Subtle East–West Phylogeographic Break of Asteropyrum (Ranunculaceae) in Subtropical China and Adjacent Areas
Abstract
1. Introduction
2. Material and Methods
2.1. Population Sampling, DNA Amplification, and Sequencing
2.2. Population Genetic Analyses
2.3. Population Divergence and Demographic Analyses
2.4. Isolation by Distance and Isolation by Environment
3. Results
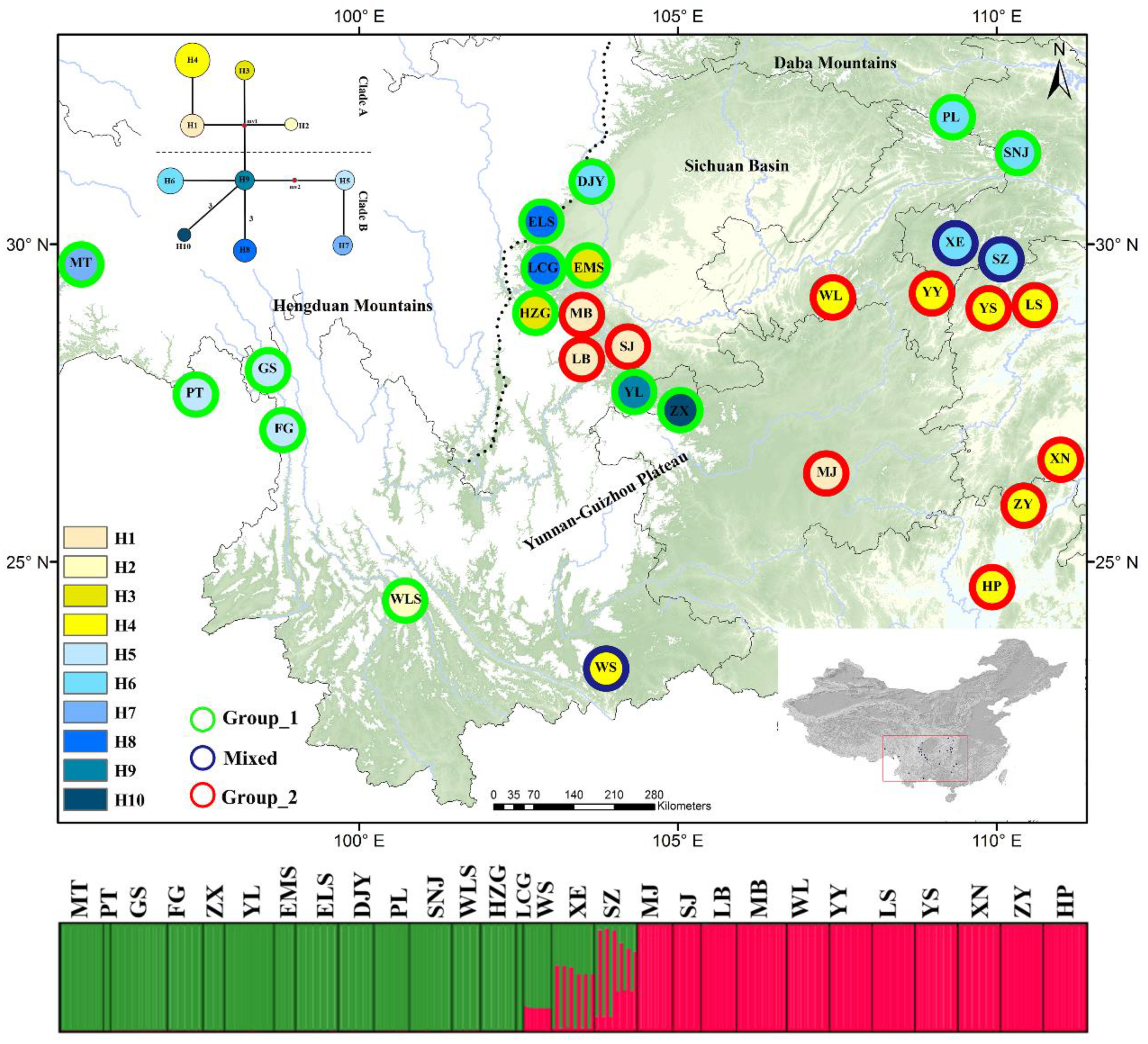
3.1. Sequence Variation and Distribution of cpDNA Haplotypes
3.2. Population Diversity and Structure for Nuclear DNA
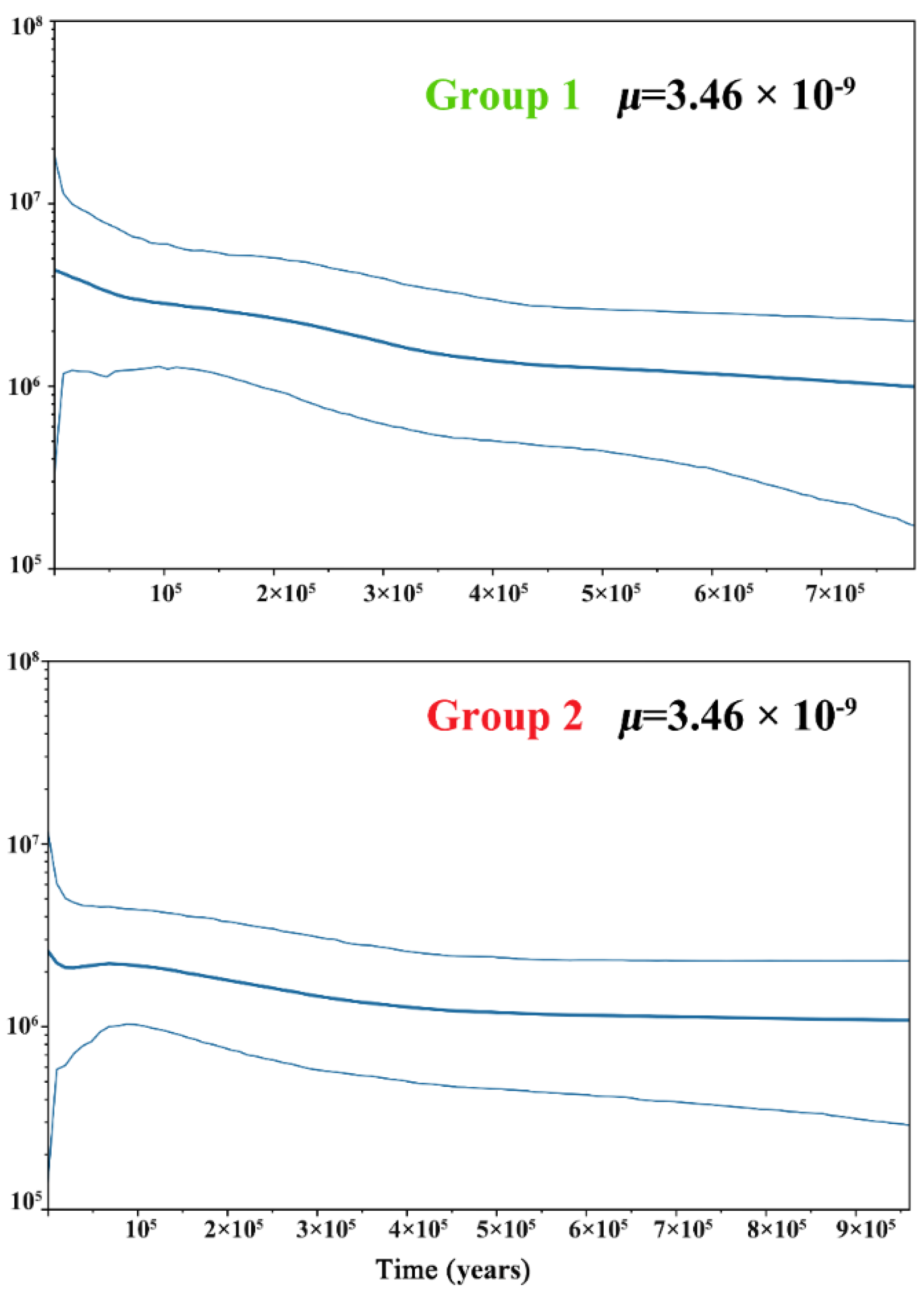
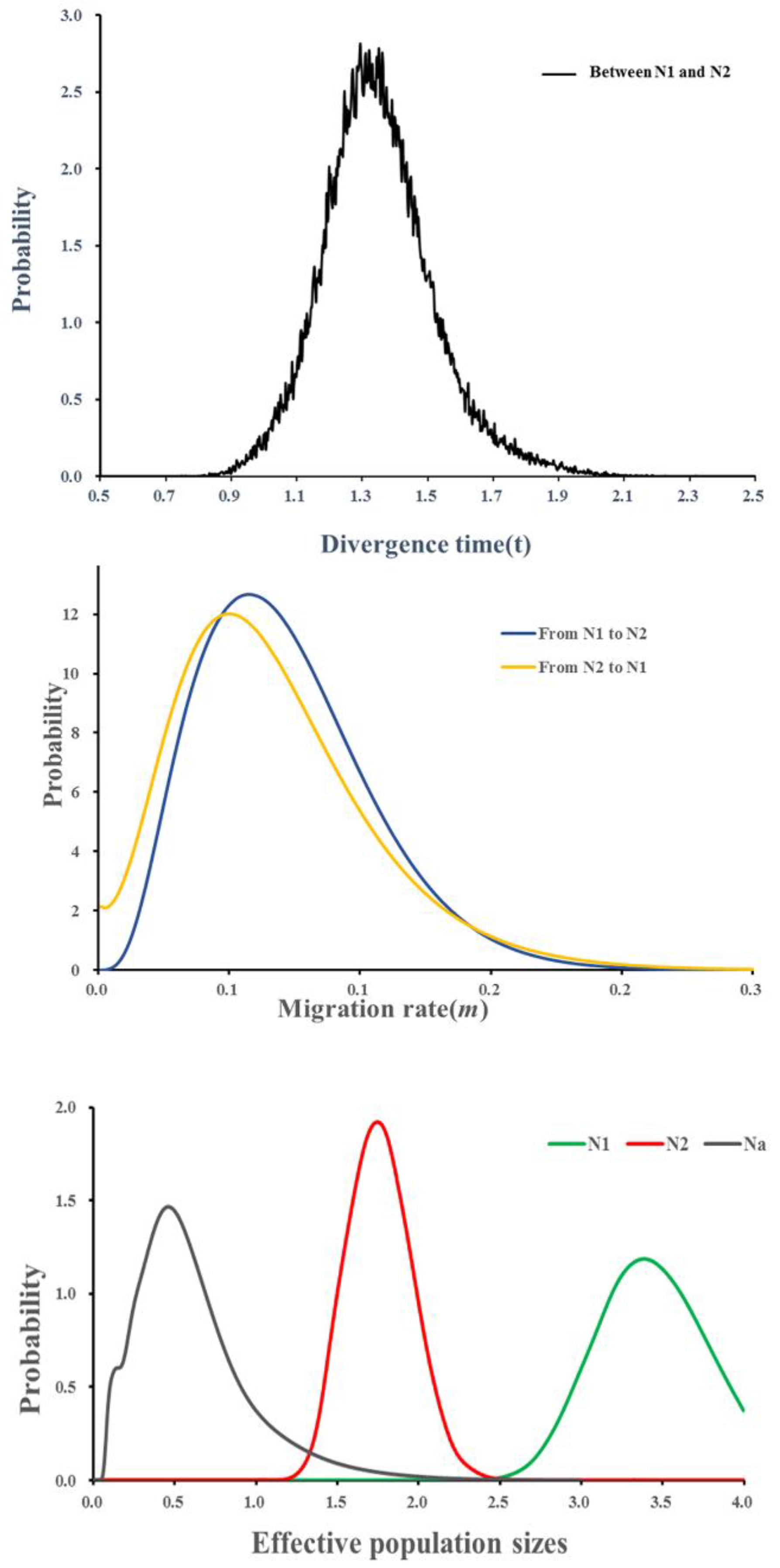
3.3. Population Divergence and Demographic History
3.4. Effects of IBD and IBE on Population Divergence
4. Discussion
4.1. Subtle East–West Differentiation in Asteropyrum
4.2. Historical Factors Responsible for Population Differentiation
4.3. Demographic History and Population Genetic Admixture
4.4. The Relative Contribution of Geographical Isolation and Environmental Heterogeneity to Population Differentiation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Qiu, Y.X.; Fu, C.X.; Comes, H.P. Plant molecular phylogeography in China and adjacent regions: Tracing the genetic imprints of Quaternary climate and environmental change in the world’s most diverse temperate flora. Mol. Phylogenet. Evol. 2011, 59, 225–244. [Google Scholar] [CrossRef]
- Fan, D.M.; Yue, J.; Nie, Z.; Li, Z.; Comes, H.P.; Sun, H. Phylogeography of Sophora davidii (Leguminosae) across the ‘Tanaka-Kaiyong Line’, an important phytogeographic boundary in southwest China. Mol. Ecol. 2013, 22, 4270–4288. [Google Scholar] [CrossRef]
- Tian, S.; Lei, S.Q.; Hu, W.; Deng, L.L.; Li, B.; Meng, Q.L.; Soltis, D.S.; Soltis, P.S.; Fan, D.M.; Zhang, Z.Y. Repeated range expansions and inter-/postglacial recolonization routes of Sargentodoxa cuneata (Oliv.) Rehd. et Wils. (Lardizabalaceae) in subtropical China revealed by chloroplast phylogeography. Mol. Phylogenet. Evol. 2015, 85, 238–246. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, Z.; Fritsch, P.W.; Tian, H.; Yang, A.; Yao, X. Phylogeography and genetic structure of a Tertiary relict tree species, Tapiscia sinensis (Tapisciaceae): Implications for conservation. Ann. Bot. 2015, 116, 727–737. [Google Scholar] [CrossRef]
