The Diversity of Culture-Dependent Gram-Negative Rhizobacteria Associated with Manihot esculenta Crantz Plants Subjected to Water-Deficit Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Drought Stress Assays
2.2. Strain Isolation
2.3. Strain Characterization and Identification
2.4. Statistical Analysis
3. Results
3.1. Strain Isolation, Morphological Characterization, and Biochemical Identification
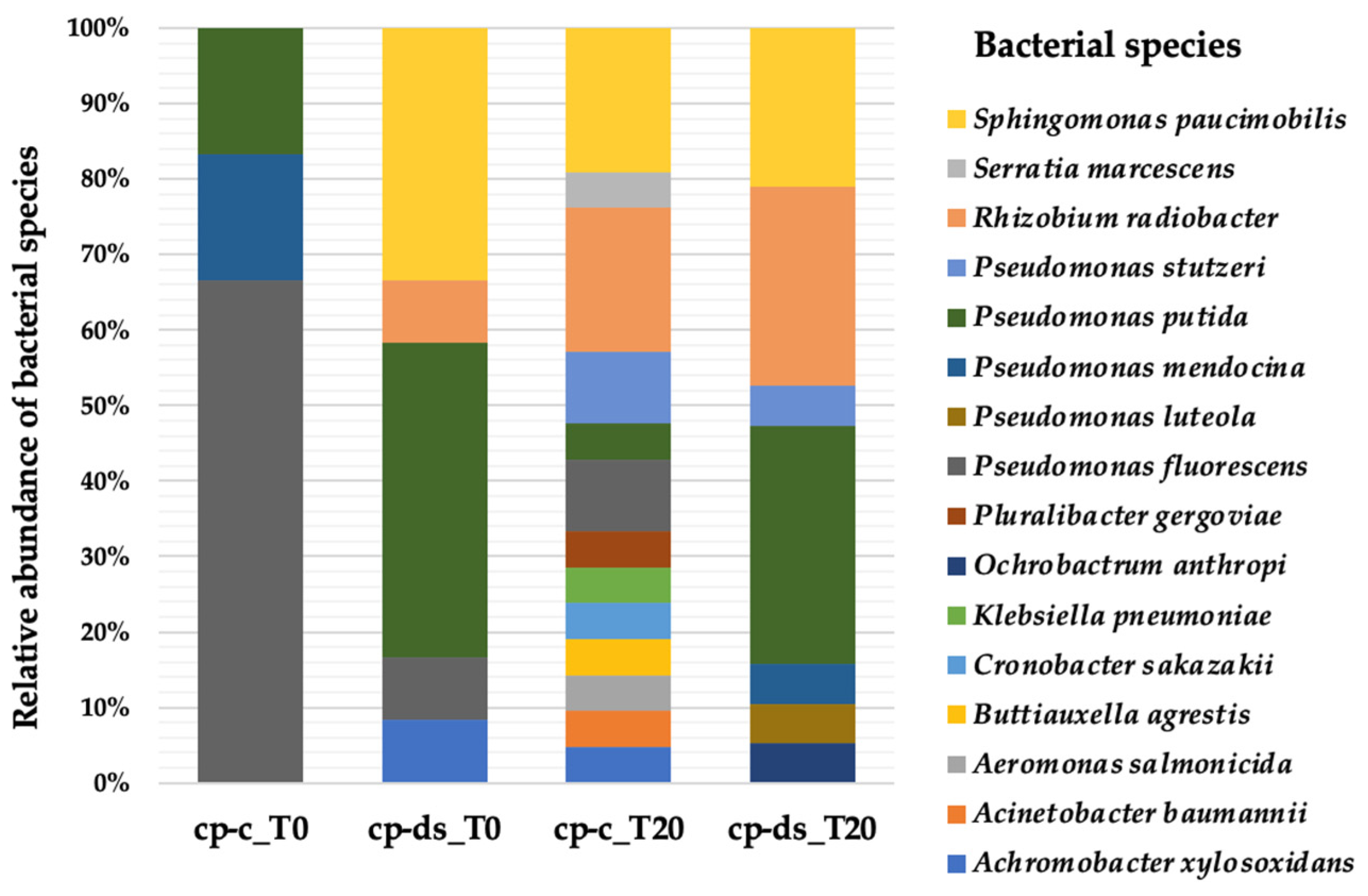
3.2. Strains Diversity at the Beginning of the Drought Stress Assay (T0)
3.3. Strains Diversity at the End of the Drought Stress Assay (T20)
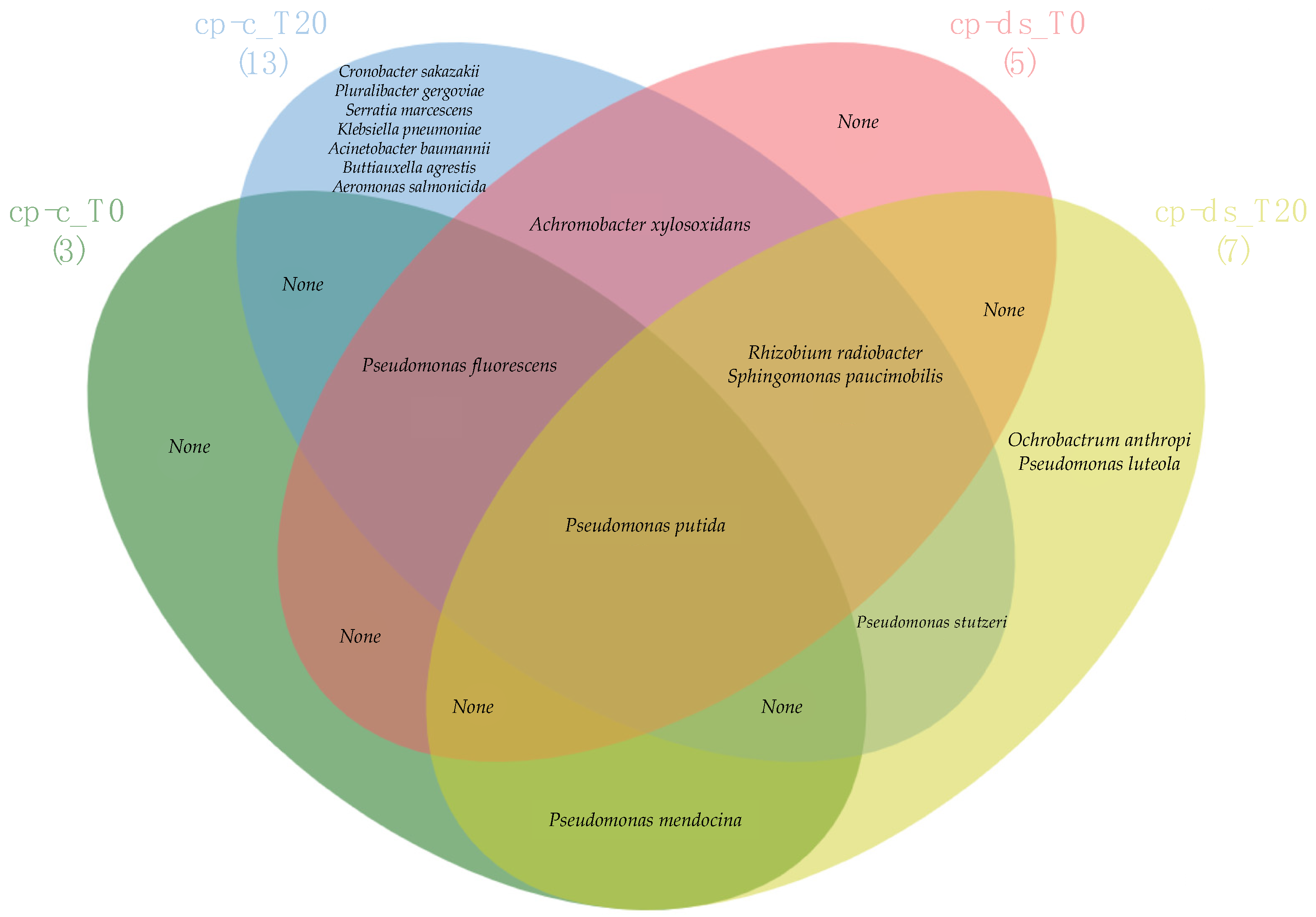
3.4. Comparison of Strain Diversity for cp-c and cp-ds Plants
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Howeler, R.; Lutaladio, N.; Thomas, G. Save and Grow: Cassava, a Guide to Sustainable Production Intensification; Food and Agriculture Organization of the United Nations (FAO), Ed.; FAO: Rome, Italy, 2013. [Google Scholar]
- Wanapat, M.; Kang, S. Cassava chip (Manihot esculenta Crantz) as an energy source for ruminant feeding. Anim. Nutr. 2015, 1, 266–270. [Google Scholar] [CrossRef]
- Santos, V.L.F.; Ferreira, M.A.; Siqueira, M.C.B.; Melo, T.T.B.; Silva, J.L.; Andrade, I.B.; Soares, A.A.; Costa, C.T.F. Rumen parameters of sheep fed cassava peel as a replacement for corn. Small Rumin. Res. 2015, 133, 88–92. [Google Scholar] [CrossRef]
- Jauhar, P.P. Modern biotechnology as an integral supplement to conventional plant breeding: The prospects and challenges. Crop Sci. 2006, 46, 1841–1859. [Google Scholar] [CrossRef]
- Tofiño, A.; Ceballos, H.; Romero, H.M. Posibilidades de expansión del cultivo de yuca (Manihot esculentum Crantz) en el Caribe eeco eolombiano a partir de investigación multidisciplinaria. Actual. Biológicas 2008, 30, 15–27. [Google Scholar]
- El-Sharkawy, M.A. Drought-tolerant Cassava for Africa, Asia, and Latin America. Bioscience 1993, 43, 441–451. [Google Scholar] [CrossRef]
- El-Sharkawy, M.A. Stress-tolerant cassava: The role of integrative ecophysiology-breeding research in crop improvement. Open J. Soil Sci. 2012, 2, 162–186. [Google Scholar] [CrossRef]
- Pardales, J.R., Jr.; Esquibel, C.B. Effect of drought during the establishment period on the root system development of cassava. Jpn. J. Crop Sci. 1996, 65, 93–97. [Google Scholar] [CrossRef]
- Hoekstra, F.A.; Golovina, E.A.; Buitink, J. Mechanisms of plant desiccation tolerance. Trends Plant Sci. 2001, 6, 431–438. [Google Scholar] [CrossRef]
- Kaushal, M.; Wani, S.P. Plant-growth-promoting rhizobacteria: Drought stress alleviators to ameliorate crop production in drylands. Ann. Microbiol. 2016, 66, 35–42. [Google Scholar] [CrossRef]
- Agili, S.M.; Pardales, J.R., Jr. Influence of moisture and allelopathic regimes in the soil on the development of cassava and mycorrhizal infection of its roots during establishment period. Philipp. J. Crop Sci. 1997, 22, 99–105. [Google Scholar]
