Zooshikella harenae sp. nov., Isolated from Pacific Oyster Crassostrea gigas, and Establishment of Zooshikella ganghwensis subsp. marina subsp. nov. and Zooshikella ganghwensis subsp. ganghwensis subsp. nov.
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation
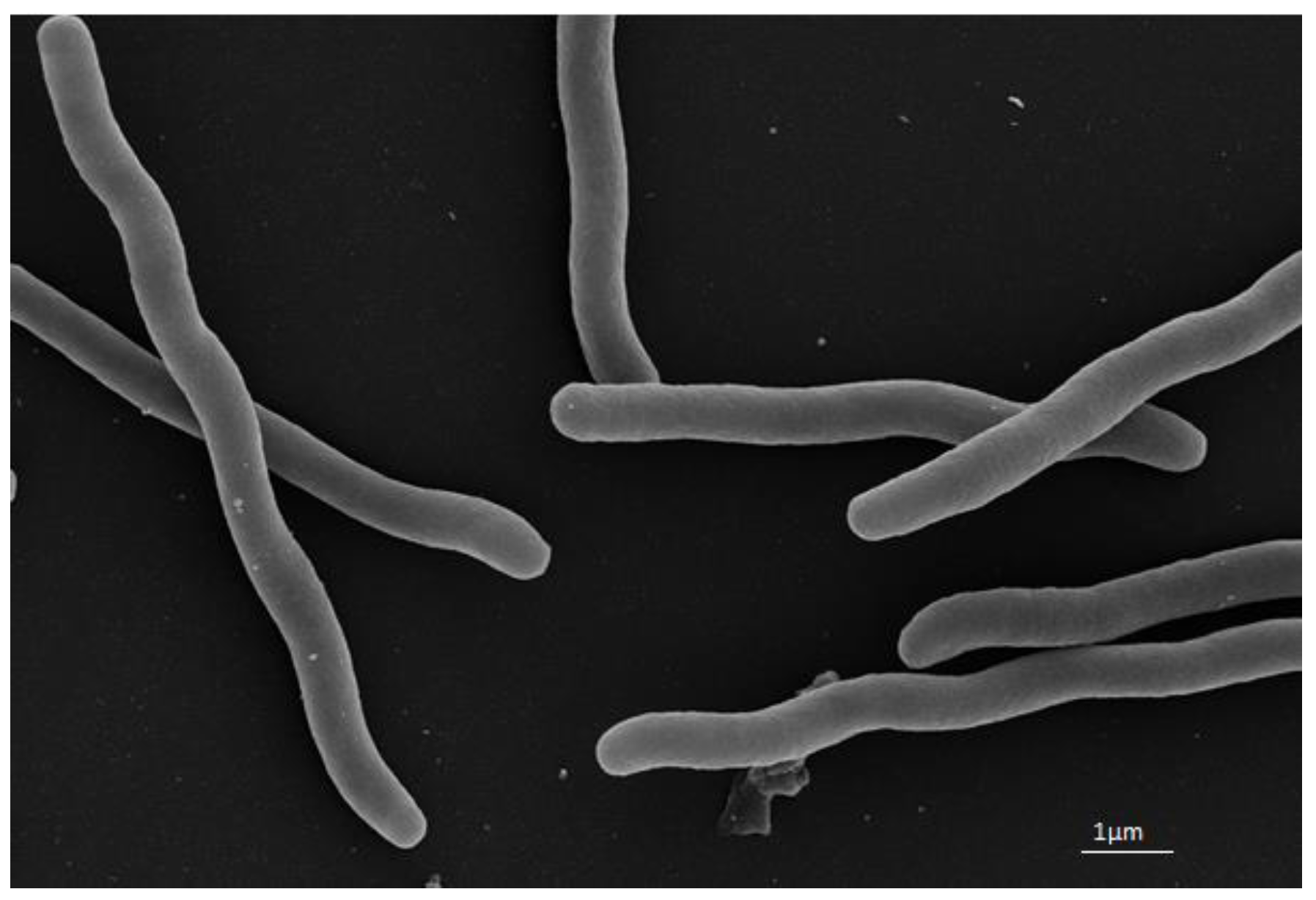
2.2. Morphological, Physiological, and Biochemical Studies
2.3. 16S rRNA Gene Analysis
2.4. Chemotaxonomy
2.5. Whole-Genome Analysis
2.6. DNA–DNA Hybridisation (DDH)
2.7. Secondary Metabolite Production and Antimicrobial Activity
3. Result and Discussion
3.1. Morphological, Physiological, and Biochemical Results
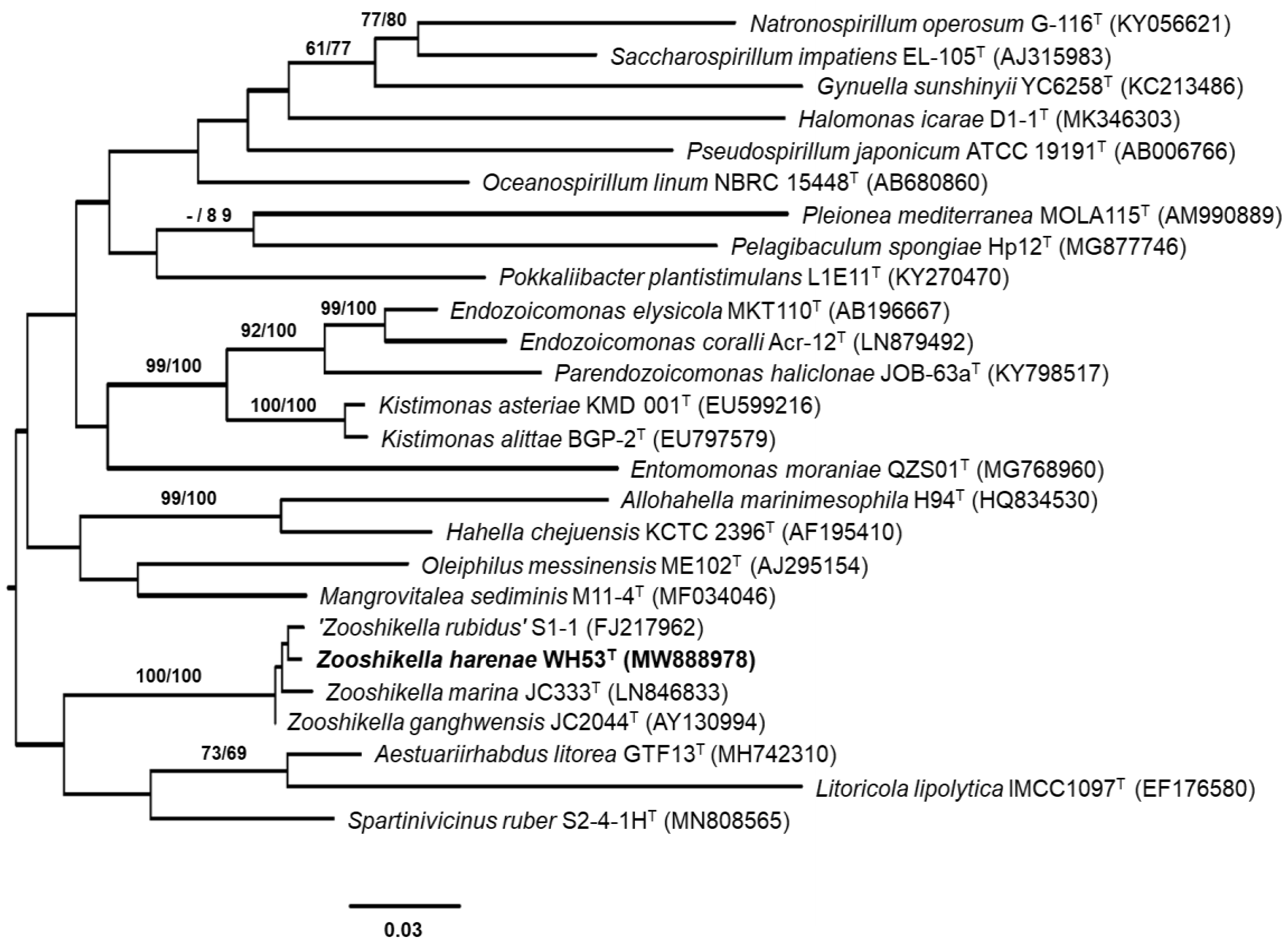
3.2. 16S rRNA Gene Analysis
3.3. Chemotaxonomic Characterization
3.4. Genomic Characteristics and Phylogenomic Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tortorella, E.; Tedesco, P.; Palma Esposito, F.; January, G.G.; Fani, R.; Jaspars, M.; De Pascale, D. Antibiotics from Deep-Sea Microorganisms: Current Discoveries and Perspectives. Mar. Drugs 2018, 16, 355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nawaz, A.; Chaudhary, R.; Shah, Z.; Dufossé, L.; Fouillaud, M.; Mukhtar, H.J.M. An Overview on Industrial and Medical Applications of Bio-Pigments Synthesized by Marine Bacteria. Microorganisms 2021, 9, 11. [Google Scholar] [CrossRef] [PubMed]
- Bruckner, A.W.J.I.I.S. Technology. Life-saving products from coral reefs. Issues Sci. Technol. 2002, 18, 39–44. [Google Scholar]
- Soliev, A.B.; Hosokawa, K.; Enomoto, K. Bioactive Pigments from Marine Bacteria: Applications and Physiological Roles. Evid.-Based Complement. Altern. Med. 2011, 2011, 670349. [Google Scholar] [CrossRef]
- Pawar, R.; Mohandass, C.; Rajasabapathy, R.; Meena, R.M. Molecular diversity of marine pigmented bacteria in the central Arabian Sea with special reference to antioxidant properties. Cah. Biol. Mar. 2018, 59, 409–420. [Google Scholar]
- Baharum, S.; Beng, E.; Mokhtar, M. Marine microorganisms: Potential application and challenges. J. Biol. Sci. 2010, 10, 555–564. [Google Scholar] [CrossRef] [Green Version]
- Podar, M.; Reysenbach, A.-L. New opportunities revealed by biotechnological explorations of extremophiles. Curr. Opin. Biotechnol. 2006, 17, 250–255. [Google Scholar] [CrossRef]
- Liao, H.; Lin, X.; Li, Y.; Qu, M.; Tian, Y. Reclassification of the Taxonomic Framework of Orders Cellvibrionales, Oceanospirillales, Pseudomonadales, and Alteromonadales in Class Gammaproteobacteria through Phylogenomic Tree Analysis. mSystems 2020, 5, e00543-20. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.; Chang, Y.-H.; Oh, H.W.; Bae, K.S.; Chun, J. Zooshikella ganghwensis gen. nov., sp. nov., isolated from tidal flat sediments. Int. J. Syst. Evol. Microbiol. 2003, 53, 1013–1018. [Google Scholar] [CrossRef]
