Application of COI Primers 30F/885R in Rotifers to Regional Species Diversity in (Sub)Tropical China
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection, Identification, and Counting of Rotifers
2.2. DNA Extraction, PCR Amplification, and Sequencing
2.3. Species Identification and Analysis Based on Molecular Methods
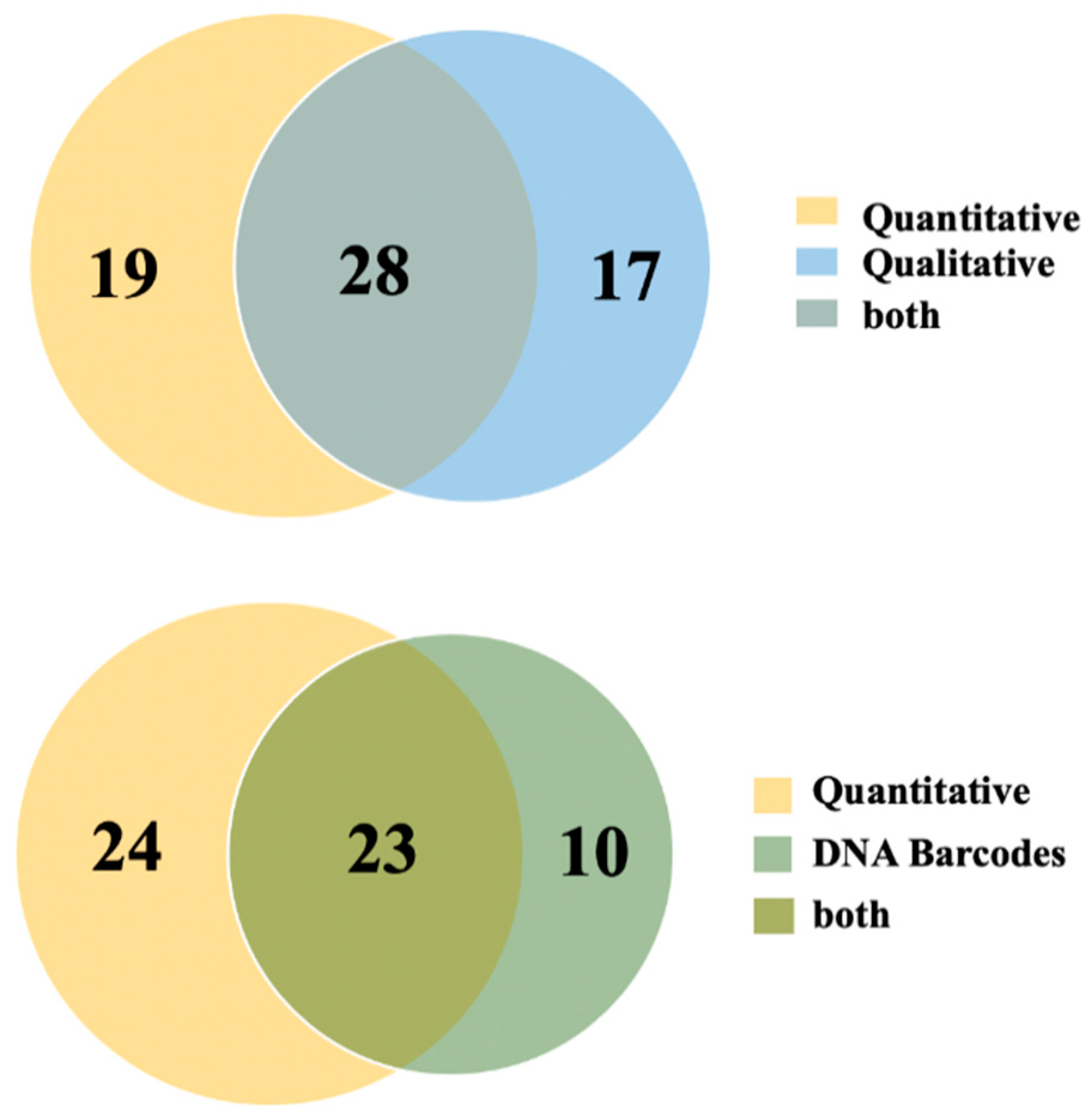
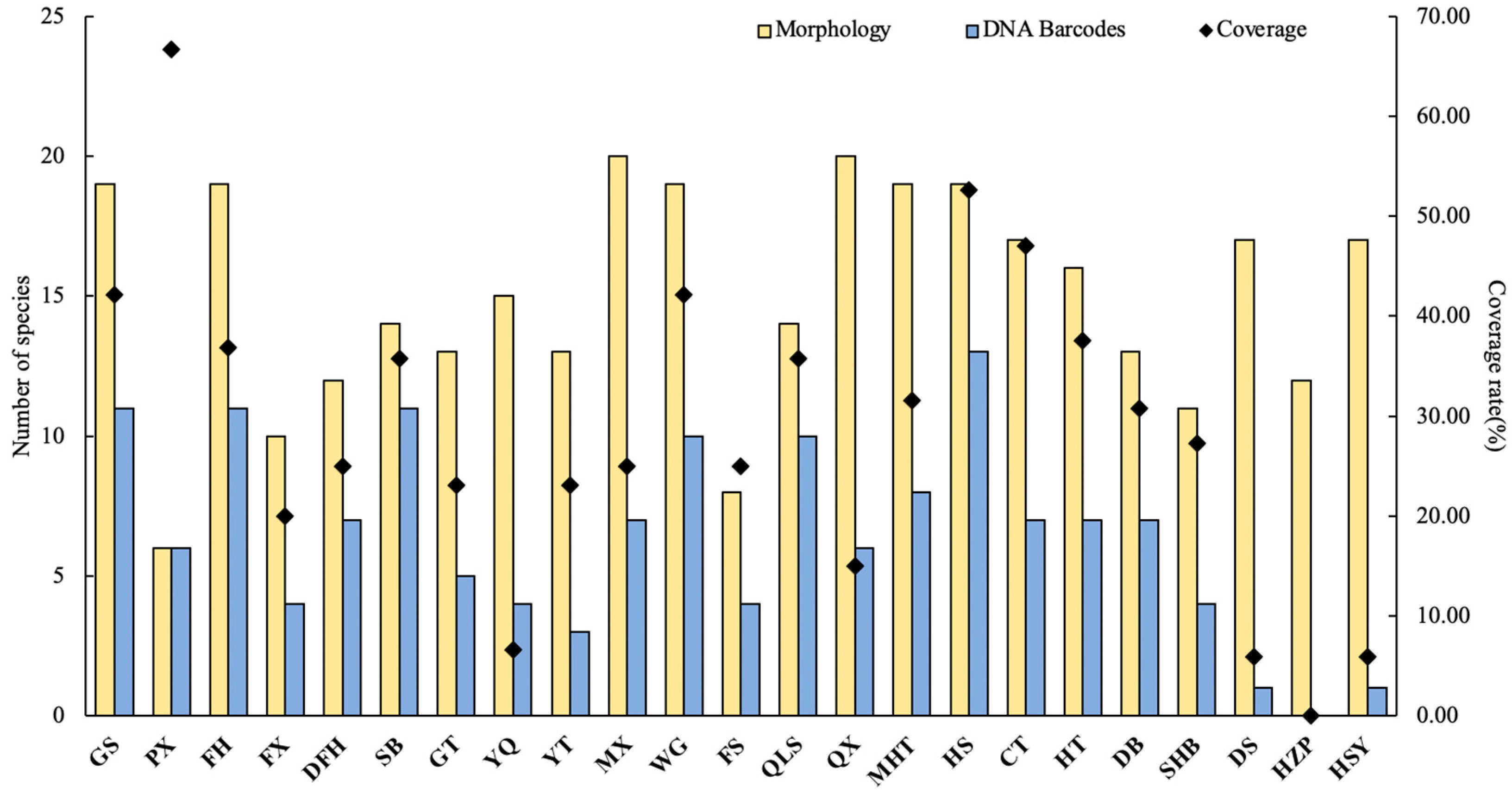
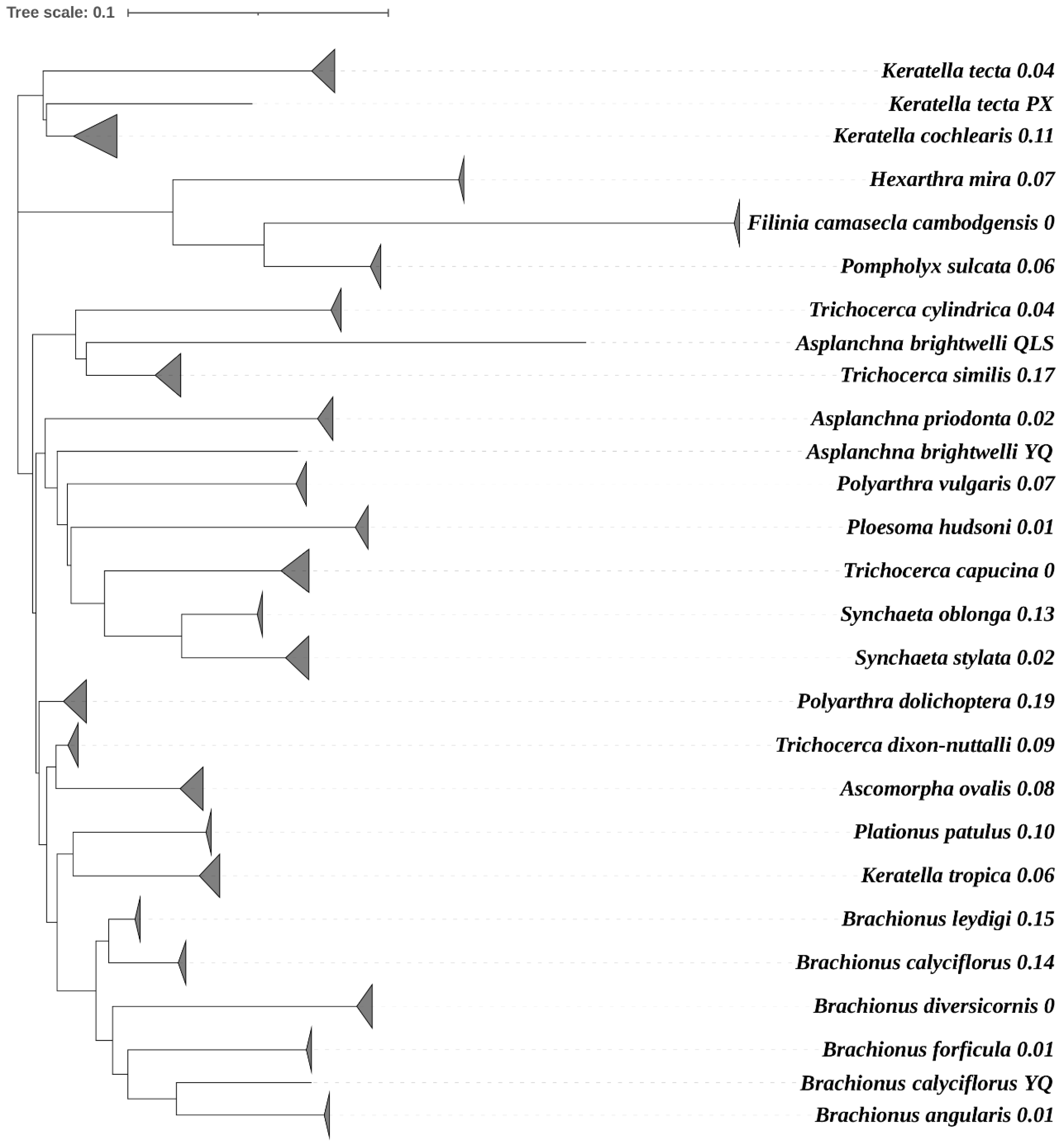
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Duggan, I.C. The ecology of periphytic rotifers. Rotifera IX 2001, 153, 139–148. [Google Scholar]
- Wallace, R.L.; Snell, T.W.; Ricci, C.; Nogrady, T. Guides to the identification of the microinvertebrates of the continental waters of the world. In Rotifera Part 1: Biology, Ecology and Systematics; Dumont, H.J.F., Ed.; Kenobi Production & Backhuys Publishers: Hague, The Netherlands, 2006; Volume 23, p. 299. [Google Scholar]
- Segers, H. Global diversity of rotifers (Rotifera) in freshwater. Hydrobiologia 2007, 595, 49–59. [Google Scholar] [CrossRef]
- Wallace, R.L. Rotifers: Exquisite metazoans. Integr. Comp. Biol. 2002, 42, 660–667. [Google Scholar] [CrossRef] [PubMed]
- Arndt, H. Rotifers as predators on components of the microbial web (bacteria, heterotrophic flagellates, ciliates)—A review. Hydrobiologia 1993, 255–256, 231–246. [Google Scholar] [CrossRef]
- Schmid-Araya, J.M.; Schmid, P.E. Trophic relationships: Integrating meiofauna into a realistic benthic food web. Freshw. Biol. 2000, 44, 149–163. [Google Scholar] [CrossRef]
- Rao, T.R.; Sarma, S.S.S. Demographic parameters of Brachionus patulus Muller (Rotifera) exposed to sublethal DDT concentrations at low and high food levels. Hydrobiologia 1986, 139, 193–200. [Google Scholar]
- Koste, W. Rotatoria. Die Rädertiere Mitteleuropas, Bd. Ⅱ.; Gebrüder: Berlin, Germany, 1978. [Google Scholar]
- Pavón-Meza, E.L.; Sarma, S.S.S.; Nandini, S. Combined effects of temperature, food (Chlorella vulgaris) concentration and predation (Asplanchna girodi) on the morphology of Brachionus havanaensis (Rotifera). Hydrobiologia 2007, 593, 95–101. [Google Scholar] [CrossRef]