- Hershey, C.H.; Álvarez, E.; Aye, T.M.; Becerra, L.A.; Bellotti, A.C.; Ceballos, H.; Fahrney, K.; Howeler, R.H.; Lefroy, R.D.B.; Ospina, B.; et al. Eco-efficient interventions to support cassava’s multiple roles in improving the lives of smallholders. In Ecoefficiency: From Vision to Reality; CIAT: Cali, Colombia, 2012. [Google Scholar]
- IWMI. Water for Food, Water for Life. A Comprehensive Assessment of Water Management in Agriculture; Molden, D., Ed.; IMWI: Colombo, Sri Lanka; Earthscan: London, UK, 2007. [Google Scholar]
- Kawano, K. Cassava. In Hybridization of Crop Plants; Fehr, W.R., Hadley, H.H., Eds.; ASA & CSSA: Madison, WI, USA, 1980; pp. 225–233. [Google Scholar]
- Kawano, K. Thirty years of cassava breeding for productivity—Biological and social factors for success. Crop Sci. 2003, 43, 1325–1335. [Google Scholar] [CrossRef]
- Ceballos, H.; Hershey, C.H.; Becerra-López-Lavalle, L.A. New Approaches to Cassava Breeding. In Plant Breeding Reviews, Volume 36; Janick, J., Ed.; Wiley-Blackwell: Hoboken, NJ, USA, 2012; pp. 427–504. [Google Scholar]
- Farooq, M.; Wahid, A.; Kobayashi, N.; Fujita, D.; Basra, S.M.A. Plant drought stress: Effects, mechanisms and management. Agron. Sustain. Dev. 2009, 29, 185–212. [Google Scholar] [CrossRef]
- Turyagyenda, L.F.; Kizito, E.B.; Ferguson, M.; Baguma, Y.; Agaba, M.; Harvey, J.J.W.; Osiru, D.S.O. Physiological and molecular characterization of drought responses and identification of candidate tolerance genes in cassava. AoB Plants 2013, 5, 1–17. [Google Scholar] [CrossRef]
- Mutumba, F.A.; Zagal, E.; Gerding, M.; Castillo-Rosales, D.; Paulino, L.; Schoebitz, M. Plant growth promoting rhizobacteria for improved water stress tolerance in wheat genotypes. J. Soil Sci. Plant Nutr. 2018, 18, 1080–1096. [Google Scholar] [CrossRef]
- Etesami, H.; Maheshwari, D.K. Use of plant growth promoting rhizobacteria (PGPRs) with multiple plant growth promoting traits in stress agriculture: Action mechanisms and future prospects. Ecotoxicol. Environ. Saf. 2018, 156, 225–246. [Google Scholar] [CrossRef]
- Etesami, H.; Alikhani, H.A.; Mirseyed Hosseini, H. Indole-3-Acetic Acid and 1-Aminocyclopropane-1-Carboxylate Deaminase: Bacterial Traits Required in Rhizosphere, Rhizoplane and/or Endophytic Competence by Beneficial Bacteria. In Bacterial Metabolites in Sustainable Agroecosystem. Sustainable Development and Biodiversity; Maheshwari, D., Ed.; Springer: Cham, Switzerland, 2015. [Google Scholar]
- Etesami, H.; Beattie, G.A. Plant-Microbe Interactions in Adaptation of Agricultural Crops to Abiotic Stress Conditions. In Probiotics and Plant Health; Kumar, V., Kumar, M., Sharma, S., Prasad, R., Eds.; Springer: Singapore, 2017. [Google Scholar]
- Vurukonda, S.S.K.P.; Vardharajula, S.; Shrivastava, M.; SkZ, A. Enhancement of drought stress tolerance in crops by plant growth promoting rhizobacteria. Microbiol. Res. 2016, 184, 13–24. [Google Scholar] [CrossRef]