- Lee, J.S.; Kim, Y.-S.; Park, S.; Kim, J.; Kang, S.-J.; Lee, M.-H.; Ryu, S.; Choi, J.M.; Oh, T.-K.; Yoon, J.-H. Exceptional Production of both Prodigiosin and Cycloprodigiosin as Major Metabolic Constituents by a Novel Marine Bacterium, Zooshikella rubidus S1-1. Appl. Environ. Microbiol. 2011, 77, 4967–4973. [Google Scholar] [CrossRef] [Green Version]
- Ramaprasad, E.V.V.; Bharti, D.; Sasikala, C.; Ramana, C.V. Zooshikella marina sp. nov. a cycloprodigiosin- and prodigiosin-producing marine bacterium isolated from beach sand. Int. J. Syst. Evol. Microbiol. 2015, 65, 4669–4673. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, C.; Takemoto, H.; Kuno, K.; Yamamoto, D.; Tsubura, A.; Kamata, K.; Hirata, H.; Yamamoto, A.; Kano, H.; Seki, T.; et al. Cycloprodigiosin hydrochloride, a new H+/Cl− symporter, induces apoptosis in human and rat hepatocellular cancer cell lines in vitro and inhibits the growth of hepatocellular carcinoma xenografts in nude mice. Hepatology 1999, 30, 894–902. [Google Scholar] [CrossRef]
- Boger, D.L.; Patel, M. Total synthesis of prodigiosin, prodigiosene, and desmethoxyprodigiosin: Diels-Alder reactions of heterocyclic azadienes and development of an effective palladium(II)-promoted 2,2′-bipyrrole coupling procedure. J. Org. Chem. 1988, 53, 1405–1415. [Google Scholar] [CrossRef]
- Castro, A.J. Antimalarial Activity of Prodigiosin. Nature 1967, 213, 903–904. [Google Scholar] [CrossRef] [PubMed]
- Beatriz, M.; Ricardo, P.-T. The Prodigiosins: A New Family of Anticancer Drugs. Curr. Cancer Drug Targets 2003, 3, 57–65. [Google Scholar] [CrossRef]
- Han, S.B.; Kim, H.M.; Kim, Y.H.; Lee, C.W.; Jang, E.-S.; Son, K.H.; Kim, S.U.; Kim, Y.K. T-cell specific immunosuppression by prodigiosin isolated from Serratia marcescens. Int. J. Immunopharmacol. 1998, 20, 1–13. [Google Scholar] [CrossRef]
- Jeong, H.; Yim, J.H.; Lee, C.; Choi, S.-H.; Park, Y.K.; Yoon, S.H.; Hur, C.-G.; Kang, H.-Y.; Kim, D.; Lee, H.H.; et al. Genomic blueprint of Hahella chejuensis, a marine microbe producing an algicidal agent. Nucleic Acids Res. 2005, 33, 7066–7073. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, T.; Miyazaki, Y.; Matsuyama, Y.; Muraoka, W.; Yamaguchi, K.; Oda, T. Producing mechanism of an algicidal compound against red tide phytoplankton in a marine bacterium γ-proteobacterium. Appl. Microbiol. Biotechnol. 2006, 73, 684–690. [Google Scholar] [CrossRef]
- Alihosseini, F.; Ju, K.-S.; Lango, J.; Hammock, B.D.; Sun, G. Antibacterial Colorants: Characterization of Prodiginines and Their Applications on Textile Materials. Biotechnol. Prog. 2008, 24, 742–747. [Google Scholar] [CrossRef] [Green Version]