- Ye, J.; Li, D.; Hampe, A. Differential Quaternary dynamics of evergreen broadleaved forests in subtropical China revealed by phylogeography of Lindera aggregate (Lauraceae). J. Biogeogr. 2019, 46, 1112–1123. [Google Scholar] [CrossRef]
- Kou, Y.X.; Cheng, S.M.; Tian, S.; Li, B.; Fan, D.M.; Chen, Y.; Soltis, D.E.; Soltis, P.S.; Zhang, Z.Y. The antiquity of Cyclocarya paliurus (Juglandaceae) provides new insights into the evolution of relict plants in subtropical China since the late Early Miocene. J. Biogeogr. 2015, 43, 351–360. [Google Scholar] [CrossRef]
- Kou, Y.X.; Zhang, L.; Fan, D.M.; Cheng, S.M.; Li, D.; Hodel, R.G.J.; Zhang, Z.Y. Evolutionary history of a relict conifer, Pseudotaxus chienii (Taxaceae), in southeast China during the late Neogene: Old lineage, young populations. Ann. Bot. 2020, 125, 105–117. [Google Scholar] [CrossRef]
- Zhong, Y.; Yang, A.; Liu, S.; Liu, L.; Li, Y.; Wu, Z.; Yu, F. RAD-Seq data point to a distinct split in Liriodendron (Magnoliaceae) and obvious east–west genetic divergence in L. chinense. Forests 2018, 10, 13. [Google Scholar] [CrossRef]
- Sun, Y.; Moore, M.; Yue, L.; Feng, T.; Chu, H.; Chen, S.; Ji, Y.; Wang, H.; Li, J. Chloroplast phylogeography of the East Asian Arcto-Tertiary relict Tetracentron sinense (Trochodendraceae). J. Biogeogr. 2014, 41, 1721–1732. [Google Scholar] [CrossRef]
- Wang, Y.H.; Jiang, W.W.; Comes, H.P.; Hu, F.S.; Qiu, Y.X.; Fu, C.X. Molecular phylogeography and ecological niche modelling of a widespread herbaceous climber, Tetrastigma hemsleyanum (Vitaceae): Insights into Plio-Pleistocene range dynamics of evergreen forest in subtropical China. New Phytol. 2015, 206, 852–867. [Google Scholar] [CrossRef]
- Fan, L.; Zheng, H.; Milne, R.I.; Zhang, L.; Mao, K. Strong population bottleneck and repeated demographic expansions of Populus adenopoda (Salicaceae) in subtropical China. Ann. Bot. 2018, 121, 665–679. [Google Scholar] [CrossRef]
- Edwards, S.; Beerli, P. Perspective: Gene divergence, population divergence, and the variance in coalescence time in phylogeographic studies. Evolution 2000, 54, 1839–1854. [Google Scholar] [CrossRef]
- Csilléry, K.; Blum, M.G.; Gaggiotti, O.E.; François, O. Approximate Bayesian Computation (ABC) in practice. Trends Ecol. Evol. 2010, 25, 410–418. [Google Scholar] [CrossRef] [PubMed]