- Berjak, P. Unifying perspectives of some mechanisms basic to desiccation tolerance across life forms. Seed Sci. Res. 2012, 16, 1–15. [Google Scholar] [CrossRef]
- Ryu, C.M.; Farag, M.A.; Hu, C.H.; Reddy, M.S.; Kloepper, J.W.; Paré, P.W. Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol. 2004, 134, 1017–1026. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.I.; Cho, W.K.; Kim, S.N.; Chu, H.; Ryu, K.Y.; Yun, J.C.; Park, C.S. Genetic diversity of cultivable plant growth-promoting rhizobacteria in Korea. J. Microbiol. Biotechnol. 2011, 21, 777–790. [Google Scholar] [CrossRef]
- Hartman, K.; Tringe, S.G. Interactions between plants and soil shaping the root microbiome under abiotic stress. Biochem. J. 2019, 476, 2705–2724. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, J.; Wei, Y.; Hu, W.; Liu, G.; Zeng, H.; Shi, H. Microbiome-wide association studies reveal correlations between the structure and metabolism of the rhizosphere microbiome and disease resistance in cassava. Plant Biotechnol. J. 2021, 19, 689–701. [Google Scholar] [CrossRef] [PubMed]
- Frediansyah, A. The Microbiome of Cassava (Manihot esculanta). In Cassava—Biology, Production, and Application; Frediansyah, A., Ed.; Headquarters IntechOpen Limited: London, UK, 2021; ISBN 978-1-83968-909-3. [Google Scholar]
- Buensateai, N.; Thumanu, K.; Sompong, M. The FTIR spectroscopy investigation of the cellular components of cassava after sensitization with plant growth promoting rhizobacteria, Bacillus subtilis CaSUT007. Afr. J. Microbiol. Res. 2012, 6, 603–610. [Google Scholar] [CrossRef]
- Suja, S.P.; Hegde, V.; Makeshkumar, T.; Anjanadevi, I.P. Screening of rhizobacteria associated with cassava for plant growth promotion and biocontrol potential. J. Root Crop. 2014, 40, 66–73. [Google Scholar]
- Li, H.; Yan, C.; Tang, Y.; Ma, X.; Chen, Y.; Chen, S.; Lin, M.; Liu, Z. Endophytic bacterial and fungal microbiota in different cultivars of cassava (Manihot esculenta Crantz). J. Microbiol. 2020, 58, 614–623. [Google Scholar] [CrossRef]
- Freitas, M.A.; Medeiros, F.H.V.; Carvalho, S.P.; Guilherme, L.R.G.; Teixeira, W.D.; Zhang, H.; Paré, P.W. Augmenting iron accumulation in cassava by the beneficial soil bacterium Bacillus subtilis (GBO3). Front. Plant Sci. 2015, 6, 596. [Google Scholar] [CrossRef]
- Zeng, H.; Xu, H.; Liu, G.; Wei, Y.; Zhang, J.; Shi, H. Physiological and metagenomic strategies uncover the rhizosphere bacterial microbiome succession underlying three common environmental stresses in cassava. J. Hazard. Mater. 2021, 411. [Google Scholar] [CrossRef]
- Barnard, R.L.; Osborne, C.A.; Firestone, M.K. Responses of soil bacterial and fungal communities to extreme desiccation and rewetting. ISME J. 2013, 7, 2229–2241. [Google Scholar] [CrossRef] [PubMed]
