Morphological and Molecular Characterization of Trichuris sp. (Nematoda: Trichuridae) in Crested Porcupines (Hystrix cristata; Rodentia: Hystricidae) from Italy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection of Material
2.2. Morphological and Morphometric Analysis
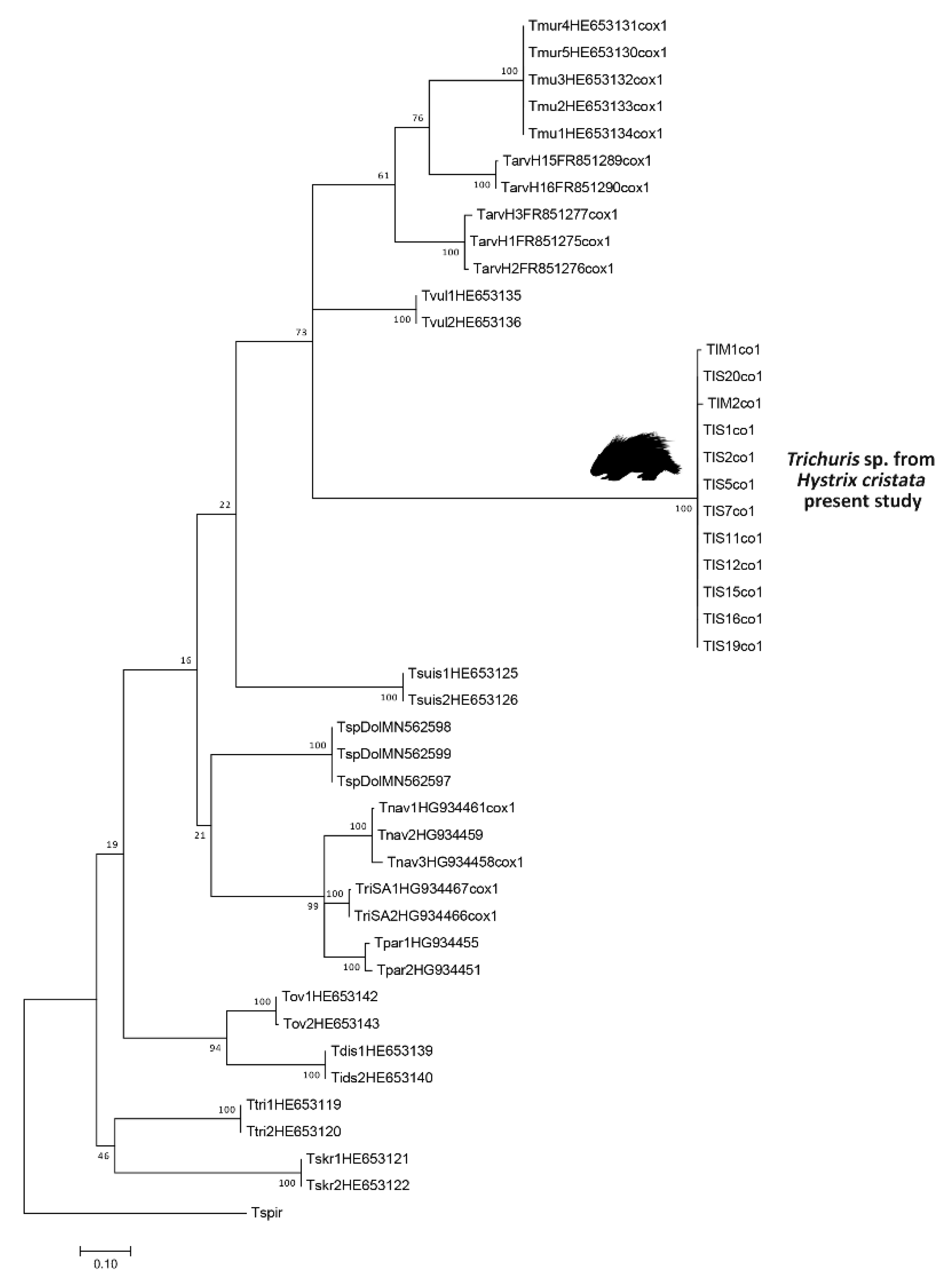
2.3. Genetic and Comparative Phylogenetic Analyses
| Parasite Species | Host Species | GenBank Accession Number | Specimen Code | References |
|---|---|---|---|---|
| Trichuris sp. | Hystrix cristata | TIS: MK779003 TIM: OK489802 | TIS and TIM | Present study |
| Trichuris sp. | Dolichotis patagonum | MN562597-99 | TspDol | [19] |
| T. colobae | Colobus guereza | HE653119-20 | Ttri1-2 | [17] |
| T. suis | Sus scrofa | HE653125-26 | S1-2 | Zhang et al. (2016) (unpublished) |
| T. ovis | Capra hircus | HE653142-43 | Tov1-2 | [17] |
| T. skrjabini | Capra hircus | HE653121-22 | Tskr1-2 | [17] |
| T. discolor | Bos taurus | HE653139-40 | Tdis1-2 | [17] |
| T. pardinasi | Phyllotis xanthopygus | HG934451,55 | Tpar1-2 | [8] |
| T. navonae | Akodon montensis | HG934458,59,61 | Tnav1-3 | [8] |
| Trichuris sp. | Sooretamys angouya | HG934465-66 | Sa1-2 | [8] |
| T. muris | Mus domesticus/musculus | HE653130-34 | M1-5 | [17] |
| T. arvicolae | Microtus agrestis | FR851275-77,89-90 | TarvH1-3, H15-16 | [17] |
| T. vulpis | Canis (lupus) familiaris | HE653135-36 | Tvul1-2 | [17] |
| Outgroup species | ||||
| Trichinella spiralis | AF293969 | Tspir | [20] | |
3. Results
3.1. Morphological and Morphometric Analysis
3.2. Molecular Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Anderson, R.C. Nematode Parasites of Vertebrates: Their Development and Transmission, 2nd ed.; CAB International: Wallingford, UK, 2000. [Google Scholar]
- Špakulová, M. Discriminant analysis as a method for the numerical evaluation of taxonomic characters in male trichurid nematodes. System. Parasitol. 1994, 29, 113–119. [Google Scholar] [CrossRef]