- Hey, J. Isolation with migration models for more than two populations. Mol. Biol. Evol. 2009, 27, 905–920. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.J.C.; Zhao, S. Physical Geography of China; Science Press: Beijing, China, 1986. [Google Scholar]
- Shi, Y.; Ren, B.; Wang, J.; Derbyshire, E. Quaternary glaciation in China. Quat. Sci. Rev. 1986, 5, 503–507. [Google Scholar] [CrossRef]
- Wu, Z.Y.; Wang, X.; Liu, F.; Zhou, F. Vegetation of China; Science Press: Beijing, China, 1980. [Google Scholar]
- Favre, A.; Päckert, M.; Pauls, S.U.; Jähnig, S.C.; Uhl, D.; Michalak, I.; Muellner-Riehl, A.N. The role of the uplift of the Qinghai-Tibetan Plateau for the evolution of Tibetan biotas. Biol. Rev. 2014, 90, 236–253. [Google Scholar] [CrossRef]
- Chen, F.; Fu, B.; Xia, J.; Wu, D.; Wu, S.; Zhang, Y.; Sun, H.; Liu, Y.; Fang, X.; Qin, B.; et al. Major advances in studies of the physical geography and living environment of China during the past 70 years and future prospects. Sci. China Earth Sci. 2019, 62, 1665–1701. [Google Scholar] [CrossRef]
- Wang, B.; Clemens, S.C.; Liu, P. Contrasting the Indian and East Asian monsoons: Implications on geologic timescales. Mar. Geol. 2003, 201, 5–21. [Google Scholar] [CrossRef]
- Wang, P.; Clemens, S.; Beaufort, L.; Braconnot, P.; Ganssen, G.; Jian, Z.; Kershaw, P.; Sarnthein, M. Evolution and variability of the Asian monsoon system: State of the art and outstanding issues. Quat. Sci. Rev. 2005, 24, 595–629. [Google Scholar] [CrossRef]
- Song, Y.C. The essential characteristics and main types of the broadleaved evergreen forest in China. Phytocoenologia 1988, 16, 105–123. [Google Scholar]
- Qiu, Y.-X.; Guan, B.-C.; Fu, C.-X.; Comes, H.P. Did glacials and/or interglacials promote allopatric incipient speciation in East Asian temperate plants? Phylogeographic and coalescent analyses on refugial isolation and divergence in Dysosma versipellis. Mol. Phylogenet. Evol. 2009, 51, 281–293. [Google Scholar] [CrossRef]
- Chen, J.M.; Zhao, S.Y.; Liao, Y.Y.; Gichira, A.W.; Gituru, R.W.; Wang, Q.F. Chloroplast DNA phylogeographic analysis reveals significant spatial genetic structure of the relictual tree Davidia involucrata (Davidiaceae). Conserv. Genet. 2014, 16, 583–593. [Google Scholar] [CrossRef]