- Bouskill, N.J.; Lim, H.C.; Borglin, S.; Salve, R.; Wood, T.E.; Silver, W.L.; Brodie, E.L. Pre-exposure to drought increases the resistance of tropical forest soil bacterial communities to extended drought. ISME J. 2013, 7, 384–394. [Google Scholar] [CrossRef]
- Naylor, D.; Coleman-Derr, D. Drought stress and root-associated bacterial communities. Front. Plant Sci. 2018, 8, 2223. [Google Scholar] [CrossRef]
- Becerra López-Lavalle, L.A.; Ovalle, T.M.; Marín, D.; Gutiérrez, J.P.; Moreno, M.P.; Moreno, M.; Calle, F.; Luna, J.; Belalcázar, J.; Labarta, R.; et al. Catálogo de Variedades de Yuca, Cauca-Colombia; Centro Internacional de Agricultura Tropical (CIAT): Cali, Colombia, 2020; p. 82. [Google Scholar]
- Cherif, H.; Marasco, R.; Rolli, E.; Ferjani, R.; Fusi, M.; Soussi, A.; Mapelli, F.; Blilou, I.; Borin, S.; Boudabous, A.; et al. Oasis desert farming selects environment-specific date palm root endophytic communities and cultivable bacteria that promote resistance to drought. Environ. Microbiol. Rep. 2015, 7, 668–678. [Google Scholar] [CrossRef]
- Naylor, D.; Degraaf, S.; Purdom, E.; Coleman-Derr, D. Drought and host selection influence bacterial community dynamics in the grass root microbiome. ISME J. 2017, 11, 2691–2704. [Google Scholar] [CrossRef] [PubMed]
- Santos-Medellín, C.; Edwards, J.; Liechty, Z.; Nguyen, B.; Sundaresan, V. Drought stress results in a compartment-specific restructuring of the rice root-associated microbiomes. MBio 2017, 8, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Adesina, M.F.; Lembke, A.; Costa, R.; Speksnijder, A.; Smalla, K. Screening of bacterial isolates from various European soils for in vitro antagonistic activity towards Rhizoctonia solani and Fusarium oxysporum: Site-dependent composition and diversity revealed. Soil Biol. Biochem. 2007, 39, 2818–2828. [Google Scholar] [CrossRef]
- Sindhu, S.S.; Gupta, S.K.; Dadarwal, K.R. Antagonistic effect of Pseudomonas spp. on pathogenic fungi and enhancement of growth of green gram (Vigna radiata). Biol. Fertil. Soils 1999, 29, 62–68. [Google Scholar] [CrossRef]
- Mercado-Blanco, J.; Rodríguez-Jurado, D.; Hervás, A.; Jiménez-Díaz, R.M. Suppression of Verticillium wilt in olive planting stocks by root-associated fluorescent Pseudomonas spp. Biol. Control 2004, 30, 474–486. [Google Scholar] [CrossRef]
- Garrity, G.; Brenner, D.J.; Krieg, N.R.; Staley, J.R. (Eds.) Bergey’s Manual of Systematic Bacteriology; Springer: New York, NY, USA, 2012; ISBN 978-0-387-28022-6. [Google Scholar]
- Madigan, M.T.; Bender, K.S.; Buckley, D.H.; Sattley, W.M.; Stahl, D.A. Brock Biology of Microorganisms; Pearson: London, UK, 2018; ISBN 9780134261928. [Google Scholar]
- Shannon, C.E. A Mathematical Theory of Communication. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef]
- Pielou, E.C. The measurement of diversity in different types of biological collections. J. Theor. Biol. 1966, 13, 131–144. [Google Scholar] [CrossRef]
