Genetic Diversity of Purple Passion Fruit, Passiflora edulis f. edulis, Based on Single-Nucleotide Polymorphism Markers Discovered through Genotyping by Sequencing
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. DNA Extraction and Quality Check
2.3. Genotyping by Sequencing
2.4. Demultiplexing and SNP Calling
2.5. Data Analysis
3. Results
3.1. Success of the GBS Technique in Purple Passion Fruit
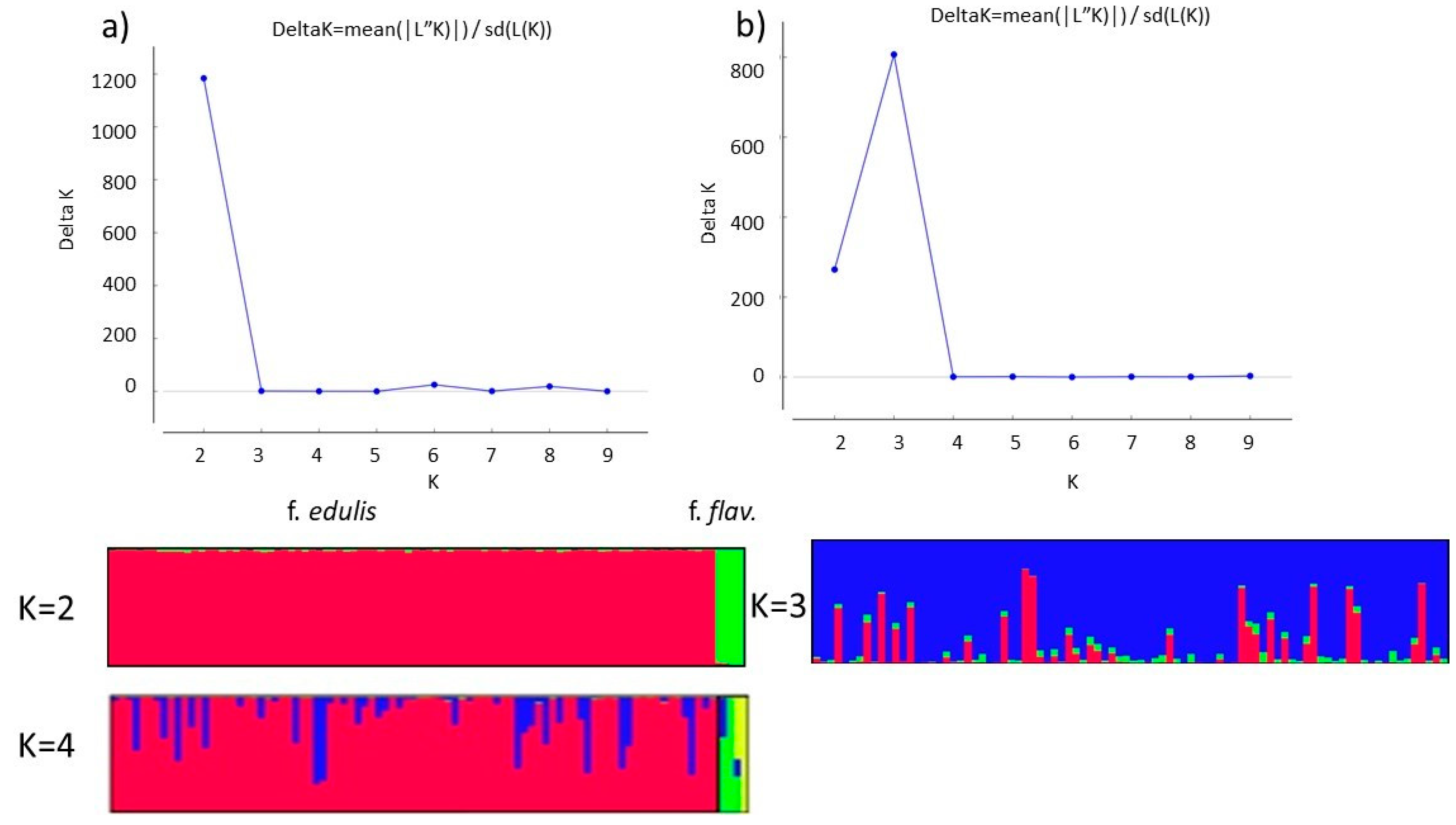
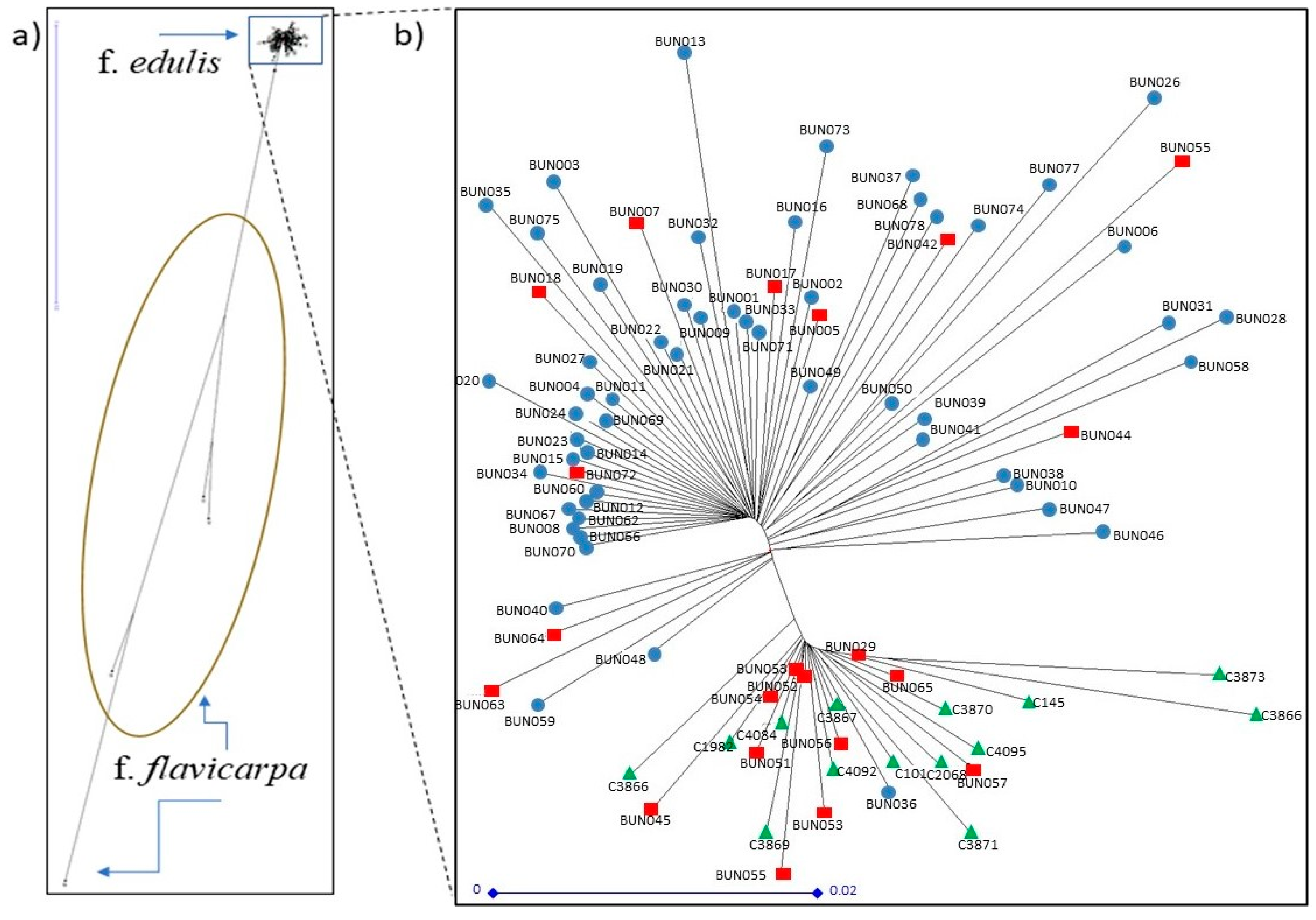
3.2. Population Structure of Purple and Yellow Passion Fruit
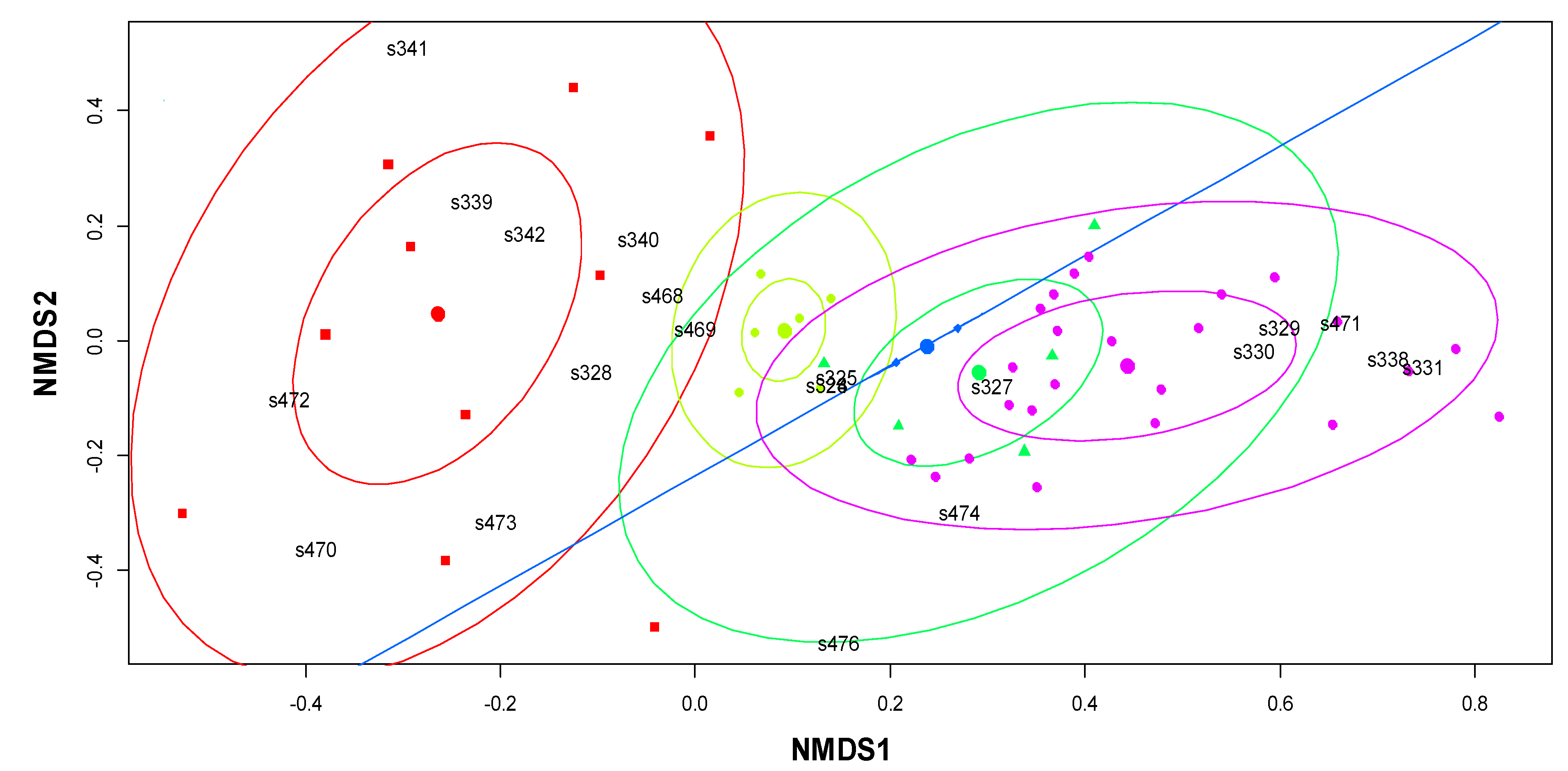
3.3. Multidimensional Scaling Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Paul, R.E.; Duarte, O. Tropical Fruits; CABI: Wallingford, UK, 2012; Volume 2, pp. 161–190. [Google Scholar]
- Patiño, V. Historia y Dispersión de los Frutales Nativos del Neotrópico. V. 326; Centro Internacional de Agricultura Tropical (CIAT): Cali, Colombia, 2002; 655p. [Google Scholar]
- Vanderplank, J. Passion Flowers and Passion Fruit; Cassell: London, UK, 1991; ISBN 0-304-34076-6. [Google Scholar]
- Krosnick, S.; Perkin, J.; Schroeder, T.; Campbell, L.; Jackson, E.; Waters, C.; Mitchell, J. New insights into floral morph variation in Passiflora incarnata L. (Passifloraceae) in Tennessee, USA. Flora 2017, 236–237, 115–125. [Google Scholar] [CrossRef]
- Cerqueira-Silva, C.; Onildo, J.; Santos, E.; Correa, R.; Souza, A. Genetic breeding and perspectives in molecular and genetics studies. Int. J. Mol. Sci. 2014, 15, 14122–14152. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, N.C.; Melgarejo, L.M.; Blair, M.W. Purple passion fruit, Passiflora edulis Sims f. edulis, variability for photosynthetic and physiological adaptation in contrasting environments. Agronomy 2019, 9, 231. [Google Scholar] [CrossRef]