- Wang, C.; Wang, T.; Su, Y. Phylogeography of Cephalotaxus oliveri (Cephalotaxaceae) in relation to habitat heterogeneity, physical barriers and the uplift of the Yungui Plateau. Mol. Phylogenet. Evol. 2014, 80, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Hao, Z.; Guang, X.; Zhao, C.; Wang, P.; Xue, L.; Zhu, Q.; Yang, L.; Sheng, Y.; Zhou, Y.; et al. Liriodendron genome sheds light on angiosperm phylogeny and species–pair differentiation. Nat. Plants 2018, 5, 18–25. [Google Scholar] [CrossRef]
- Ye, J.W.; Zhang, Y.; Wang, X.J. Phylogeographic breaks and the mechanisms of their formation in the Sino-Japanese floristic region. Chin. J. Plant Ecol. 2017, 41, 1003–1019. [Google Scholar]
- Wang, I.J.; Bradburd, G.S. Isolation by environment. Mol. Ecol. 2014, 23, 5649–5662. [Google Scholar] [CrossRef]
- Wang, I.J. Examining the full effects of landscape heterogeneity on spatial genetic variation: A multiple matrix regression approach for quantifying geographic and ecological isolation. Evolution 2013, 67, 3403–3411. [Google Scholar] [CrossRef]
- Yuan, Q.; Yang, Q.-E. Cytology, palynology, and taxonomy of Asteropyrum and four other genera of Ranunculaceae. Bot. J. Linn. Soc. 2006, 152, 15–26. [Google Scholar] [CrossRef][Green Version]
- Fu, D.Z.; Robinson, O.R.; Asteropyrum, J.R. Drummond & Hutchinson, in Flora of China; Wu, Z.Y., Raven, P.H., Eds.; Science Press: Beijing, China, 2001; Volume 6. [Google Scholar]
- Cheng, S.; Zeng, W.; Wang, J.; Liu, L.; Liang, H.; Kou, Y.; Wang, H.; Fan, D.; Zhang, Z. Species delimitation of Asteropyrum (Ranunculaceae) based on morphological, molecular, and ecological variation. Front. Plant Sci. 2021, 12. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nuclc. Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Hubisz, M.J.; Falush, D.; Stephens, M.; Pritchard, J.K. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Resour. 2009, 9, 1322–1332. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Earl, D.A.; vonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2011, 4, 359–361. [Google Scholar] [CrossRef]
- Rosenberg, N.A. Distruct: A program for the graphical display of population structure. Mol. Ecol. Notes 2004, 4, 137–138. [Google Scholar] [CrossRef]
- Excoffier, L.; Laval, G.; Schneider, S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol. Bioinform. 2005, 1, 47–50. [Google Scholar] [CrossRef]
- Cornuet, J.-M.; Pudlo, P.; Veyssier, J.; Dehne-Garcia, A.; Gautier, M.; Leblois, R.; Marin, J.-M.; Estoup, A. DIYABC v2.0: A software to make approximate Bayesian computation inferences about population history using single nucleotide polymorphism, DNA sequence and microsatellite data. Bioinformatics 2014, 30, 1187–1189. [Google Scholar] [CrossRef]