- Haddix, P.L.; Jones, S.; Patel, P.; Burnham, S.; Knights, K.; Powell, J.N.; LaForm, A. Kinetic Analysis of Growth Rate, ATP, and Pigmentation Suggests an Energy-Spilling Function for the Pigment Prodigiosin of Serratia marcescens. J. Bacteriol. 2008, 190, 7453–7463. [Google Scholar] [CrossRef] [Green Version]
- Gerber, N.N.; Gauthier, M.J. New prodigiosin-like pigment from Alteromonas rubra. Appl. Environ. Microbiol. 1979, 37, 1176–1179. [Google Scholar] [CrossRef] [Green Version]
- Kawauchi, K.; Shibutani, K.; Yagisawa, H.; Kamata, H.; Nakatsuji, S.I.; Anzai, H.; Yokoyama, Y.; Ikegami, Y.; Moriyama, Y.; Hirata, H. A Possible Immunosuppressant, Cycloprodigiosin Hydrochloride, Obtained from Pseudoalteromonas denitrificans. Biochem. Biophys. Res. Commun. 1997, 237, 543–547. [Google Scholar] [CrossRef]
- Bennett, J.W.; Bentley, R. Seeing Red: The Story of Prodigiosin. In Advances in Applied Microbiology; Academic Press: Cambridge, MA, USA, 2000; Volume 47, pp. 1–32. [Google Scholar] [CrossRef]
- Yamamoto, D.; Uemura, Y.; Tanaka, K.; Nakai, K.; Yamamoto, C.; Takemoto, H.; Kamata, K.; Hirata, H.; Hioki, K. Cycloprodigiosin hydrochloride, H+/CL– symporter, induces apoptosis and differentiation in HL-60 cells. Int. J. Cancer 2000, 88, 121–128. [Google Scholar] [CrossRef]
- Yamamoto, D.; Tanaka, K.; Nakai, K.; Baden, T.; Inoue, K.; Yamamoto, C.; Takemoto, H.; Kamato, K.; Hirata, H.; Morikawa, S.; et al. Synergistic Effects Induced by Cycloprodigiosin Hydrochloride and Epirubicin on Human Breast Cancer Cells. Breast Cancer Res. Treat. 2002, 72, 1–10. [Google Scholar] [CrossRef]
- Kamata, K.; Okamoto, S.; Oka, S.-I.; Kamata, H.; Yagisawa, H.; Hirata, H. Cycloprodigiosin hydrocloride suppresses tumor necrosis factor (TNF) α-induced transcriptional activation by NF-κB. FEBS Lett. 2001, 507, 74–80. [Google Scholar] [CrossRef] [Green Version]
- Landwehr, W.; Kämpfer, P.; Glaeser, S.P.; Rückert, C.; Kalinowski, J.; Blom, J.; Goesmann, A.; Mack, M.; Schumann, P.; Atasayar, E.; et al. Taxonomic analyses of members of the Streptomyces cinnabarinus cluster, description of Streptomyces cinnabarigriseus sp. nov. and Streptomyces davaonensis sp. nov. Int. J. Syst. Evol. Microbiol. 2018, 68, 382–393. [Google Scholar] [CrossRef]
- Kutzner, H.J.T.P. The Family Streptomycetaceae; Springer: New York, NY, USA, 1981. [Google Scholar] [CrossRef]
- Shirling, E.B.; Gottlieb, D. Methods for characterization of Streptomyces species1. Int. J. Syst. Bacteriol. 1966, 16, 313–340. [Google Scholar] [CrossRef] [Green Version]
- Humble, M.W.; King, A.; Phillips, I. API ZYM: A simple rapid system for the detection of bacterial enzymes. J. Clin. Pathol. 1977, 30, 275–277. [Google Scholar] [CrossRef] [Green Version]