- Hutcheson, K. A test for comparing diversities based on the shannon formula. J. Theor. Biol. 1970, 29, 151–154. [Google Scholar] [CrossRef]
- Niu, X.; Song, L.; Xiao, Y.; Ge, W.; Job, D. Drought-tolerant plant growth-promoting rhizobacteria associated with foxtail millet in a semi-arid agroecosystem and their potential in alleviating drought stress. Front. Microbiol. 2018, 8, 2580. [Google Scholar] [CrossRef]
- Mulissa, J.M.; Carolin, R.L.; Ruth, A.S.; Fassil, A. Characterization of phosphate solubilizing rhizobacteria isolated from lentil growing areas of Ethiopia. Afr. J. Microbiol. Res. 2015, 9, 1637–1648. [Google Scholar] [CrossRef]
- Teshome, B.; Wassie, M.; Abatneh, E. Isolation, screening and biochemical characterization of phosphate solubilizing rhizobacteria associated with Coffea arabica L. J. Fertil. Pestic. 2017, 8. [Google Scholar] [CrossRef]
- Alibrandi, P.; Schnell, S.; Perotto, S.; Cardinale, M. Diversity and structure of the endophytic bacterial communities associated with three terrestrial orchid species as revealed by 16S rRNA gene metabarcoding. Front. Microbiol. 2020, 11, 3207. [Google Scholar] [CrossRef] [PubMed]
- Hallmann, J.; Berg, G.; Kloepper, J.W.; Ryu, C.-M.; Saad, M.M.; Broughton, W.J.; Deakin, W.J.; Anand, R.; Paul, L.; Chanway, C. Endophytic Bacteria. In Microbial Root Endophytes; Schulz, B.J.E., Boyle, C.J.C., Sieber, T.N., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; pp. 15–106. ISBN 978-3-540-33526-9. [Google Scholar]
- Bresson, J.; Varoquaux, F.; Bontpart, T.; Touraine, B.; Vile, D. The PGPR strain Phyllobacterium brassicacearum STM196 induces a reproductive delay and physiological changes that result in improved drought tolerance in Arabidopsis. New Phytol. 2013, 200, 558–569. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Hilson, P.; Sedbrook, J.; Rosen, E.; Caspar, T.; Masson, P.H. The Arabidopsis thaliana AGRAVITROPIC 1 gene encodes a component of the polar-auxin-transport efflux carrier. Proc. Natl. Acad. Sci. USA 1998, 95, 15112–15117. [Google Scholar] [CrossRef]
- Kohler, J.; Hernández, J.A.; Caravaca, F.; Roldán, A. Plant-growth-promoting rhizobacteria and arbuscular mycorrhizal fungi modify alleviation biochemical mechanisms in water-stressed plants. Funct. Plant Biol. 2008, 35, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Sandhya, V.; Ali, S.Z.; Grover, M.; Reddy, G.; Venkateswarlu, B. Effect of plant growth promoting Pseudomonas spp. on compatible solutes, antioxidant status and plant growth of maize under drought stress. Plant Growth Regul. 2010, 62, 21–30. [Google Scholar] [CrossRef]
- Kurek, E.; Ozimek, E.; Sobiczewski, P.; Słomka, A.; Jaroszuk-Ściseł, J. Effect of Pseudomonas luteola on mobilization of phosphorus and growth of young apple trees (Ligol)-Pot experiment. Sci. Hortic. 2013, 164, 270–276. [Google Scholar] [CrossRef]
- Glick, B.R. Plant growth-promoting bacteria: Mechanisms and applications. Scientifica 2012, 2012. [Google Scholar] [CrossRef]