- Rodríguez, N.C.; Ambachew, D.; Melgarejo, L.M.; Blair, M.W. Morphological and Agronomic Variability among Cultivars, Landraces, and Genebank Accessions of Purple Passion Fruit, Passiflora edulis f. edulis. HortScience 2020, 55, 768–777. [Google Scholar]
- Ocampo, J.; D’Eeckenbrugge, G.; Jarvis, A. Distribution of genus Passiflora L. in Colombia and its potential as an indicator for biodiversity management in the coffee growing zone. Diversity 2010, 2, 1158–1180. [Google Scholar] [CrossRef]
- Fischer, G.; Melgarejo, L.M.; Cutler, J. Pre-harvest factors that influence the quality of passion fruit: A review. Agron. Colomb. 2018, 36, 217–226. [Google Scholar] [CrossRef]
- Ortiz, D.C.; Bohórquez, A.; Duque, M.C.; Tohme, J.; Cuéllar, D.; Vásquez, D.M. Evaluating purple passion fruit (Passiflora edulis Sims f. edulis) genetic variability in individuals from commercial plantations in Colombia. Genet. Res. Crop Evol. 2012, 59, 1089–1099. [Google Scholar] [CrossRef]
- Araya, A.; Martins, A.M.; Junqueira, N.T.V.; Costa, A.M.; Faleiro, F.G.; Ferreira, M.E. Microsatellite marker development by partial sequencing of the sour passion fruit genome (Passiflora edulis Sims). Genomics 2017, 18, 549. [Google Scholar] [CrossRef] [PubMed]
- Santos, L.F.; Oliveira, E.J.; Santos Silva, A.; Carvalho, F.M.; Costa, J.L.; Pádua, J.G. ISSR markers as a tool for the assessment of genetic diversity in Passiflora. Biochem. Genet. 2011, 49, 540–554. [Google Scholar] [CrossRef]
- Moreno, C.; Gelape, F.; Nunes, O.; Lisboa, E.; Pereira, A.; de Souza, P. The Genetic Diversity, Conservation, and Use of Passion Fruit (Passiflora spp.). In Genetic Diversity and Erosion in Plants; Springer: Cham, Switzerland, 2015; pp. 215–231. [Google Scholar]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Harriman, J.V.; Buckler, E.S.; Mitchell, S.E. A Robust, Simple Genotyping-by-Sequencing (GBS) Approach for High Diversity Species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef]