- Segers, H.; De Smet, W.H. Diversity and endemism in Rotifera: A review, and Keratella Bory de St Vincent. Biodivers. Conserv. 2007, 8, 69–82. [Google Scholar]
- Fontaneto, D.; Boschetti, C.; Ricci, C. Cryptic diversification in ancient asexuals: Evidence from the bdelloid rotifer Philodina flaviceps. J. Evol. Biol. 2008, 21, 580–587. [Google Scholar] [CrossRef]
- Fontaneto, D. Molecular phylogenies as a tool to understand diversity in rotifers. Int. Rev. Hydrobiol. 2014, 99, 178–187. [Google Scholar] [CrossRef]
- Mills, S.; Alcántara-Rodríguez, J.A.; Ciros-Pérez, J.; Gómez, A.; Hagiwara, A.; Galindo, K.H.; Jersabek, C.D.; Malekzadeh-Viayeh, R.; Leasi, F.; Lee, J.-S.; et al. Fifteen species in one: Deciphering the Brachionus plicatilis species complex (Rotifera, Monogononta) through DNA taxonomy. Hydrobiologia 2017, 796, 39–58. [Google Scholar] [CrossRef]
- Michaloudi, E.; Papakostas, S.; Stamou, G.; Neděla, V.; Tihlaříková, E.; Zhang, W.; Declerck, S.A.J. Reverse taxonomy applied to the Brachionus calyciflorus cryptic species complex: Morphometric analysis confirms species delimitations revealed by molecular phylogenetic analysis and allows the (re) description of four species. PLoS ONE 2018, 13, e0203168. [Google Scholar] [CrossRef]
- García-Morales, A.E.; Domínguez-Domínguez, O. Cryptic species within the rotifer Lecane bulla (Rotifera: Monogononta: Lecanidae) from North America based on molecular species delimitation. Rev. Mex. Biodiv. 2020, 91, 1–12. [Google Scholar] [CrossRef]
- John, G.; Elizabeth, W. Brachionus calyciflorus is a species complex: Mating behavior and genetic differentiation among four geographically isolated strains. Hydrobiologia 2005, 546, 257–265. [Google Scholar]
- Gómez, A.; Temprano, M.; Serra, M. Ecological genetics of a cyclical parthenogen in temporary habitats. J. Evol. Biol. 1995, 8, 601–622. [Google Scholar] [CrossRef]