- Soto, A.; Zapardiel, J.; Soriano, F. Evaluation of API Coryne system for identifying coryneform bacteria. J. Clin. Pathol. 1994, 47, 756–759. [Google Scholar] [CrossRef] [Green Version]
- Primahana, G.; Risdian, C.; Mozef, T.; Sudarman, E.; Köck, M.; Wink, J.; Stadler, M. Nonocarbolines A–E, β-Carboline Antibiotics Produced by the Rare Actinobacterium Nonomuraea sp. from Indonesia. Antibiotics 2020, 9, 126. [Google Scholar] [CrossRef] [Green Version]
- Chaiya, L.; Matsumoto, A.; Wink, J.; Inahashi, Y.; Risdian, C.; Pathom-aree, W.; Lumyong, S. Amycolatopsis eburnea sp. nov., an actinomycete associated with arbuscular mycorrhizal fungal spores. Int. J. Syst. Evol. Microbiol. 2019, 69, 3603–3608. [Google Scholar] [CrossRef]
- Hall, T.A. Bioedit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meier-Kolthoff, J.P.; Hahnke, R.L.; Petersen, J.; Scheuner, C.; Michael, V.; Fiebig, A.; Rohde, C.; Rohde, M.; Fartmann, B.; Goodwin, L.A.; et al. Complete genome sequence of DSM 30083T, the type strain (U5/41T) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Stand. Genom. Sci. 2014, 9, 2. [Google Scholar] [CrossRef] [Green Version]
- Meier-Kolthoff, J.P.; Göker, M.; Spröer, C.; Klenk, H.P. When should a DDH experiment be mandatory in microbial taxonomy? Arch. Microbiol. 2013, 195, 413–418. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Goloboff, P.A.; Farris, J.S.; Nixon, K.C. TNT, a free program for phylogenetic analysis. Cladistics 2008, 24, 774–786. [Google Scholar] [CrossRef]
- Pattengale, N.D.; Alipour, M.; Bininda-Emonds, O.R.; Moret, B.M.; Stamatakis, A. How many bootstrap replicates are necessary? J. Comput. Biol. A J. Comput. Mol. Cell Biol. 2010, 17, 337–354. [Google Scholar] [CrossRef]
- Swofford, D. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods); Version 4.0b10; Sinauer Associates: Sunderland, MA, USA, 2002. [Google Scholar]
- Minnikin, D.E.; O’Donnell, A.G.; Goodfellow, M.; Alderson, G.; Athalye, M.; Schaal, A.; Parlett, J.H. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 1984, 2, 233–241. [Google Scholar] [CrossRef]
- Risdian, C.; Landwehr, W.; Rohde, M.; Schumann, P.; Hahnke, R.L.; Spröer, C.; Bunk, B.; Kämpfer, P.; Schupp, P.J.; Wink, J. Streptomyces bathyalis sp. nov., an actinobacterium isolated from the sponge in a deep sea. Antonie Van Leeuwenhoek 2021, 114, 425–435. [Google Scholar] [CrossRef] [PubMed]
- Sasser, M. Identification of Bacteria by Gas Chromatography of Cellular Fatty Acids; MIDI Tech. Note 101; MIDI Inc.: Newark, DE, USA, 1990. [Google Scholar]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [Green Version]