- Liu, S.; Li, A.D.; Chen, C.; Cai, G.; Zhang, L.; Guo, C.; Xu, M. De novo transcriptome sequencing in Passiflora edulis Sims to identify genes and signaling pathways involved in cold tolerance. Forests 2017, 8, 435. [Google Scholar] [CrossRef]
- Goncalves, G.; Viana, A.; Gonzaga, M.; Valdevino, F.; Teixeira, A.; Santana, T.; Menezes, T. Genetic parameter estimates in yellow passion fruit based on design. J. Braz. Arch. Biol. Technol. 2014, 52, 523–530. [Google Scholar] [CrossRef]
- Rodríguez, N.C.; Melgarejo, L.M.; Blair, M.W. Seed Structural Variability and Germination Capacity in Passiflora edulis Sims f. edulis. Front. Plant Sci. 2020, 11, 498. [Google Scholar]
- Vega, N.E.; Chacon, M.I. Isolation of high-quality DNA in 16 aromatic and medicinal Colombian species using silica-based extraction columns. Agron. Colomb. 2011, 29, 349–357. [Google Scholar]
- Catchen, J.; Hohenlohe, P.; Bassham, S.; Amores, S.; Cresko, W. Stacks: An analysis tool set for population genomics. Mol. Ecol. 2013, 22, 3124–3140. [Google Scholar] [CrossRef] [PubMed]
- Paris, J.; Stevens, J.; Catchen, J. Lost in parameter space: A road map for STACKS. Methods Ecol. Evol. 2017, 8, 1360–1373. [Google Scholar] [CrossRef]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar] [PubMed]
- Liu, K.; Muse, S. PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.; Stephens, M.; Donelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 2012, 155, 945–959. [Google Scholar]
- Rosenberg, N.A. DISTRUCT: A program for the graphical display of population structure. Mol. Ecol. Notes 2004, 4, 137–138. [Google Scholar] [CrossRef]