- Ortells, R.; Snell, T.W.; Gómez, A.; Serra, M. Patterns of genetic differentiation in resting egg banks of a rotifer species complex in Spain. Hydrobiologia 2000, 149, 529–551. [Google Scholar] [CrossRef]
- Gómez, A. Allozyme electrophoresis: Its application to rotifers. Hydrobiologia 1998, 387/388, 385–393. [Google Scholar] [CrossRef]
- Kordbacheh, A.; Wallace, R.L.; Walsh, E.J. Evidence supporting cryptic species within two sessile microinvertebrates, Limnias melicerta and L. ceratophylli (Rotifera, Gnesiotrocha). PLoS ONE 2018, 13, e0205203. [Google Scholar] [CrossRef]
- Fontaneto, D.; Jondelius, U. Broad taxonomic sampling of mitochondrial cytochrome c oxidase subunit I does not solve the relationships between Rotifera and Acanthocephala. Zool. Anz. 2011, 250, 80–85. [Google Scholar] [CrossRef]
- Hwang, D.S.; Dahms, H.U.; Park, H.G.; Lee, J.S. A new intertidal Brachionus and intrageneric phylogenetic relationships among Brachionus as revealed by allometry and CO1-ITS1 gene analysis. Zool. Stud. 2013, 52, 1–10. [Google Scholar] [CrossRef]
- Birky, C.W.; Wolf, C.; Maughan, H.; Herbertson, L.; Henry, E. Speciation and selection without sex. Hydrobiologia 2005, 546, 29–45. [Google Scholar] [CrossRef][Green Version]
- García-Morales, A.E.; Elías-Gutiérrez, M. DNA barcoding of freshwater Rotifera in Mexico: Evidence of cryptic speciation in common rotifers. Mol. Ecol. Resour. 2013, 13, 1097–1107. [Google Scholar] [CrossRef] [PubMed]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Meyer, C.P.; Geller, J.B.; Paulay, G. Fine scale endemism on coral reefs: Archipelagic differentiation in turbinid gastropods. Evolution 2005, 59, 113–125. [Google Scholar] [CrossRef]
- Rico-Martínez, R.; Snell, T.W.; Shearer, T.L. Synergistic toxicity of Macondo crude oil and dispersant Corexit 9500A® to the Brachionus plicatilis species complex (Rotifera). Environ. Pollut. 2013, 173, 5–10. [Google Scholar] [CrossRef]
- Wilts, E.F.; Bruns, D.; Fontaneto, D.; Ahlrich, W.H. Phylogenetic study on Proales daphnicola Thompson, 1892 (Proalidae) and its relocation to Epiphanes (Rotifera: Epiphanidae). Zool. Anz. 2012, 251, 180–196. [Google Scholar] [CrossRef]