- Cornuet, J.-M.; Ravigné, V.; Estoup, A. Inference on population history and model checking using DNA sequence and microsatellite data with the software DIYABC (v1.0). BMC Bioinform. 2010, 11, 401. [Google Scholar] [CrossRef]
- Hey, J. The Divergence of Chimpanzee Species and Subspecies as Revealed in Multipopulation Isolation-with-Migration Analyses. Mol. Biol. Evol. 2009, 27, 921–933. [Google Scholar] [CrossRef]
- Hey, J.; Nielsen, R. Integration within the Felsenstein equation for improved Markov chain Monte Carlo methods in population genetics. Proc. Natl. Acad. Sci. USA 2007, 104, 2785–2790. [Google Scholar] [CrossRef] [PubMed]
- Richardson, J.E.; Pennington, R.T.; Pennington, T.D.; Hollingsworth, P.M. Rapid Diversification of a Species-Rich Genus of Neotropical Rain Forest Trees. Science 2001, 293, 2242–2245. [Google Scholar] [CrossRef]
- Wang, L.; Abbott, R.J.; Zheng, W.; Chen, P.; Wang, Y.; Liu, J. History and evolution of alpine plants endemic to the Qinghai-Tibetan Plateau: Aconitum gymnandrum (Ranunculaceae). Mol. Ecol. 2009, 18, 709–721. [Google Scholar] [CrossRef] [PubMed]
- Suchard, M.A.; Lemey, P.; Baele, G.; Ayres, D.L.; Drummond, A.J.; Rambaut, A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018, 4, vey016. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarisation in bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Hijmans, R.J.; Guarino, L.; Cruz, M.; Rojas, E. Computer tools for spatial analysis of plant genetic resources data: 1. DIVA-GIS. Plant Genet. Resour. Newsl. 2001, 127, 15–19. [Google Scholar]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Fan, D.M.; Hu, W.; Li, B.; Morris, A.B.; Zheng, M.; Soltis, U.E.; Soltis, P.S.; Zhang, Z.Y. Idiosyncratic responses of evergreen broad-leaved forest constituents in China to the late Quaternary climate changes. Sci. Rep. 2016, 6, 31044. [Google Scholar] [CrossRef]
- Tzedakis, P.; Raynaud, D.; McManus, J.F.; Berger, A.; Brovkin, V.; Kiefer, T. Interglacial diversity. Nat. Geosci. 2009, 2, 751–755. [Google Scholar] [CrossRef]
- Beaumont, M.A. Approximate bayesian computation in evolution and ecology. Annu. Rev. Ecol. Evol. Syst. 2010, 41, 379–406. [Google Scholar] [CrossRef]