- Perrier, X.; Flori, A.; Bonnot, F. Data analysis methods. In Genetic Diversity of Cultivated Tropical Plants; Hamon, P., Seguin, M., Perrier, X., Glaszmann, J.C., Eds.; Scientific editors: CIRAD SPI, 2003; pp. 43–76. [Google Scholar]
- Guisande, C. Rwizard Software. University of Vigo. Spain. 2014. Available online: http://www.ipez.es/Rwizard (accessed on 27 March 2021).
- Gower, J.C. A general coefficient of similarity and some of its properties. Biometrics 1971, 27, 857–871. [Google Scholar] [CrossRef]
- Wright, S. Evolution and the Genetics of Populations. Variability Within and Among Natural Populations; University of Chicago Press: Chicago, IL, USA, 1978. [Google Scholar]
- Gürcan, K.; Teber, S.; Ercisli, S.; Ugurtan, K. Genotyping by Sequencing (GBS) in Apricots and Genetic Diversity Assessment with GBS-Derived Single-Nucleotide Polymorphisms (SNPs). Biochem. Genet. 2017, 54, 854–885. [Google Scholar] [CrossRef]
- He, J.F.; Zhao, X.; Laroche, A.; Lu, Z.X.; Liu, H.K.; Li, Z. Genotyping-by-sequencing (GBS), an ultimate marker-assisted selection (MAS) tool to accelerate plant breeding. Front. Plant Sci. 2014, 5, 484. [Google Scholar] [CrossRef] [PubMed]
- Pádua, J.G.; Oliveira, E.J.; Zucchi, M.I.; Oliveira, G.C.X.; Camargo, L.E.A.; Vieira, M.L.C. Isolation and characterization of microsatellite markers from the sweet passion fruit (Passiflora alata Curtis: Passifloraceae). Mol. Ecol. Notes 2005, 5, 863–865. [Google Scholar] [CrossRef]
- Paiva, C.; Viana, A.; Azevedo Santos, E.; de Oliveira Freitas, J.C.; Oliveira Silva, R.N.; de Oliveira, E.J. Genetic variability assessment in the genus Passiflora by SSR markers. Chilean J. Agric. Res. 2014, 74, 355–360. [Google Scholar] [CrossRef]