- Robles, M.D.R.; Cutillas, C.; Panei, C.J.; Callejòn, R. Morphological and molecular characterization of a new Trichuris species (Nematoda- Trichuridae), and phylogenetic relationships of Trichuris species of Cricetid rodents from Argentina. PLoS ONE 2014, 9, e112069. [Google Scholar]
- Tenora, F.; Kamiya, M.; Spakulová, M.; Stanek, M.; Ooi, H.K. Scanning electron microscopy of Trichuris suis and Trichuris vulpis from Slovakia and Japan. Helminthologia 1993, 30, 93–98. [Google Scholar]
- Cutillas, C.; German, P.; Arias, P.; Guevara, D. Trichuris ovis and Trichuris globulosa: Morphological, biometrical, and genetic studies. Exp. Parasitol. 1995, 81, 621–625. [Google Scholar] [CrossRef] [PubMed]
- Feliu, C.; Spakulová, M.; Casanova, J.C.; Renaud, F.; Morand, S.; Hugot, J.P.; Santalla, F.; Durand, P. Genetic and morphological heterogeneity in small rodent whipworms in south western Europe: Characterization of Trichuris muris and description of Trichuris arvicolae n. sp. (Nematoda: Trichuridae). J. Parasitol. 2000, 86, 442–449. [Google Scholar] [CrossRef]
- Cutillas, C.; Oliveros, R.; de Rojas, M.; Guevara, D.C. Determination of Trichuris skrjabini by sequencing of the ITS1-5.8S-ITS2 segment of the ribosomal DNA: Comparative molecular study of different species of trichurids. J. Parasitol. 2004, 90, 648–652. [Google Scholar] [CrossRef]
- Callejón, R.; Robles, M.D.R.; Panei, C.J.; Cutillas, C. Molecular diversification of Trichuris spp. from Sigmodontinae (Cricetidae) rodents from Argentina based on mitochondrial DNA sequences. Parasitol. Res. 2016, 115, 2933–2945. [Google Scholar] [CrossRef]
- Hall, M.C. Nematode Parasites of Mammals of the Orders Rodentia, Lagomorpha, and Hyracoidea; US Government Printing Office: New York, NY, USA, 1916; Volume 50.