- Lee, I.; Chalita, M.; Ha, S.-M.; Na, S.-I.; Yoon, S.-H.; Chun, J. ContEst16S: An algorithm that identifies contaminated prokaryotic genomes using 16S RNA gene sequences. Int. J. Syst. Evol. Microbiol. 2017, 67, 2053–2057. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [Green Version]
- Medema, M.H.; Blin, K.; Cimermancic, P.; de Jager, V.; Zakrzewski, P.; Fischbach, M.A.; Weber, T.; Takano, E.; Breitling, R. antiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011, 39, W339–W346. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [Green Version]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Carbasse, J.S.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acids Res. 2021, 10, 2182. [Google Scholar] [CrossRef]
- Lefort, V.; Desper, R.; Gascuel, O. FastME 2.0: A Comprehensive, Accurate, and Fast Distance-Based Phylogeny Inference Program. Mol. Biol. Evol. 2015, 32, 2798–2800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arkin, A.P.; Cottingham, R.W.; Henry, C.S.; Harris, N.L.; Stevens, R.L.; Maslov, S.; Dehal, P.; Ware, D.; Perez, F.; Canon, S.; et al. KBase: The United States Department of Energy Systems Biology Knowledgebase. Nat. Biotechnol. 2018, 36, 566–569. [Google Scholar] [CrossRef] [Green Version]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.-H.; Ha, S.-m.; Lim, J.; Kwon, S.; Chun, J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 2017, 110, 1281–1286. [Google Scholar] [CrossRef] [PubMed]
- Qin, Q.-L.; Xie, B.-B.; Zhang, X.-Y.; Chen, X.-L.; Zhou, B.-C.; Zhou, J.; Oren, A.; Zhang, Y.-Z. A Proposed Genus Boundary for the Prokaryotes Based on Genomic Insights. J. Bacteriol. 2014, 196, 2210–2215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez-R, L.M.; Konstantinidis, K.J.M.M. Bypassing Cultivation To Identify Bacterial Species: Culture-independent genomic approaches identify credibly distinct clusters, avoid cultivation bias, and provide true insights into microbial species. ASM 2014, 9, 111–118. [Google Scholar] [CrossRef]
- Ziemke, F.; Höfle, M.G.; Lalucat, J.; Rossellö-Mora, R. Reclassification of Shewanella putrefaciens Owens genomic group II as Shewanella baltica sp. nov. Int. J. Syst. Bacteriol. 1998, 48, 179–186. [Google Scholar] [CrossRef] [Green Version]
- Khosravi Babadi, Z.; Ebrahimipour, G.; Wink, J.; Narmani, A.; Risdian, C. Isolation and identification of Streptomyces sp. Act4Zk, a good producer of Staurosporine and some derivatives. Lett. Appl. Microbiol. 2021, 72, 206–218. [Google Scholar] [CrossRef]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; da Costa, M.S.; Rooney, A.P.; Yi, H.; Xu, X.-W.; De Meyer, S.; et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef]