- Oliveira, E.J.; Pádua, J.G.; Zucchi, M.I.; Camargo, L.E.A.; Fungaro, M.H.P.; Vieira, M.L.C. Development and characterization of microsatellite markers from the yellow passion fruit (Passiflora edulis f. flavicarpa). Mol. Ecol. Notes 2006, 5, 331–333. [Google Scholar] [CrossRef]
- Junqueira, N.T.V.; Braga, M.F.; Faleiro, F.G.; Peixoto, J.R.; Bernacci, L.C. Potencial de especies silvestres de maracujazeiro como fonte de resistência a doenças. In Maracujá: Germoplasma e Melhoramento Genético; Faleiro, F.G., Junqueira, N.T.V., Braga, M.F., Eds.; EMBRAPA Cerrados: Livro cientifico (ALICE): Planaltina, Brazil, 2005; pp. 80–108. [Google Scholar]
- Ocampo, J.; Coppens dÉeckenbrugge, G.; Restrepo, M.; Jarvis, A.; Salazar, M.; Caetano, C. Diversity of Colombian passifloraceae biogeography and a update list for conservation. Biota Colombiana 2007, 8, 1–45. [Google Scholar]
- Cerqueira-Silva, C.B.M.; Santos, E.S.L.; Souza, A.M.; Mori, G.M.; Oliveira, E.J.; Corrêa, R.X.; Souza, A.P. Development and characterization of microsatellite markers for the wild South American Passiflora cincinnata (Passifloraceae). Am. J. Bot. 2012, 99, 170–172. [Google Scholar] [CrossRef]
- Mäder, G.; Zamberlan, P.M.; Fagundes, N.J.R.; Magnus, T.; Salzano, F.M.; Bonatto, S.L.; Freitas, L.B. The use and limits of ITS data in the analysis of intraspecific variation in Passiflora L. (Passifloraceae). Genet. Mol. Biol. 2010, 33, 99–108. [Google Scholar] [CrossRef]
- Souza, M.; Pereira, T.; Viana, A.; Pereira, M.; Amaral, A.; Madureira, H. Flower receptivity and fruit characteristics associated to time of pollination in the yellow passion fruit Passiflora edulis Sims f. flavicarpa Degener (Passifloraceae). Sci. Hort. 2004, 101, 373–385. [Google Scholar] [CrossRef]
- Cerqueira-Silva, C.B.M.; Santos, E.S.L.; Vieira, J.G.P.; Mori, G.M.; Jesus, O.N.; Correa, R.X.; Souza, A.P. New microsatellite markers for wild and commercial species of Passiflora (Passifloraceae) and cross-amplification. Appl. Plant Sci. 2014, 2, 1–5. [Google Scholar] [CrossRef]