- Kreis, H.A. Beitrage zur Kenntuis parasitischer Nematoden VII. Neue parasitische Nematoden aus dem Naturhistorischen Museum Basel. Zentralbl. Bakt. Abt. Orig. 1938, 142, 329–352. [Google Scholar]
- Skrjabin, K.I.; Shikhobalova, N.P.; Orlov, I.V. Trichocephalidae and diseases caused by them. In Trichocephalidae and Capillariidae of Animals and Man and the Diseases Caused by Them; Skrjabin, K.I., Shikhobalova, N.P., Orlov, I.V., Eds.; Academy of Sciences of the USSR: Moscow, Russia, 1957; pp. 26–223. [Google Scholar]
- Yamaguti, S. The Nematodes of Vertebrates; Systema helminthum; Interscience Publishers: New York, NY, USA, 1961; Volume III. [Google Scholar]
- Youssefi, M.R.; Hoseini, S.H.; Rahimi, M.T.; Esfandiari, B. Trichuris hystricis, a whipworm from Hystrix indica in Iran. World J. Zool. 2010, 5, 244–245. [Google Scholar]
- Petriov, A.M.; Sadikhov, I.A. Trichuris lenkorani n. sp. from the intestine of Hystrix hirsutirostris in Azerbaidzhan. Dokl. Akad. Nauk. Azerb. SSR 1961, 7, 631–634. [Google Scholar]
- Purwaningsih, E. The first report of new species: Trichuris landak n. sp. Asian Pac. J. Trop. Biomed. 2013, 3, 85–88. [Google Scholar] [CrossRef] [Green Version]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R package for multivariate analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Callejón, R.; Nadler, S.; De Rojas, M.; Zurita, A.; Petrášová, J.; Cutillas, C. Molecular characterization and phylogeny of whipworm nematodes inferred from DNA sequences of COX1 mtDNA and 18S rDNA. Parasitol. Res. 2013, 112, 3933–3949. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Di Filippo, M.M.; Berrilli, F.; De Liberato, C.; Di Giovanni, V.; D’Amelio, S.; Friedrich, K.G.; Cavallero, S. Molecular characterization of Trichuris spp. from captive animals based on mitochondrial markers. Parasitol. Int. 2020, 75, 102043. [Google Scholar] [CrossRef]
- Lavrov, D.V.; Brown, W.M. Trichinella spiralis mtDNA: A nematode mitochondrial genome that encodes a putative ATP8 and normally structured tRNAs and has a gene arrangement relatable to those of coelomate metazoans. Genetics 2001, 157, 621–637. [Google Scholar] [CrossRef] [PubMed]
- Monetti, L.; Massolo, A.; Sforzi, A.; Lovari, S. Site selection and fidelity by crested porcupine for denning. Ethol. Ecol. Evol. 2005, 17, 149–159. [Google Scholar] [CrossRef]
- Mori, E.; Lovari, S. Sexual size monomorphism in the crested porcupine (Hystrix cristata). Mamm. Biol. 2014, 79, 157–160. [Google Scholar] [CrossRef]
- Viviano, A.; Amori, G.; Luiselli, L.; Oebel, H.; Bahleman, F.; Mori, E. Blessing the rains down in Africa: Spatiotemporal behaviour of the crested porcupine Hystrix cristata (Mammalia: Rodentia) in the rainy and dry seasons, in the African savannah. Trop. Zool. 2020, 33, 113–124. [Google Scholar] [CrossRef]
- Amori, G.; De Smet, K. Hystrix cristata. The IUCN Red List of Threatened Species 2016: E.T10746A22232484. 2016. Available online: http://dx.doi.org/10.2305/IUCN.UK.2016-2.RLTS.T10746A22232484.en (accessed on 25 October 2021).