- Prosser, S.; Martínez-Arce, A.; Elías-Gutiérrez, M. A new set of primers for COI amplification from freshwater microcrustaceans. Mol. Ecol. Resour. 2013, 13, 1151–1155. [Google Scholar] [CrossRef]
- Elías-Gutiérrez, M.; Valdez-Moreno, M.; Topan, J.; Young, M.R.; Cohuo-Colli, J.A. Improved protocols to accelerate the assembly of DNA barcode reference libraries for freshwater zooplankton. Ecol. Evol. 2018, 8, 3002–3018. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.N.; Xu, S.L.; Sun, C.H.; Dumont, H.; Han, B.P. A new set of highly efficient primers for COI amplification in rotifers. Mitochondrial DNA Part B 2021, 6, 636–640. [Google Scholar] [CrossRef]
- Lan, Z.H.; Du, L.M. Studies on zooplankton in the Hanjiang River. J. Hanshan Norm. Univ. 1996, 97–104. [Google Scholar]
- Lin, Q.Q.; Zhao, S.Y.; Han, B.P. Rotifer distribution in tropical reservoirs, Guangdong Province, China. Acta Ecol. Sin. 2005, 25, 1123–1131. [Google Scholar]
- Zhang, Z.X.; Huang, X.F. Freshwater Plankton Research Method; Science Press: Beijing, China, 1991. [Google Scholar]
- Hillebrand, H.; Dürselen, C.D.; Kirschtel, D. Biovolume calculation for pelagic and benthic microalgae. J. Phycol. 1999, 35, 403–424. [Google Scholar] [CrossRef]
- Xu, Z.L.; Chen, Y.Q. Aggregated intensity of dominant species of zooplankton in autumn in the East China Sea and Yellow Sea. Chin. J. Ecol. 1989, 8, 13–15. [Google Scholar]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Ranwez, V.; Douzery, E.J.P.; Cambon, C.; Chantret, N.; Delsuc, F. MACSE v2: Toolkit for the alignment of coding sequences accounting for frameshifts and stop codons. Mol. Biol. Evol. 2018, 35, 2582–2584. [Google Scholar] [CrossRef]
- Henzinger, T.A.; Jhala, R.; Majumdar, R.; Sutre, G. Software verification with BLAST. In International SPIN Workshop on Model Checking of Software; Ball, T., Rajamani, S.R., Eds.; Springer: Berlin/Heidelberg, Germany, 2003; pp. 235–239. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Leutbecher, C. A routine method of DNA-extraction from extremely small metazoans, eg single rotifer specimens for RAPD-PCR analyses. Hydrobiologia 2000, 437, 133–137. [Google Scholar] [CrossRef]