- Trujillo, N.; Márquez, M.P.; Moreno, J.H.; Terán, W.; Schuler, I. Caracterización molecular de materiales cultivados de gulupa (Passiflora edulis f. edulis). Universitas Scientarum 2010, 14, 3. [Google Scholar]
- Hansen, A.K.; Escobar, L.K.; Gilbert, L.E.; Jansen, R.K. Paternal, maternal, and biparental inheritance of the chloroplast genome in Passiflora (Passifloraceae): Implications for phylogenetic studies. Am. J. Bot. 2007, 94, 42–46. [Google Scholar] [CrossRef] [PubMed]
- Souza, M.; Palomino, G.; Pereira, T.N.S.; Pereira, M.G.; Viana, A.P. Flow cytometric analysis of genome size variation in some Passiflora species. Hereditas 2004, 141, 31–38. [Google Scholar] [CrossRef]
- Angel, C.; Nates, G.; Ospina, R.; Melo, C.; Amaya, M. Biología floral y reproductiva de la gulupa, Passiflora edulis Sims f. Edulis. Caldasia 2011, 33, 433–451. [Google Scholar]
- Medina, J.; Ospina, R.; Nates, G. Efectos de la variación altitudinal sobre la polinización en cultivos de gulupa (Passiflora edulis f. Edulis). Acta Biol. Colomb. 2012, 7, 379–394. [Google Scholar]
- Kishore, K.; Pathak, K.A.; Shukla, R.; Bharali, R. Studies on floral biology of passion fruit (Passiflora spp.). Pak. J. Bot. 2010, 42, 21–29. [Google Scholar]
- Rendon, J.S.; Ocampo, J.; Urrea, R. Estudio sobre polinización y biología floral en Passiflora edulis f. edulis Sims, como base para el premejoramiento genético. Acta Agron. 2013, 62, 232–241. [Google Scholar]
| SNP Type | Number of SNPs | Frequency (%) * |
|---|---|---|
| Transition (subtotal) | 2568 | 67.23 |
| A/G | 1210 | 31.68 |
| C/T | 1358 | 35.55 |
| Transversions (subtotal) | 1252 | 32.77 |
| A/C | 294 | 7.70 |
| A/T | 368 | 9.63 |