- Masseti, M.; Albarella, U.; De Grossi Mazzorin, J. The crested porcupine, Hystrix cristata L., 1758, in Italy. Anthropozoologica 2010, 45, 27–42. [Google Scholar] [CrossRef]
- Trucchi, E.; Sbordoni, V. Unveiling an ancient biological invasion: Molecular analysis of an old European alien, the crested porcupine (Hystrix cristata). BMC Evol. Biol. 2009, 9, 109. [Google Scholar] [CrossRef] [Green Version]
- Mori, E.; Ficetola, G.F.; Bartolomei, R.; Capobianco, G.; Varuzza, P.; Falaschi, M. How the South was won: Current and potential range expansion of the crested porcupine in Southern Italy. Mamm. Biol. 2021, 101, 11–19. [Google Scholar] [CrossRef]
- Mori, E.; Sforzi, A.; Bogliani, G.; Milanesi, P. Range expansion and redefinition of a crop-raiding rodent associated with global warming and temperature increase. Clim. Chang. 2018, 150, 319–331. [Google Scholar] [CrossRef]
- Torretta, E.; Orioli, V.; Bani, L.; Mantovani, S.; Dondina, O. En route to the North: Modelling crested porcupine habitat suitability and dispersal flows across a highly anthropized area in northern Italy. Mamm. Biol. 2021, 1–11. [Google Scholar] [CrossRef]
- Bailly-Choumaira, H.; Morel, P.C.; Rageau, J. Sommaire des donnees actuelles sur les tiques du Maroc (Acari, Ixodoidea). Bull. Inst. Sci. 1976, 1, 102–117. [Google Scholar]
- Beaucournu, J.C.; Launay, H. Les Puces de France et du Bassin Méditerranéen Occidental. Faune de France 76; Fédération Française des Sociétés de Sciences Naturelles: Paris, France, 1990. [Google Scholar]
- Kolonin, G.V. Haemaphysalis lobachovi sp. n. (Acharina: Ixodidae) from porcupine (Hystrix cristata) from Ethiopia. Folia Parasitol. 1995, 42, 239. [Google Scholar]
- Mori, E.; Sforzi, A.; Menchetti, M.; Mazza, G.; Lovari, S.; Pisanu, B. Ectoparasite load in the crested porcupine Hystrix cristata Linnaeus, 1758 in Central Italy. Parasitol. Res. 2015, 114, 2223–2229. [Google Scholar] [CrossRef]