- Wayne, L.G.; Brenner, D.J.; Colwell, R.R.; Grimont, P.A.D.; Kandler, O.; Krichevsky, M.I.; Moore, L.H.; Moore, W.E.C.; Murray, R.G.E.; Stackebrandt, E.; et al. Report of the Ad Hoc Committee on Reconciliation of Approaches to Bacterial Systematics. Int. J. Syst. Evol. Microbiol. 1987, 37, 463–464. [Google Scholar] [CrossRef] [Green Version]
- International Code of Nomenclature of Prokaryotes. Int. J. Syst. Evol. Microbiol. 2019, 69, S1–S111. [CrossRef] [PubMed]


| Enzyme | Observation | Enzyme | Observation |
|---|---|---|---|
| Phosphatase alkaline | ++ | Naphtol-AS-BI-phosphohydrolase | ++ |
| Esterase (C4) | + | α-galactosidase | (+) |
| Esterase lipase (C8) | + | β-galactosidase | (+) |
| Lipase (C14) | (+) | β-glucuronidase | - |
| Leucin arylamidase | ++ | α-glucosidase | ++ |
| Valine arylamidase | (+) | β-glucosidase | (+) |
| Cystine arylamidase | (+) | N-acetyl-beta- glucosaminidase | ++ |
| Trypsin | (+) | α-mannosidase | + |
| Chymotrypsin | - | α-fucosidase | (+) |
| Phosphatase acid | ++ |
| Strain | Zooshikella harenae WH53T (JASOY000000000) | Zooshikella marina LMG 28823T (JAGSGA000000000) | ||
|---|---|---|---|---|
| OrthoANIu (%) | dDDH (%) | OrthoANIu (%) | dDDH (%) | |
| Zooshikella harenae WH53T (JAGSOY000000000) | 100 | 100 | 82.74 | 26.10 |
| Zooshikella marina LMG 28823T (JAGSGA000000000) | 82.74 | 26.10 | 100 | 100 |
| Zooshikella ganghwensis DSM 15267T (AUAF01000000) | 82.78 | 26.30 | 97.84 | 79.90 |
| Endozoicomonas montiporae CL-33T (CP013251) | 66.94 | 24.30 | 66.76 | 28.50 |
| Endozoicomonas elysicola (JOJP01000000) | 66.93 | 27.50 | 67.44 | 23.30 |
| Endozoicomonas arenosclerae (LASA01000000) | 66.88 | 23.10 | 67.26 | 22.30 |
| Endozoicomonas atrinae (LUKQ02000000) | 67.25 | 23.50 | 67.17 | 22.50 |
| Hahella chejuensis KCTC 2396T (CP000155) | 66.79 | 38.60 | 66.43 | 29.00 |
| Marinobacter lutaoensis (MSCW01000000) | 66.03 | 23.20 | 65.40 | 21.50 |
| Mangrovitalea sediminis (NTLB01000000) | 66.08 | 32.80 | 65.95 | 30.80 |
| Aestuariirhabdus litorea (QWEZ01000000) | 66.59 | 19.30 | 66.88 | 18.60 |
| Kistimonas asteriae JCM 15607T (JAEVHF000000000) | 66.84 | 20.00 | 67.08 | 34.40 |
| Azomonas agilis DSM375T (NZ_VLKG00000000) | 67.62 | 31.80 | 66.35 | 20.40 |
| Hydrocarboniclastica marina Soil36-7T (PRJNA479718) | 68.07 | 38.60 | 68.17 | 39.50 |
| Kangiella spongicola ATCC BAA-2076T (PRJNA473557) | 67.74 | 32.90 | 67.63 | 34.10 |
| Spartinivicinus ruber KCTC 72148T (PRJNA607118) | 68.91 | 21.30 | 69.06 | 22.80 |
| Characteristic | 1 | 2 | 3 | 4 † |
|---|---|---|---|---|
| pH range for growth | 6–8 | 6–9 | 5–8 | 4.5–9.5 |
| NaCl tolerance (w/v) | 7.5% | 5.0% | 5.0% | 10% |
| Color of colony | Red | Red | Red | Dark red |
| Butyrate esterase (C4) | + | (+) | (+) | - |
| Caprylate esterase lipase (C8) | + | (+) | (+) | + |
| Myristate lipase (C14) | (+) | - | - | - |
| Cystine arylamidase | (+) | - | - | - |