- Rascovan, N.; Carbonetto, B.; Perrig, D.; Díaz, M.; Canciani, W.; Abalo, M.; Alloati, J.; González-Anta, G.; Vazquez, M.P. Integrated analysis of root microbiomes of soybean and wheat from agricultural fields. Sci. Rep. 2016, 6, 28084. [Google Scholar] [CrossRef]
- Suárez, R.; Wong, A.; Ramírez, M.; Barraza, A.; Orozco, M.D.C.; Cevallos, M.A.; Lara, M.; Hernández, G.; Iturriaga, G. Improvement of drought tolerance and grain yield in common bean by overexpressing trehalose-6-phosphate synthase in rhizobia. Mol. Plant. Microbe. Interact. 2008, 21, 958–966. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Zhang, X.; Cao, Z.; Zhao, K.; Wang, S.; Chen, M.; Hu, X. Growth-promoting sphingomonas paucimobilis ZJSH1 associated with Dendrobium officinale through phytohormone production and nitrogen fixation. Microb. Biotechnol. 2014, 7, 611–620. [Google Scholar] [CrossRef]
- Takeuchi, M.; Sakane, T.; Yanagi, M.; Yamasato, K.; Hamana, K.; Yokota, A. Taxonomic study of bacteria isolated from plants: Proposal of Sphingomonas rosa sp. nov., Sphingomonas pruni sp. nov., Sphingomonas asaccharolytica sp. nov., and Sphingomonas mali sp. nov. Int. J. Syst. Bacteriol. 1995, 45, 334–341. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Luo, Y.; Wang, F.; Huang, Y.; Zhou, M.; Gao, J.; Yan, T.; Sheng, H.; An, L. Sphingomonas sp. Cra20 increases plant growth rate and alters rhizosphere microbial community structure of Arabidopsis thaliana under drought stress. Front. Microbiol. 2019, 10, 1221. [Google Scholar] [CrossRef] [PubMed]
- Rolli, E.; Marasco, R.; Vigani, G.; Ettoumi, B.; Mapelli, F.; Deangelis, M.L.; Gandolfi, C.; Casati, E.; Previtali, F.; Gerbino, R.; et al. Improved plant resistance to drought is promoted by the root-associated microbiome as a water stress-dependent trait. Environ. Microbiol. 2015, 17, 316–331. [Google Scholar] [CrossRef]
- Wang, C.J.; Yang, W.; Wang, C.; Gu, C.; Niu, D.D.; Liu, H.X.; Wang, Y.P.; Guo, J.H. Induction of drought tolerance in cucumber plants by a consortium of three plant growth-promoting rhizobacterium strains. PLoS ONE 2012, 7, e52565. [Google Scholar] [CrossRef] [PubMed]
- Ojuederie, O.B.; Olanrewaju, O.S.; Babalola, O.O. Plant growth promoting rhizobacterial mitigation of drought stress in crop plants: Implications for sustainable agriculture. Agronomy 2019, 9, 712. [Google Scholar] [CrossRef]
- Mishra, S.K.; Khan, M.H.; Misra, S.; Dixit, V.K.; Khare, P.; Srivastava, S.; Chauhan, P.S. Characterisation of Pseudomonas spp. and Ochrobactrum sp. isolated from volcanic soil. Antonie van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2017, 110, 253–270. [Google Scholar] [CrossRef]
| Plant ID | Strain ID | Gram Reaction | Colony Characteristics at Several Culture Media | Biochemical Analysis | |||||
|---|---|---|---|---|---|---|---|---|---|
| NA | AMA | KAB | EMB | Catalase Test | Oxidase Test | Identification (VITEK® 2) | |||