- Scaravelli, D.; Senini, C.; Bonacci, T. First case of traumatic myiasis caused by Calliphora vicina in a crested porcupine Hystrix cristata L. in Italy. J. Entomol. Acarol. Res. 2017, 49, 6823. [Google Scholar] [CrossRef] [Green Version]
- Cilia, G.; Bertelloni, F.; Coppola, F.; Turchi, B.; Biliotti, C.; Poli, A.; Parisi, F.; Felicioli, A.; Cerri, D.; Fratini, F. Isolation of Leptospira serovar Pomona from a crested porcupine (Hystrix cristata, L., 1758). Vet. Med. Sci. 2020, 6, 985–991. [Google Scholar] [CrossRef] [PubMed]
- Coppola, F.; Maestrini, M.; Berrilli, F.; Procesi, I.G.; Felicioli, A.; Perrucci, S. First report of Giardia duodenalis infection in the crested porcupine (Hystrix cristata L., 1758). Int. J. Parasitol. Parasites Wildl. 2020, 11, 108–113. [Google Scholar] [CrossRef] [PubMed]
- Coppola, F.; Cilia, G.; Bertelloni, F.; Casini, L.; D’Addio, E.; Fratini, F.; Cerri, D.; Felicioli, A. Crested porcupine (Hystrix cristata L.): A new potential host for pathogenic Leptospira among semi-fossorial mammals. Comp. Immunol. Microbiol. Infect. Dis. 2020, 70, 101472. [Google Scholar] [CrossRef]
- Harrison, T.M.; Moorman, J.B.; Bolin, S.R.; Grosjean, N.L.; Lim, A.; Fitzgerald, S.D. Toxoplasma gondii in an African porcupine. J. Vet. Diagn. Investig. 2007, 19, 191–194. [Google Scholar] [CrossRef] [Green Version]
- Robles, M.D.R.; Navone, G.T. Redescription of Trichuris laevitestis (Nematoda: Trichuridae) from Akodon azarae and Scapteromys aquaticus (Sigmodontinae: Cricetidae) in Buenos Aires province, Argentina. J. Parasitol. 2006, 92, 1053–1057. [Google Scholar] [CrossRef]