| Trypsin | (+) | - | - | - |
| Acid phosphatase | ++ | (+) | + | + |
| Naphtol-AS-BI-phosphohydrolase | ++ | - | - | - |
| α-Glucosidase | + | - | - | - |
| β-Glucosidase | (+) | - | - | - |
| N-acetyl-β-glucosaminidase | ++ | ++ | - | - |
| Phosphatase alkaline | ++ | ++ | ++ | + |
| Valine arylamidase | (+) | (+) | (+) | - |
| α-galactosidase | (+) | - | - | - |
| β-galactosidase | (+) | - | - | - |
| β-glucuronidase | - | - | - | - |
| α-mannosidase | + | + | - | - |
| α-fucosidase | (+) | - | - | - |
| Glucose fermentation | - | + | - | NR |
| Sucrose fermentation | - | + | - | NR |
| Glycogen fermentation | - | + | - | NR |
| Polar lipids | DPG–PG–PE–APL–PL | DPG–PG–PE–APL–PL | DPG–PG–PE–AL–PL | NR |
| Major fatty acid | C16:0 C16:1ω7c C18:1ω7c | C12:0 3-OH C14:0 C16:0 C16:1ω7c C18:1ω7c | C12:0 3-OH C16:0 C16:1ω7c C18:1ω7c | C12:0 3-OH C16:0 C18:1ω7c |
| Major quinone | Ubiquinone-9 | Ubiquinone-9 | Ubiquinone-9 | Ubiquinone-9 |
| Ampicillin susceptibility | - | NR | NR | - |
| Tetracycline susceptibility | + | NR | NR | - |
| Total sequence length (bp) | 5,914,969 | 6,060,265 | 5,798,664 | NR |
| Contigs | 377 | 83 | 414 | NR |
| No. of protein | 5,120 | 5,083 | 4,943 | NR |
| rRNA | 3 | 3 | 10 | NR |
| tRNA | 54 | 55 | 55 | NR |
| No. of Gene | 5241 | 5160 | 5164 | NR |
| Other RNA | 4 | 4 | 4 | NR |
| Pseudogene | 60 | 15 | 152 | NR |
| G + C content (mol%) | 40.08% | 40.94% | 41.02% | 41% |
| Strain | 1 | 2 | 3 | |||
|---|---|---|---|---|---|---|
| POCP (%) | AAI (%) | POCP (%) | AAI (%) | POCP (%) | AAI (%) | |
| Zooshikella harenae WH53T (JAGSOY000000000) | 76.08 | 87.01 | 77.37 | 86.73 | 100 | 100 |
| Zooshikella marina LMG 28823T (JAGSGA000000000) | 76.08 | 87.01 | 100 | 100 | 77.37 | 86.73 |
| Zooshikella ganghwensis DSM 15267T (AUAF01000000) | 100 | 100 | 76.08 | 87.01 | 76.08 | 87.01 |
| Mangrovitalea sediminis(= MCCC 1K03312T = JCM 32104T) (NTLB01000000) | 29.76 | 49.90 | 30.51 | 49.76 | 31.26 | 49.79 |
| Endozoicomonas acroporae strain Acr-14T (PRJNA422318) | 28.44 | 51.65 | 28.40 | 51.37 | 28.58 | 51.20 |
| Endozoicomonas arenosclerae (PRJNA279233) | 29.50 | 50.97 | 29.08 | 50.74 | 29.40 | 50.58 |
| Endozoicomonas ascidiicola (NZ_LUTW00000000) | 30.17 | 51.00 | 30.26 | 51.06 | 29.82 | 51.32 |
| Endozoicomonas atrinae (NZ_LUKQ00000000) | 26.35 | 50.80 | 26.53 | 50.72 | 26.32 | 50.72 |
| Endozoicomonas elysicola (NZ_JOJP00000000) | 30.89 | 51.22 | 31.17 | 51.11 | 31.27 | 51.19 |
| Endozoicomonas montiporae Strain LMG 24815T (NZ_JOKG00000000) | 30.03 | 51.25 | 30.26 | 51.24 | 29.86 | 51.37 |
| Endozoicomonas numazuensis (NZ_JOKH00000000) | 29.82 | 51.41 | 29.94 | 51.11 | 30.08 | 51.02 |
| Hahella chejuensis KCTC 2396T (PRJNA16064) | 30.68 | 49.72 | 31.48 | 49.62 | 31.44 | 49.73 |