- Clark, M.; House, M.; Royden, L.; Whipple, K.; Burchfiel, B.; Zhang, X.; Tang, W. Late Cenozoic uplift of southeastern Tibet. Geology 2005, 33, 525. [Google Scholar] [CrossRef]
- Wang, E.; Kirby, E.; Furlong, K.P.; Van Soest, M.; Xu, G.; Shi, X.; Kamp, P.; Hodges, K. Two-phase growth of high topography in eastern Tibet during the Cenozoic. Nat. Geosci. 2012, 5, 640–645. [Google Scholar] [CrossRef]
- Thomson, J.R.; Holden, P.B.; Anand, P.; Edwards, N.R.; Porchier, C.A.; Harris, N.B.W. Tectonic and climatic drivers of Asian monsoon evolution. Nat. Commun. 2021, 12, 4022. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.T.; Sun, B.; Zhang, Z.S.; Peng, S.Z.; Xiao, G.Q.; Ge, J.Y.; Hao, Q.Z.; Qiao, Y.S.; Liang, M.Y.; Liu, J.F.; et al. A major reorganization of Asian climate by the early Miocene. Clim. Past 2008, 4, 153–174. [Google Scholar] [CrossRef]
- Clift, P.D.; Hodges, K.; Heslop, D.; Hannigan, R.; Van Long, H.; Calvès, G. Correlation of Himalayan exhumation rates and Asian monsoon intensity. Nat. Geosci. 2008, 1, 875–880. [Google Scholar] [CrossRef]
- Sun, X.; Wang, P. How old is the Asian monsoon system?—Palaeobotanical records from China. Palaeogeogr. Palaeoclim. Palaeoecol. 2005, 222, 181–222. [Google Scholar] [CrossRef]
- Zheng, B.; Xu, Q.; Shen, Y. The relationship between climate change and Quaternary glacial cycles on the Qinghai–Tibetan Plateau: Review and speculation. Quat. Int. 2002, 97, 93–101. [Google Scholar] [CrossRef]
- Cui, Z.J.; Chen, Y.X.; Zhang, W.; Zhou, S.Z.; Zhou, L.P.; Zhang, M.; Li, C.C. Research history, glacial chronology and origins of Quaternary glaciations in China. Quatern. Sci. 2011, 31, 749–764. [Google Scholar]
- Harrison, S.P.; Yu, G.; Takahara, H.; Prentice, I.C. Palaeovegetation (Communications arising): Diversity of temperate plants in East Asia. Nature 2001, 413, 129–130. [Google Scholar] [CrossRef]
- Guan, B.-C.; Fu, C.-X.; Qiu, Y.-X.; Zhou, S.-L.; Comes, H.P. Genetic structure and breeding system of a rare understory herb, Dysosma versipellis (Berberidaceae), from temperate deciduous forests in China. Am. J. Bot. 2010, 97, 111–122. [Google Scholar] [CrossRef]
- An, Z.S.; Kutzbach, J.E.; Prell, W.L.; Porter, S.C. Evolution of Asian monsoons and phased uplift of the Himalaya–Tibetan plateau since Late Miocene times. Nature 2001, 411, 62–66. [Google Scholar] [CrossRef]