- Vejl, P.; Nechybová, S.; Peřinková, P.; Melounová, M.; Sedláková, V.; Vašek, J.; Čílová, D.; Rylková, K.; Jankovská, I.; Vadlejch, J.; et al. Reliable molecular differentiation of Trichuris ovis and Trichuris discolor from sheep (Ovis orientalis aries) and roe deer (Capreolus capreolus) and morphological characterisation of their females: Morphology does not work sufficiently. Parasitol. Res. 2017, 116, 2199–2210. [Google Scholar] [CrossRef] [PubMed]
- Ribas, A.; López, S.; Makundi, R.H.; Leirs, H.; de Bellocq, J.G. Trichuris spp. (Nematoda: Trichuridae) from two rodents, Mastomys natalensis and Gerbilliscus vicinus in Tanzania. J. Parasitol. 2013, 99, 868–875. [Google Scholar] [CrossRef] [PubMed]
- Bernard, J. Faune des nematodes parasites des mammiferes de Tunisie et des contrees voisines. Arch. L’inst. Pasteur Tunis 1987, 64, 265–319. [Google Scholar]
- Poglayen, G.; Scaravelli, D.; Tampieri, M.P.; Galuppi, R.; Nuti, C.; Gaglio, G.; Abbene, S. Fauna parassitaria dell’istrice Hystrix cristata in Italia. Hystrix J. Mammal. 2005, 1, 106. [Google Scholar]
- Callejón, R.; De Rojas, M.; Feliú, C.; Balao, F.; Marrugal, A.; Henttonen, H.; Guevara, D.; Cutillas, C. Phylogeography of Trichuris populations isolated from different Cricetidae rodents. Parasitology 2012, 39, 1795–1812. [Google Scholar] [CrossRef] [Green Version]
- Milazzo, C.; de Bellocq, J.G.; Cagnin, M.; Casanova, J.C.; di Bella, C.; Feliu, C.; Morand, S.; Santalla, F. Helminths and ectoparasites of Rattus rattus and Mus musculus from Sicily, Italy. Comp. Parasitol. 2003, 70, 199–204. [Google Scholar] [CrossRef]
- Iglesias, R.; D’Amelio, S.; Ingrosso, S.; Farjallah, S.; Martínez-Cedeira, J.A.; García-Estévez, J.M. Molecular and morphological evidence for the occurrence of Anisakis sp. A (Nematoda, Anisakidae) in the Blainville’s beaked whale Mesoplodon densirostris. J. Helminthol. 2008, 82, 305–308. [Google Scholar] [CrossRef]