- Makino, W.; Maruoka, N.; Nakagawa, M.; Takamura, N. DNA barcoding of freshwater zooplankton in Lake Kasumigaura, Japan. Ecol. Res. 2017, 32, 481–493. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; Dewaard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. B 2003, 270, 313–321. [Google Scholar] [CrossRef]
- Leray, M.; Yang, J.Y.; Meyer, C.P.; Mills, S.C.; Agudelo, N.; Ranwez, V.; Boehm, J.T.; Machida, R.J. A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity: Application for characterizing coral reef fish gut contents. Front. Zool. 2013, 10, 34. [Google Scholar] [CrossRef]
- Lim, N.K.M.; Tay, Y.C.; Srivathsan, A.; Yeo, D.C.J. Next-generation freshwater bioassessment: eDNA metabarcoding with a conserved metazoan primer reveals species-rich and reservoir-specific communities. R. Soc. Open Sci. 2016, 3, 160635. [Google Scholar] [CrossRef]
- Yang, J.H.; Zhang, X.W.; Zhang, W.W.; Sun, J.Y.; Xie, Y.W.; Zhang, Y.M.; Burton, G.A.; Yu, H.X. Indigenous species barcode database improves the identification of zooplankton. PLoS ONE 2017, 12, e0185697. [Google Scholar] [CrossRef]
- Crampton-Platt, A.; Timmermans, M.J.T.N.; Gimmel, M.L.; Kutty, S.N.; Cockerill, T.D.; Khen, C.V.; Vogler, A.P. Soup to tree: The phylogeny of beetles inferred by mitochondrial metagenomics of a Bornean rainforest sample. Mol. Biol. Evol. 2015, 32, 2302–2316. [Google Scholar] [CrossRef]
- Gómez-Rodríguez, C.; Crampton-Platt, A.; Timmermans, M.J.T.N.; Baselga, A.; Vogler, A.P. Validating the power of mitochondrial metagenomics for community ecology and phylogenetics of complex assemblages. Methods Ecol. Evol. 2015, 6, 883–894. [Google Scholar] [CrossRef]
- Derry, A.M.N.; Hebert, P.D.; Prepas, E.E. Evolution of rotifers in saline and subsaline lakes: A molecular phylogenetic approach. Limnol. Oceanogr. 2003, 48, 675–685. [Google Scholar] [CrossRef]