- Wang, H.-W.; Ge, S. Phylogeography of the endangered Cathaya argyrophylla (Pinaceae) inferred from sequence variation of mitochondrial and nuclear DNA. Mol. Ecol. 2006, 15, 4109–4122. [Google Scholar] [CrossRef]
- Lei, M.; Wang, Q.; Wu, Z.J.; López-Pujol, J.; Li, D.Z.; Zhang, Z.Y. Molecular phylogeography of Fagus engleriana (Fagaceae) in subtropical China: Limited admixture among multiple refugia. Tree Genet. Genomes 2012, 8, 1203–1212. [Google Scholar] [CrossRef]
- Wang, J.; Gao, P.; Kang, M.; Lowe, A.J.; Huang, H. Refugia within refugia: The case study of a canopy tree Eurycorymbus cavaleriei in subtropical China. J. Biogeogr. 2009, 36, 2156–2164. [Google Scholar] [CrossRef]
- Zhang, Z.Y.; Wu, R.; Wang, Q.; Zhang, Z.R.; López-Pujol, J.; Fan, D.M.; Li, D.Z. Comparative phylogeography of two sympatric beeches in subtropical China: Species-specific geographic mosaic of lineages. Ecol. Evol. 2013, 3, 4461–4472. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.H.; Wang, I.; Comes, H.P.; Peng, H.; Qiu, Y.X. Contributions of historical and contemporary geographic and environmental factors to phylogeographic structure in a Tertiary relict species, Emmenopterys henryi (Rubiaceae). Sci. Rep. 2016, 6, 24041. [Google Scholar] [CrossRef]
- Boufford, D. Biodiversity hotspot: China’s Hengduan Mountains. Arnoldia 2014, 72, 24–35. [Google Scholar]
- Cao, Y.; Zhu, S.; Chen, J.; Comes, H.P.; Wang, I.J.; Chen, L.; Sakaguchi, S.; Qiu, Y. Genomic insights into historical population dynamics, local adaptation, and climate change vulnerability of the East Asian Tertiary relict Euptelea (Eupteleaceae). Evol. Appl. 2020, 13, 2038–2055. [Google Scholar] [CrossRef] [PubMed]

| Primers | Sequences(5′–3′) | N | L | S | π | θw | Nh | Hd | D | Rm |
|---|---|---|---|---|---|---|---|---|---|---|
| psbA- trnH | F:GTTATGCATGAACGTAATGCTC | 323 | 219 | 13 | 0.011 | 0.009 | 10 | 0.849 | 0.491 | 1 |
| R:CGCGCATGGTGGATTCACAAATC | ||||||||||
| X14 | F:GTTTCGGGTGTTCTTGTT | 162 | 388 | 19 | 0.005 | 0.008 | 27 | 0.807 | −0.837 | 4 |
| R:CATCTTCTTGGCTCGTAG | ||||||||||
| X17 | F:AACCTTAGAACTCCGATTA | 146 | 368 | 49 | 0.016 | 0.021 | 40 | 0.953 | −0.747 | 7 |
| R:CCTGATGATACCCTACTTT | ||||||||||
| X23 | F:TGAGCGGTTCCTTGTCCC | 156 | 411 | 33 | 0.007 | 0.013 | 28 | 0.914 | −1.316 | 1 |
| R:CAGCGTGATGTCTTACGATGC | ||||||||||