- Mattiucci, S.; Paoletti, M.; Webb, S.C. Anisakis nascettii n. sp. (Nematoda: Anisakidae) from beaked whales of the southern hemisphere: Morphological description, genetic relationships between congeners and ecological data. Syst. Parasitol. 2009, 74, 199–217. [Google Scholar] [CrossRef] [PubMed]
- Blouin, M.S. Molecular prospecting for cryptic species of nematodes: Mitochondrial DNA versus internal transcribed spacer. Int. J. Parasitol. 2002, 32, 527–531. [Google Scholar] [CrossRef]
- Callejón, R.; Cutillas, C.; Nadler, S.A. Nuclear and mitochondrial genes for inferring Trichuris phylogeny. Parasitol. Res. 2015, 114, 4591–4599. [Google Scholar] [CrossRef] [PubMed]
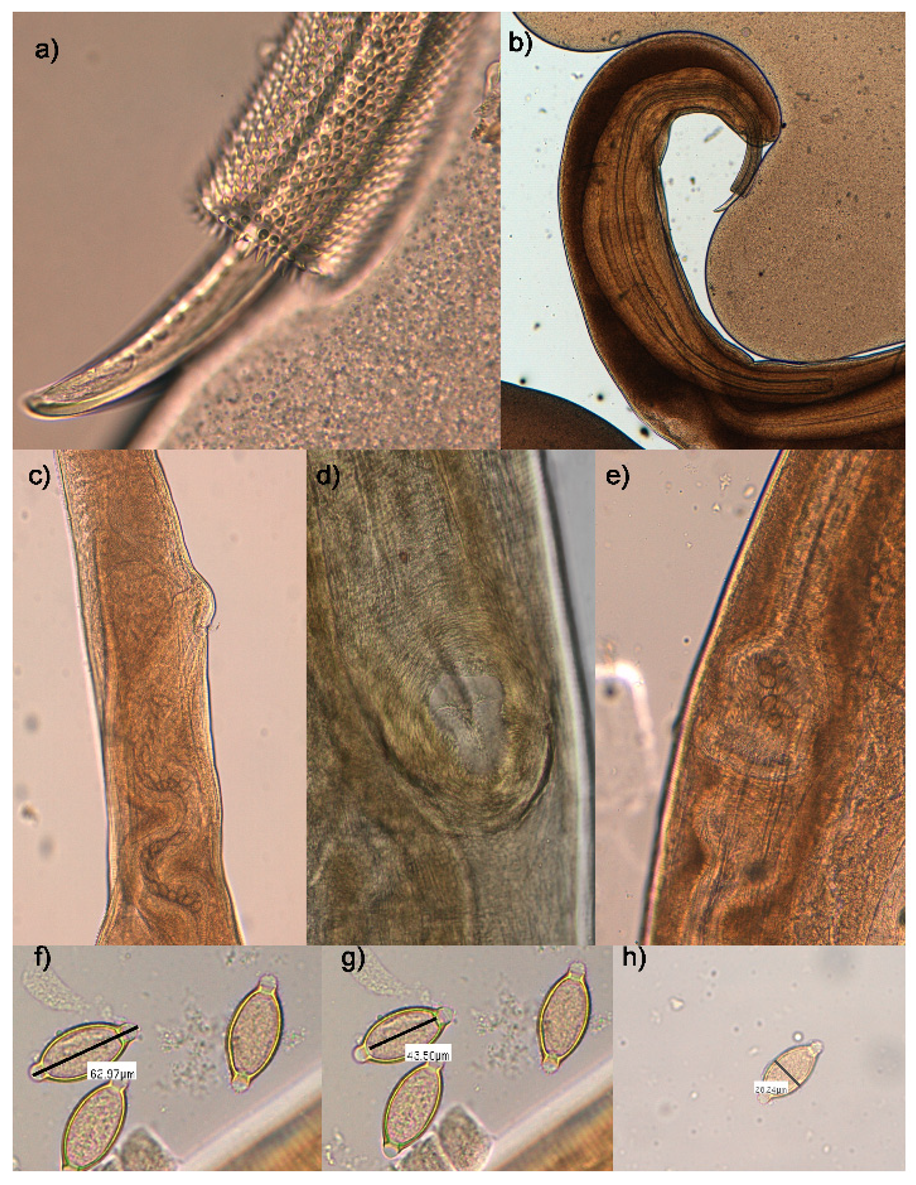
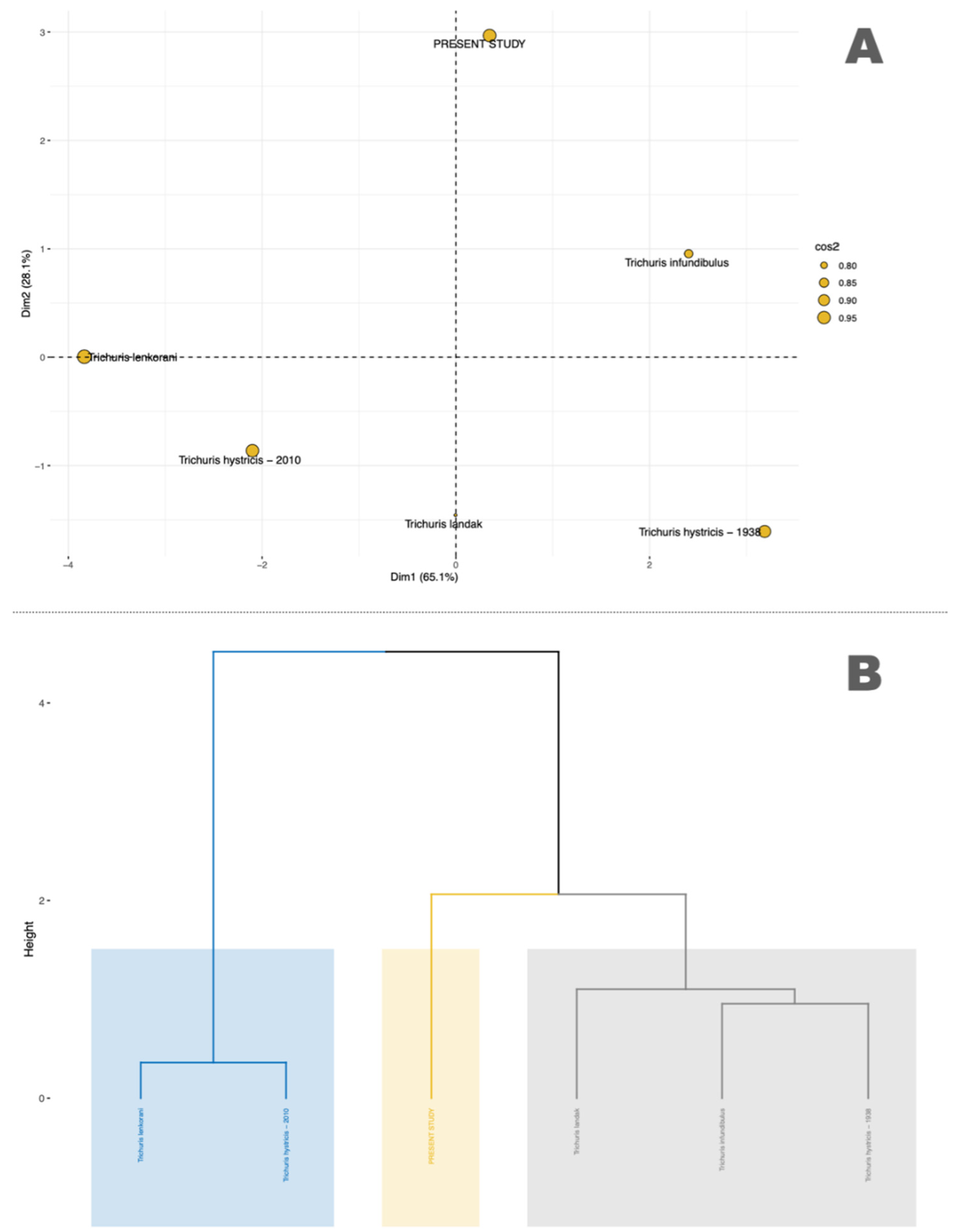
| Trichuris sp. from Hystrix cristata Present Study | Trichuris landak from Hystrix javanica [15] | Trichuris hystricis from Hystrix indica [11] | Trichuris hystricis from Hystrix cristata [10] | Trichuris infundibulus from Hystrix cristata [9] | Trichuris lenkorani from Hystrix indica [14] | |
|---|---|---|---|---|---|---|
| M1 | 30.70 (25–35) ± 5.0 | 39.34 (36.05–42.64) | 25.60 (23.76–27.45) | 42.56 (39.42–45.70) | 44.60 | 14.21–16.72 |
| M2 | 0.20 (0.08–0.31) ± 0.1 | 0.286 (0.282–0.29) | - | 0.37 (0.34–0.41) | - | |
| M3 | 0.56 (0.46–0.63) ± 0.08 | 0.74 (0.71–0.77) | - | 0.20 (0.19–0.21) | - | |
| M4 | 3.6 (3.18–4.08) ± 0.45 | - | 1.92 (1.87–1.98) | 1.05 (0.89–1.21) | 1.94 | 2.42–2.74 |
| M5 | 0.07 (0.06–0.09) ± 0.01 | 0.04 (0.03–0.05) | 0.00076 | 0.08 | ||
| M6 | 18.80 (15.0–21.0) ± 3 | - | - | - | - | |
| M7 | 0.04 (0.03–0.04) ± 0.005 | - | - | - | - | |
| M9 | 11.97 (10.0–14.8) ± 2.4 | |||||
| M10 | 18.80 (15.0–21.0) ± 3 | |||||
| F1 | 42.34 (30.00–49.30) ± 9.65 | 36.40 (36.00–41.90) | 34.40 (32.90–35.90) | 48.40 (47.20–49.50) | 52.10 | 20.00–27.70 |
| F2 | 25.00 (20–28) ± 4 | 23.09 (22.20–23.80) | 13.80 (12.32–15.21) | - | - | |