- Koste, W.; Shiel, R.J. Rotifera from Australian inland waters. II. Epiphanidae and Brachionidae (Rotifera: Monogononta). Invertebr. Syst. 1987, 1, 949–1021. [Google Scholar] [CrossRef]
- Xiang, X.L.; Xi, Y.L.; Wen, X.L.; Zhang, J.Y.; Ma, Q. Spatial patterns of genetic differentiation in Brachionus calyciflorus species complex collected from East China in summer. Hydrobiologia 2010, 638, 67–83. [Google Scholar] [CrossRef]

| Species | Number of Specimens | Number of Sequences | PCR Success Rate (%) |
|---|---|---|---|
| Keratella cochlearis | 24 | 17 | 71 |
| Keratella tropica | 15 | 8 | 53 |
| Keratella tecta | 24 | 10 | 42 |
| Brachionus diversicornis | 8 | 6 | 67 |
| Brachionus calyciflorus | 5 | 4 | 80 |
| Brachionus caudatus | 2 | 1 | 50 |
| Brachionus urceolaris | 2 | 1 | 50 |
| Brachionus angularis | 3 | 2 | 67 |
| Brachionus quadridentatus | 3 | 1 | 33 |
| Brachionus forficula | 3 | 2 | 67 |
| Brachionus leydigi | 2 | 2 | 100 |
| Brachionus budapestinensis | 1 | 1 | 100 |
| Asplanchna priodonta | 9 | 6 | 67 |
| Asplanchna brightwelli | 2 | 2 | 100 |
| Polyarthra vulgaris | 14 | 4 | 29 |
| Polyarthra dolichoptera | 9 | 9 | 100 |
| Ploesoma hudsoni | 7 | 5 | 71 |
| Ploesoma truncatum | 1 | 1 | 100 |
| Pompholyx sulcata | 5 | 4 | 80 |
| Trichocerca dixonnuttalli | 4 | 4 | 100 |
| Trichocerca capucina | 23 | 11 | 48 |
| Trichocerca cylindrica | 18 | 4 | 22 |
| Trichocerca similis | 16 | 10 | 63 |
| Filinia opoliensis | 1 | 1 | 100 |
| Filinia camaseclacambodgensis | 2 | 2 | 100 |
| Trichotria pocillum | 1 | 1 | 100 |
| Trichotria tetractis similis | 3 | 1 | 33 |
| Synchaeta oblonga | 3 | 2 | 67 |
| Synchaeta stylata | 10 | 9 | 90 |
| Lecane bulla | 3 | 1 | 33 |
| Ascomorpha ovalis | 11 | 9 | 82 |
| Hxarthra mira | 8 | 2 | 25 |
| Plationus patulus | 3 | 2 | 67 |
| Species | Numbers of Sequences | Min Distance | Max Distance | Mean Distance |
|---|---|---|---|---|
| Keratella cochlearis | 17 | 0.00 | 0.19 | 0.11 |
| Keratella tecta | 10 | 0.00 | 0.18 | 0.04 |
