | MW567823 | ZMKU AM 01565 |
| Limnonectes pseudodoriae | MW567824 | ZMKU AM 01567 |
| Limnonectes pseudodoriae | MW567825 | ZMKU AM 01577 |
| Limnonectes pseudodoriae | MW567826 | ZMKU AM 01579 |
| Limnonectes pseudodoriae | MW567827 | ZMKU AM 01580 |
| Limnonectes pseudodoriae | MW567828 | ZMKU AM 01582 |
| Limnonectes pseudodoriae | MW567829 | ZMKU AM 01583 |
| Limnonectes pseudodoriae | MW567830 | ZMKU AM 01585 |
| Limnonectes savan | MK688602 | NCSM 84943 |
| Limnonectes savan | MK688603 | NUOL 00061 |
| Limnonectes taylori | KU599866 | KIZ 022399 |
References
- Frost, D.R. Amphibian Species of the World: An Online Reference. Version 6.0. Available online: http://research.amnh.org/herpetology/amphibia/index.html (accessed on 14 July 2021).
- Köhler, G.; Mogk, L.; Khaing, K.P.P.; Than, N.L. The genera Fejervarya and Minervarya in Myanmar: Description of a new species, new country records, and taxonomic notes (Amphibia, Anura, Dicroglossidae). Vertebr. Zool. 2019, 69, 183–226. [Google Scholar]
- Raj, P.; Dinesh, K.P.; Das, A.; Dutta, S.K.; Kar, N.B.; Mohapatra, P.P. Two new species of Cricket Frogs of the genus Fejervarya Bolkay, 1915 (Anura: Dicroglossidae) from the Peninsular India. Rec. Zool. Surv. India 2018, 118, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Dehling, J.M. A new wide-headed Fanged Frog of the Limnonectes kuhlii group (Anura: Dicroglossidae) from western Borneo with a redescription of Rana conspicillata Günther, 1872. Zootaxa 2017, 4317, 291. [Google Scholar] [CrossRef]
- Köhler, G.; Vargas, J.; Than, N.L.; Schell, T.; Janke, A.; Pauls Steffen, U.; Thammachoti, P. A taxonomic revision of the genus Phrynoglossus in Indochina with the description of a new species and comments on the classification within Occidozyginae (Amphibia, Anura, Dicroglossidae). Vertebr. Zool. 2021, 71, 1–26. [Google Scholar] [CrossRef]
- Stuart, B.L.; Schoen, S.N.; Nelson, E.E.; Maher, H.; Neang, T.; Rowley, J.J.; McLeod, D.S. A new fanged frog in the Limnonectes kuhlii complex (Anura: Dicroglossidae) from northeastern Cambodia. Zootaxa 2020, 4894, 451–473. [Google Scholar] [CrossRef]
- Pham, C.T.; Le, M.D.; Ngo, H.T.; Ziegler, T.; Nguyen, T.Q. A new species of Limnonectes (Amphibia: Anura: Dicroglossidae) from Vietnam. Zootaxa 2018, 4508, 115–130. [Google Scholar] [CrossRef]
- Pham, C.T.; Le, M.D.; Nguyen, T.T.; Ziegler, T.; Wu, Z.J.; Nguyen, T.Q. A new species of Limnonectes (Amphibia: Anura: Dicroglossidae) from Vietnam. Zootaxa 2017, 4269, 545–558. [Google Scholar] [CrossRef]
- Inger, R.F.; Stuart, B.L. Systematics of Limnonectes (Taylorana) Dubois. Curr. Herpetol. 2010, 29, 51–68. [Google Scholar] [CrossRef]
- Fea, L. Viaggio di Leonardo Fea in Birmania e regione vicine. LXXVI Riassunto generale dei risultati zoologici. Ann. Mus. Civ. Stor. Nat. Genova 1897, 37, 385–658. [Google Scholar]
- Hallermann, J.; Ananjeva, N.; Orlov, N.; Tillack, F. Leonardo Fea’s historical collections of Amphibia and Reptilia from Burma deposited at the Zoologisches Museum Hamburg. Mitt. Aus Dem Hambg. Zool. Mus. Und Inst. 2002, 99, 139–153. [Google Scholar]
- Dubois, A. Miscellanea taxinomica batrachologica (I). Alytes 1987, 5, 7–95. [Google Scholar]
- Frost, D.; Grant, T.; Faivovich, J.; Bain, R.H.; Haas, A.; Haddad, C.F.; De Sá, R.O.; Channing, A.; Wilkinson, M.; Donnellan, S.; et al. THE AMPHIBIAN TREE OF LIFE. Bull. Am. Mus. Nat. Hist. 2006, 297, 1–291. [Google Scholar] [CrossRef]
- Pyron, R.A.; Wiens, J.J. A large-scale phylogeny of Amphibia including over 2800 species, and a revised classification of extant frogs, salamanders, and caecilians. Mol. Phylogenetics Evol. 2011, 61, 543–583. [Google Scholar] [CrossRef]