| Hahella ganghwensis DSM 17046T (NZ_AQXX00000000) | 30.01 | 49.56 | 30.54 | 49.41 | 30.67 | 49.17 |
| Kistimonas asteriae (NZ_JAEVHF000000000) | 33.61 | 53.09 | 33.94 | 52.56 | 34.09 | 52.75 |
| Parendozoicomonas haliclonae (NZ_FWPT00000000) | 31.66 | 51.49 | 32.19 | 51.20 | 32.17 | 50.91 |
| Pseudomonas xiamenensis (NZ_JACLGO000000000) | 30.30 | 51.18 | 30.28 | 51.06 | 30.28 | 50.89 |
| Kistimonas asteriae JCM 15607 T (JAEVHF000000000) | 33.61 | 53.09 | 33.92 | 52.55 | 34.09 | 52.75 |
| Spartinivicinus ruber KCTC 72148T (PRJNA607118) | 49.90 | 58.67 | 49.94 | 58.38 | 49.97 | 58.69 |
| Pseudomonas pertucinogena JCM 11590T (BMNN00000000) | 32.11 | 52.18 | 32.53 | 52.10 | 32.18 | 51.93 |
| Pseudomonas yangmingesis DSM 24213T (NZ_FOUI00000000) | 31.78 | 52.27 | 32.05 | 52.20 | 31.74 | 51.95 |
| Pseudomonas abyssi Strain MT5T (PRJNA406957) | 31.34 | 51.28 | 32.07 | 51.16 | 31.90 | 50.99 |
| Azomonasa gilis DSM 375T (NZ_VLKG00000000) | 27.95 | 50.87 | 28.25 | 51.03 | 28.53 | 50.95 |
| Hydrocarboniclastica marina Soil36-7T (PRJNA479718) | 28.46 | 49.31 | 28.65 | 49.18 | 28.97 | 49.10 |
| Kangiella spongicola ATCC BAA-2076T (PRJNA473557) | 25.06 | 47.38 | 25.60 | 47.41 | 24.74 | 47.11 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pira, H.; Risdian, C.; Kämpfer, P.; Müsken, M.; Schupp, P.J.; Wink, J. Zooshikella harenae sp. nov., Isolated from Pacific Oyster Crassostrea gigas, and Establishment of Zooshikella ganghwensis subsp. marina subsp. nov. and Zooshikella ganghwensis subsp. ganghwensis subsp. nov. Diversity 2021, 13, 641. https://doi.org/10.3390/d13120641
Pira H, Risdian C, Kämpfer P, Müsken M, Schupp PJ, Wink J. Zooshikella harenae sp. nov., Isolated from Pacific Oyster Crassostrea gigas, and Establishment of Zooshikella ganghwensis subsp. marina subsp. nov. and Zooshikella ganghwensis subsp. ganghwensis subsp. nov. Diversity. 2021; 13(12):641. https://doi.org/10.3390/d13120641
Chicago/Turabian StylePira, Hani, Chandra Risdian, Peter Kämpfer, Mathias Müsken, Peter J. Schupp, and Joachim Wink. 2021. "Zooshikella harenae sp. nov., Isolated from Pacific Oyster Crassostrea gigas, and Establishment of Zooshikella ganghwensis subsp. marina subsp. nov. and Zooshikella ganghwensis subsp. ganghwensis subsp. nov." Diversity 13, no. 12: 641. https://doi.org/10.3390/d13120641
APA StylePira, H., Risdian, C., Kämpfer, P., Müsken, M., Schupp, P. J., & Wink, J. (2021). Zooshikella harenae sp. nov., Isolated from Pacific Oyster Crassostrea gigas, and Establishment of Zooshikella ganghwensis subsp. marina subsp. nov. and Zooshikella ganghwensis subsp. ganghwensis subsp. nov. Diversity, 13(12), 641. https://doi.org/10.3390/d13120641