| C/G | 278 | 7.28 |
| T/G | 312 | 8.17 |
| Grand total SNP verified | 3820 | 100.00 |
| Number of independent loci | 1706 | 44.65 |
| Number of polymorphic loci in P. edulis | 966 | 25.28 |
| Type of Accession | Exp. Het. | Obs. Het. | PIC |
|---|---|---|---|
| Landrace | 0.78 ± 0.54 a | 0.45 ± 0.53 a | 0.75 ± 0.12 a |
| Commercial cultivar | 0.41 ± 0.56 b | 0.18 ± 0.48 b | 0.38 ± 0.95 b |
| Genebank entry | 0.43 ± 0.47 b | 0.22 ± 0.87 b | 0.41 ± 0.87 b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodriguez Castillo, N.C.; Wu, X.; Chacón, M.I.; Melgarejo, L.M.; Blair, M.W. Genetic Diversity of Purple Passion Fruit, Passiflora edulis f. edulis, Based on Single-Nucleotide Polymorphism Markers Discovered through Genotyping by Sequencing. Diversity 2021, 13, 144. https://doi.org/10.3390/d13040144
Rodriguez Castillo NC, Wu X, Chacón MI, Melgarejo LM, Blair MW. Genetic Diversity of Purple Passion Fruit, Passiflora edulis f. edulis, Based on Single-Nucleotide Polymorphism Markers Discovered through Genotyping by Sequencing. Diversity. 2021; 13(4):144. https://doi.org/10.3390/d13040144
Chicago/Turabian StyleRodriguez Castillo, Nohra Cecilia, Xingbo Wu, María Isabel Chacón, Luz Marina Melgarejo, and Matthew Wohlgemuth Blair. 2021. "Genetic Diversity of Purple Passion Fruit, Passiflora edulis f. edulis, Based on Single-Nucleotide Polymorphism Markers Discovered through Genotyping by Sequencing" Diversity 13, no. 4: 144. https://doi.org/10.3390/d13040144
APA StyleRodriguez Castillo, N. C., Wu, X., Chacón, M. I., Melgarejo, L. M., & Blair, M. W. (2021). Genetic Diversity of Purple Passion Fruit, Passiflora edulis f. edulis, Based on Single-Nucleotide Polymorphism Markers Discovered through Genotyping by Sequencing. Diversity, 13(4), 144. https://doi.org/10.3390/d13040144