| cp-c-1 | cp-c-1.1 | –ve |  |  |  |  | + | + | Pseudomonas putida |
| cp-c-1.2 | –ve |  |  |  |  | + | + | Pseudomonas fluorescens | |
| cp-c-1.3 | –ve |  |  |  |  | + | + | Pseudomonas fluorescens | |
| cp-c-2 | cp-c-2.1 | –ve |  |  |  |  | + | + | Pseudomonas fluorescens |
| cp-c-2.2 | –ve |  |  |  |  | + | + | Pseudomonas fluorescens | |
| cp-c-2.3 | –ve |  |  |  |  | + | + | Pseudomonas mendocina | |
| cp-ds-1 | cp-ds-1.1 | –ve |  |  |  |  | + | + | Pseudomonas putida |
| cp-ds-1.2 | –ve |  |  |  |  | + | + | Pseudomonas putida | |
| cp-ds-1.3 | –ve |  |  |  |  | + | + | Pseudomonas fluorescens | |
| cp-ds-2 | cp-ds-2.1 | –ve |  |  |  |  | + | + | Pseudomonas putida |
| cp-ds-2.2 | –ve |  |  |  |  | + | + | Pseudomonas putida | |
| cp-ds-2.3 | –ve |  |  |  |  | + | + | Pseudomonas putida | |
| cp-ds-3 | cp-ds-3.1 | –ve |  |  |  |  | + | + | Achromobacter xylosoxidans |
| cp-ds-3.2 | –ve |  |  |  |  | + | + | Sphingomonas paucimobilis | |
| cp-ds-4 | cp-ds-4.1 | –ve |  |  |  |  | + | + | Sphingomonas paucimobilis |
| cp-ds-4.2 | –ve |  |  |  |  | + | + | Sphingomonas paucimobilis | |
| cp-ds-5 | cp-ds-5.1 | –ve |  |  |  |  | + | + | Sphingomonas paucimobilis |
| cp-ds-5.2 | –ve |  |  |  |  | + | + | Rhizobium radiobacter | |
| Plant ID | Strain ID | Gram Reaction | Colony Characteristics at Several Culture Media | Biochemical Analysis | |||||
|---|---|---|---|---|---|---|---|---|---|
| NA | AMA | KAB | EMB | Catalase Test | Oxidase Test | Identification (VITEK® 2) | |||
| cp-c-1 | cp-c-1.4 | –ve |  |  |  |  | + | + | Rhizobium radiobacter |
| cp-c-1.5 | –ve |  |  |  |  | + | – | Sphingomonas paucimobilis | |
| cp-c-1.6 | –ve |  |  |  |  | + | – | Sphingomonas paucimobilis | |
| cp-c-1.7 | –ve |  |  |  |  | – | + | Cronobacter sakazakii | |
| cp-c-1.8 | –ve |  |  |  |  | + | + | Rhizobium radiobacter | |
| cp-c-1.9 | –ve |  |  |  |  | + | + | Pluralibacter gergoviae | |
| cp-c-1.10 | –ve |  |  |  |  | - | + | Serratia marcescens | |
| cp-c-1.11 | –ve |  |  |  |  | + | + | Pseudomonas putida | |
| cp-c-1.12 | –ve |  |  |  |  | – | + | Klebsiella pneumoniae | |
| cp-c-1.13 | –ve |  |  |  |  | + | + | Pseudomonas stutzeri | |
| cp-c-2 | cp-c-2.4 | –ve |  |  |  | no growth | + | + | Pseudomonas stutzeri |
| cp-c-2.5 | –ve |  |  |  |  | + | + | Acinetobacter baumannii | |
| cp-c-2.6 | –ve |  |  |  |  | + | + | Rhizobium radiobacter | |
| cp-c-2.7 | –ve |  |  |  |  | - | + | Achromobacter xylosoxidans | |
| cp-c-2.8 | –ve |  |  |  |  | - | + | Sphingomonas paucimobilis | |
| cp-c-2.9 | –ve |  |  |  |  | + | + | Rhizobium radiobacter | |
| cp-c-2.10 | –ve |  |  |  |  | + | + | Sphingomonas paucimobilis | |
| cp-c-2.11 | –ve |  |  |  | no growth | - | + | Buttiauxella agrestis | |
| cp-c-2.12 | –ve |  |  |  |  | + | + | Aeromonas salmonicida | |
| cp-c-2.13 | –ve |  |  |  |  | + | + | Pseudomonas fluorescens | |