- Borah, M.M.; BordoloI, S.; Purkayastha, J.; Das, M.; Dubois, A.; Ohler, A. Limnonectes (Taylorana) medogensis (FEI, YE & HUANG, 1997) from Arunachal Pradesh (India), and on the identity of some diminutive ranoid frogs. Herpetozoa 2013, 26, 39–48. [Google Scholar]
- Boulenger, G.A. An account of the reptiles and batrachians obtained in Tenasserim by M. L. Fea of the Genoa Civic Museum. Ann. Mus. Civ. Stor. Nat. Genova 1887, 5, 474–486. [Google Scholar]
- Ohler, A.; Dubois, A. The identity of Elachyglossa gyldenstolpei Andersson 1916 (Amphibia, Ranidae), with comments on some aspects of statistical support to taxonomy. Zool. Scr. 1999, 28, 269–279. [Google Scholar] [CrossRef]
- Smith, M.A. The frogs allied to Ranadoriae. J. Nat. Hist. Soc. Siam. 1922, 4, 215. [Google Scholar]
- Lambertz, M.; Hartmann, T.; Walsh, S.; Geissler, P.; McLeod, D.S. Anatomy, histology, and systematic implications of the head ornamentation in the males of four species of Limnonectes (Anura: Dicroglossidae). Zool. J. Linn. Soc. 2014, 172, 117–132. [Google Scholar] [CrossRef]
- Phimmachak, S.; Richards, S.J.; Sivongxay, N.; Seateun, S.; Chuaynkern, Y.; Makchai, S.; Som, H.E.; Stuart, B.L. A new caruncle-bearing fanged frog (Limnonectes, Dicroglossidae) from Laos and Thailand. ZooKeys 2019, 846, 133–156. [Google Scholar] [CrossRef]
- Yodthong, S.; Rujirawan, A.; Stuart, B.; Aowphol, A. A new Limnonectes (Anura: Dicroglossidae) from southern Thailand. Animals 2021, 11, 566. [Google Scholar] [CrossRef]
- Köhler, G.; Khaing, K.P.P.; Than, N.L.; Baranski, D.; Schell, T.; Greve, C.; Janke, A.; Pauls, S.U. A new genus and species of mud snake from Myanmar (Reptilia, Squamata, Homalopsidae). Zootaxa 2021, 4915, 301–325. [Google Scholar] [CrossRef] [PubMed]
- Köhler, G. Color Catalogue for Field Biologists; Herpeton: Offenbach, Germany, 2012. [Google Scholar]
- Simpson, G.G. The species concept. Evolution 1951, 5, 285–298. [Google Scholar] [CrossRef]
- Wiley, E.O. The Evolutionary Species Concept Reconsidered. Syst. Zool. 1978, 27, 17–26. [Google Scholar] [CrossRef] [Green Version]
- Savage, J.M.; Heyer, W.R. Digital webbing formulae for anurans: A refinement. Herpetol. Rev. 1997, 28, 131. [Google Scholar]
- Heyer, W.R.; Rand, A.S.; Goncalves da Cruz, C.A.; Peixoto, O.L.; Nelson, C.E. Frogs of Boraceia. Arq. Zool. 1990, 31, 235–410. [Google Scholar]
- Köhler, J.; Jansen, M.; Rodríguez, A.; Kok, P.; Toledo, L.F.; Emmrich, M.; Glaw, F.; Haddad, C.F.B.; Rödel, M.-O.; Vences, M. The use of bioacoustics in anuran taxonomy: Theory, terminology, methods and recommendations for best practice. Zootaxa 2017, 4251, 1–124. [Google Scholar] [CrossRef] [Green Version]
- Ivanova, N.V.; Dewaard, J.R.; Hebert, P. An inexpensive, automation-friendly protocol for recovering high-quality DNA. Mol. Ecol. Notes 2006, 6, 998–1002. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatic 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Glez-Peña, D.; Gómez-Blanco, D.; Reboiro-Jato, M.; Fdez-Riverola, F.; Posada, D. ALTER: Program-oriented conversion of DNA and protein alignments. Nucleic Acids Res. 2010, 38, 14–18. [Google Scholar] [CrossRef] [PubMed]
- Lanfear, R.; Frandsen, P.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef] [Green Version]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate Maximum-Likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [Green Version]
- Burnham, K.P.; Anderson, D.R. Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach, 2nd ed.; Springer: New York, NY, USA, 2002; ISBN 9780387953649. [Google Scholar]
- Ronquist, F.; Teslenko, M.; Mark, P.; Ayres, D.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.; Huelsenbeck, J. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [Green Version]