| Keratella tropica | 8 | 0.00 | 0.12 | 0.06 |
| Brachionus diversicornis | 6 | 0.00 | 0.00 | 0.00 |
| Brachionus leydigi | 2 | N/A | N/A | 0.15 |
| Brachionus forficula | 4 | 0.00 | 0.01 | 0.01 |
| Brachionus angularis | 2 | N/A | N/A | 0.01 |
| Brachionus calyciflorus | 4 | 0.04 | 0.18 | 0.14 |
| Plationus patulus | 2 | N/A | N/A | 0.10 |
| Asplanchna priodonta | 7 | 0.00 | 0.03 | 0.02 |
| Asplanchna brightwelli | 2 | N/A | N/A | 0.31 |
| Polyarthra vulgaris | 4 | 0.06 | 0.25 | 0.07 |
| Polyarthra dolichoptera | 9 | 0.00 | 0.25 | 0.19 |
| Ascomorpha ovalis | 11 | 0.00 | 0.14 | 0.08 |
| Trichocercasimilis | 10 | 0.00 | 0.32 | 0.17 |
| Trichocerca cylindrica | 5 | 0.00 | 0.07 | 0.04 |
| Trichocerca dixon-nuttalli | 4 | 0.00 | 0.18 | 0.09 |
| Trichocerca capucina | 13 | 0.00 | 0.01 | 0.00 |
| Synchaeta stylata | 9 | 0.00 | 0.06 | 0.02 |
| Synchaeta oblonga | 2 | N/A | N/A | 0.13 |
| Ploesoma hudsoni | 5 | 0.02 | 0.00 | 0.01 |
| Pompholyx sulcata | 4 | 0.00 | 0.12 | 0.06 |
| Filiniacamasecla cambodgensis | 2 | N/A | N/A | 0.00 |
| Hexarthramira | 2 | N/A | N/A | 0.07 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.-N.; Xu, S.-L.; Huang, Q.; Liu, P.; Han, B.-P. Application of COI Primers 30F/885R in Rotifers to Regional Species Diversity in (Sub)Tropical China. Diversity 2021, 13, 390. https://doi.org/10.3390/d13080390
Zhang Y-N, Xu S-L, Huang Q, Liu P, Han B-P. Application of COI Primers 30F/885R in Rotifers to Regional Species Diversity in (Sub)Tropical China. Diversity. 2021; 13(8):390. https://doi.org/10.3390/d13080390
Chicago/Turabian StyleZhang, Ya-Nan, Shao-Lin Xu, Qi Huang, Ping Liu, and Bo-Ping Han. 2021. "Application of COI Primers 30F/885R in Rotifers to Regional Species Diversity in (Sub)Tropical China" Diversity 13, no. 8: 390. https://doi.org/10.3390/d13080390
APA StyleZhang, Y.-N., Xu, S.-L., Huang, Q., Liu, P., & Han, B.-P. (2021). Application of COI Primers 30F/885R in Rotifers to Regional Species Diversity in (Sub)Tropical China. Diversity, 13(8), 390. https://doi.org/10.3390/d13080390