| cp-c-2.14 | –ve |  |  |  |  | + | + | Pseudomonas fluorescens | |
| cp-ds-1 | cp-ds-1.4 | –ve |  |  |  |  | + | + | Pseudomonas putida |
| cp-ds-1.5 | –ve |  |  |  |  | + | + | Pseudomonas putida | |
| cp-ds-2 | cp-ds-2.4 | –ve |  |  |  |  | + | + | Pseudomonas putida |
| cp-ds-2.5 | –ve |  |  |  |  | + | + | Pseudomonas mendocina | |
| cp-ds-2.6 | –ve |  |  |  |  | + | + | Ochrobactrum anthropi | |
| cp-ds-2.7 | –ve |  |  |  |  | + | + | Sphingomonas paucimobilis | |
| cp-ds-3 | cp-ds-3.3 | –ve |  |  |  |  | + | + | Rhizobium radiobacter |
| cp-ds-3.4 | –ve |  |  |  |  | + | + | Sphingomonas paucimobilis | |
| cp-ds-3.5 | –ve |  |  |  |  | - | + | Pseudomonas stutzeri | |
| cp-ds-4 | cp-ds-4.3 | –ve |  |  |  |  | + | + | Pseudomonas putida |
| cp-ds-4.4 | –ve |  |  |  |  | - | + | Rhizobium radiobacter | |
| cp-ds-4.5 | –ve |  |  |  |  | + | + | Pseudomonas putida | |
| cp-ds-4.6 | –ve |  |  |  |  | + | + | Rhizobium radiobacter | |
| cp-ds-4.7 | –ve |  |  |  |  | + | + | Sphingomonas paucimobilis | |
| cp-ds-5 | cp-ds-5.3 | –ve |  |  |  |  | + | + | Pseudomonas putida |
| cp-ds-5.4 | –ve |  |  |  |  | + | + | Pseudomonas luteola | |
| cp-ds-5.5 | –ve |  |  |  |  | - | + | Rhizobium radiobacter | |
| cp-ds-5.6 | –ve |  |  |  |  | + | + | Sphingomonas paucimobilis | |
| cp-ds-5.7 | –ve |  |  |  |  | + | + | Rhizobium radiobacter | |
| Cassava Plants Treatments | Abundance | Species Richness | Shannon Diversity Index (H′) | p-Value 1 | Evenness | p-Value 2 |
|---|---|---|---|---|---|---|
| cp-c_T0 | 6 | 3 | 0.868 | 0.0019 | 0.789 | 0.7070 |
| cp-c_T20 | 21 | 13 | 2.384 | 0.929 | ||
| cp-ds_T0 | 12 | 5 | 1.350 | 0.2899 | 0.840 | 0.9678 |
| cp-ds_T20 | 19 | 7 | 1.663 | 0.855 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zapata, T.; Galindo, D.M.; Corrales-Ducuara, A.R.; Ocampo-Ibáñez, I.D. The Diversity of Culture-Dependent Gram-Negative Rhizobacteria Associated with Manihot esculenta Crantz Plants Subjected to Water-Deficit Stress. Diversity 2021, 13, 366. https://doi.org/10.3390/d13080366
Zapata T, Galindo DM, Corrales-Ducuara AR, Ocampo-Ibáñez ID. The Diversity of Culture-Dependent Gram-Negative Rhizobacteria Associated with Manihot esculenta Crantz Plants Subjected to Water-Deficit Stress. Diversity. 2021; 13(8):366. https://doi.org/10.3390/d13080366
Chicago/Turabian StyleZapata, Tatiana, Diana Marcela Galindo, Alba Rocío Corrales-Ducuara, and Iván Darío Ocampo-Ibáñez. 2021. "The Diversity of Culture-Dependent Gram-Negative Rhizobacteria Associated with Manihot esculenta Crantz Plants Subjected to Water-Deficit Stress" Diversity 13, no. 8: 366. https://doi.org/10.3390/d13080366
APA StyleZapata, T., Galindo, D. M., Corrales-Ducuara, A. R., & Ocampo-Ibáñez, I. D. (2021). The Diversity of Culture-Dependent Gram-Negative Rhizobacteria Associated with Manihot esculenta Crantz Plants Subjected to Water-Deficit Stress. Diversity, 13(8), 366. https://doi.org/10.3390/d13080366