- Gelman, A.; Rubin, D.B. Inference from Iterative Simulation Using Multiple Sequences. Stat. Sci. 1992, 7, 457–472. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.-T.; Von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minh, B.Q.; Nguyen, M.A.T.; Von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Ogilvie, H.A.; Bouckaert, R.R.; Drummond, A. StarBEAST2 brings faster species tree inference and accurate estimates of substitution rates. Mol. Biol. Evol. 2017, 34, 2101–2114. [Google Scholar] [CrossRef]
- Bouckaert, R.; Vaughan, T.; Barido-Sottani, J.; Duchene, S.; Fourment, M.; Gavryushkina, A.; Heled, J.; Jones, G.; Kuhnert, D.; de Maio, N.; et al. BEAST 2.5: An advanced Ssftware platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019, 15, e1006650. [Google Scholar] [CrossRef] [Green Version]
- Bouckaert, R.; Heled, J. DensiTree 2: Seeing trees through the forest. BioRxiv 2014, 012401. [Google Scholar] [CrossRef] [Green Version]
- Sambrook, J.; Russell, W.D. (Eds.) Protocol 1: DNA isolation from mammalian tissue. In Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001; pp. 623–627. [Google Scholar]
- Marçais, G.; Kingsford, C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 2011, 27, 764–770. [Google Scholar] [CrossRef] [Green Version]
- Vurture, G.W.; Sedlazeck, F.; Nattestad, M.; Underwood, C.; Fang, H.; Gurtowski, J.; Schatz, M.C. GenomeScope: Fast reference-free genome profiling from short reads. Bioinformatic 2017, 33, 2202–2204. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatic 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–4777. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Handsaker, R.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011, 27, 2987–2993. [Google Scholar] [CrossRef] [Green Version]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De novoassembly of organelle genomes from whole genome data. Nucleic Acids Res. 2016, 45, e18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef] [PubMed]
- Donath, A.; Jühling, F.; Al-Arab, M.; Bernhart, S.H.; Reinhardt, F.; Stadler, P.F.; Middendorf, M.; Bernt, M. Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 2019, 47, 10543–10552. [Google Scholar] [CrossRef] [PubMed]
- Emerson, S.B.; Inger, R.F.; Iskandar, D. Molecular systematics and biogeography of the fanged frogs of Southeast Asia. Mol. Phylogenetics Evol. 2000, 16, 131–142. [Google Scholar] [CrossRef] [PubMed]
- IUCN. IUCN Red List Categories and Criteria, Version 3. 1. Second Edition. Available online: https://portals.ucn.org/library/node/10315 (accessed on 17 November 2018).
- Simao, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stuart, B.L.; Som, H.E.; Neang, T.; Hoang, H.D.; Le, D.T.T.; Dau, V.Q.; Potter, K.; Rowley, J.J.L. Integrative taxonomic analysis reveals a new species of Leptobrachium (Anura: Megophryidae) from north-eastern Cambodia and central Vietnam. J. Nat. Hist. 2020, 54, 225–255. [Google Scholar] [CrossRef]
- Ziegler, T. Die Amphibien Und Reptilien Eines Tieflandfeuchtwald-Schutzgebietes in Vietnam; Natur und Tier-Verlag: Münster, Germany, 2002. [Google Scholar]
- Vineeth, K.K.; Radhakrishna, U.K.; Godwin, R.D.; Anwesha, S.; Rajashekhar, K.P.; Aravind, N.A. A new species of Microhyla Tschudi, 1838 (Anura: Microhylidae) from West Coast of India: An integrative taxonomic approach. Zootaxa 2018, 4420, 151–179. [Google Scholar] [CrossRef]
- Priti, H.; Roshmi, R.S.; Ramya, B.; Sudhira, H.S.; Ravikanth, G.; Aravind, N.A.; Gururaja, K.V. Integrative taxonomic approach for describing a new cryptic species of Bush Frog (Raorchestes: Anura: Rhacophoridae) from the Western Ghats, India. PLoS ONE 2016, 11, e0149382. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).