| F3 | 0.15 (0.13–0.19) ± 0.03 | 0.30 (0.30–0.31) | - | - | - | |
| F4 | 0.80 (0.44–1.03) ± 0.3 | 0.80 (0.80–0.81) | - | - | - | |
| F5 | 0.06 (0.06–0.07) ± 0.04 | 0.05 (0.05–0.06) | 0.05 (0.04–0.06) | 0.06 | ||
| F6 | 0.03 (0.02–0.03) ± 0.02 | 0.02 (0.02–0.03) | 0.03 (0.02–0.03) | 0.03 | ||
| F7 | 0.42 (0.26–0.58) ± 0.16 | - | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cavallero, S.; Montalbano Di Filippo, M.; Mori, E.; Viviano, A.; De Liberato, C.; Sforzi, A.; D’Amelio, S.; Berrilli, F. Morphological and Molecular Characterization of Trichuris sp. (Nematoda: Trichuridae) in Crested Porcupines (Hystrix cristata; Rodentia: Hystricidae) from Italy. Diversity 2021, 13, 628. https://doi.org/10.3390/d13120628
Cavallero S, Montalbano Di Filippo M, Mori E, Viviano A, De Liberato C, Sforzi A, D’Amelio S, Berrilli F. Morphological and Molecular Characterization of Trichuris sp. (Nematoda: Trichuridae) in Crested Porcupines (Hystrix cristata; Rodentia: Hystricidae) from Italy. Diversity. 2021; 13(12):628. https://doi.org/10.3390/d13120628
Chicago/Turabian StyleCavallero, Serena, Margherita Montalbano Di Filippo, Emiliano Mori, Andrea Viviano, Claudio De Liberato, Andrea Sforzi, Stefano D’Amelio, and Federica Berrilli. 2021. "Morphological and Molecular Characterization of Trichuris sp. (Nematoda: Trichuridae) in Crested Porcupines (Hystrix cristata; Rodentia: Hystricidae) from Italy" Diversity 13, no. 12: 628. https://doi.org/10.3390/d13120628
APA StyleCavallero, S., Montalbano Di Filippo, M., Mori, E., Viviano, A., De Liberato, C., Sforzi, A., D’Amelio, S., & Berrilli, F. (2021). Morphological and Molecular Characterization of Trichuris sp. (Nematoda: Trichuridae) in Crested Porcupines (Hystrix cristata; Rodentia: Hystricidae) from Italy. Diversity, 13(12), 628. https://doi.org/10.3390/d13120628