| X35 | F:CCGCTTTGCCACAGATTA | 162 | 253 | 14 | 0.009 | 0.009 | 15 | 0.820 | −0.047 | 1 |
| R:TGCTTTACCAGCCGTTGA | ||||||||||
| X47 | F:ACAACATCCCAATCAGCA | 160 | 231 | 23 | 0.017 | 0.017 | 23 | 0.881 | −0.267 | 0 |
| R:ACAACCCACAACACCAGA | ||||||||||
| X63 | F:CGTCGCCCAGTAGTATCTT | 159 | 389 | 23 | 0.004 | 0.009 | 16 | 0.807 | −1.485 | 0 |
| R:ACATTCATCGTTCGCTTG | ||||||||||
| X82 | F:ATGAGAAGGGCACCAAAT | 159 | 308 | 17 | 0.007 | 0.009 | 18 | 0.867 | −0.431 | 1 |
| R:AAGCAAACAGAGGGAAGC | ||||||||||
| X89 | F:CGAGTAGCCTTTACGACG | 157 | 432 | 39 | 0.010 | 0.014 | 35 | 0.927 | −0.881 | 3 |
| R:GAGTGGAACCCAACCTTT | ||||||||||
| X125 | F:CAGGTGCGGTCATAGTTG | 134 | 206 | 26 | 0.008 | 0.020 | 23 | 0.794 | −1.802 * | 1 |
| R:CGAGTCGCCTTGAGTTTT | ||||||||||
| X130 | F:GGGAAGCCGTAGACTCAC | 160 | 275 | 21 | 0.006 | 0.012 | 21 | 0.780 | −1.419 | 0 |
| R:CCCGACAAGGCATAGAAC |
| Group | π | Hd | Hs | Fst | Nm | D | Fs |
|---|---|---|---|---|---|---|---|
| Group_1 | 0.005 | 0.976 | 0.704 | 0.531 | 0.630 | 0.332 | −1.222 |
| Group_2 | 0.005 | 0.957 | 0.634 | 0.730 | 0.490 | 0.617 | −0.840 |
| Asteropyrum | 0.007 | 0.988 | 0.689 | 0.872 | 0.570 | 0.599 | −0.927 |
| Source of Variation | d.f. | Sum of Squares | Variance Components | Percentage of Variation | Fst |
|---|---|---|---|---|---|
| Among groups | 1 | 2.3370 | 0.0001 | 0.18 | |
| Among populations within groups | 23 | 46.5690 | 0.1668 | 33.36 | |
| Within populations | 231 | 76.7670 | 0.3323 | 66.46 | |
| Total | 255 | 125.6730 | 0.5000 | 0.3342 * |
| Group | Parameter | Mean | Median | Mode | 95% HPD |
|---|---|---|---|---|---|
| Na | 1.57 × 106 | 1.59 × 106 | 1.59 × 106 | 0.47–2.61 × 106 | |
| N1 | 2.32 × 106 | 2.40 × 106 | 2.55 × 106 | 1.48–2.86 × 106 | |
| N2 | 1.61 × 106 | 1.63 × 106 | 1.85 × 106 | 0.70–2.51 × 106 | |
| 3 | N3 | 3.18 × 105 | 2.21 × 105 | 1.65 × 105 | 0.71–9.81 × 105 |
| T2 (years) | 1.23 × 106 | 1.23 × 106 | 1.20 × 106 | 0.48–1.90 × 106 | |
| T1 (years) | 4.77 × 105 | 4.11 × 105 | 2.38 × 105 | 0.15–1.08 × 106 | |
| μ | 3.47 × 10−9 | 2.84 × 10−9 | 1.93 × 10−9 | 1.21–7.56 × 10−9 | |
| ra | 2.97 × 10−1 | 2.37 × 10−1 | 1.49 × 10−1 | 0.35–7.63 × 10−1 |
| Par. | θ1 | θ2 | θa | m1 | m2 | t | N1 | N2 | Na | 2N1m1 | 2N2m2 | T |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | 3.398 | 1.761 | 0.615 | 0.072 | 0.067 | 1.349 | 7.53 × 105 | 3.90 × 105 | 1.36 × 105 | 0.122 | 0.059 | 1.20 × 106 |
| HPD95Lo | 2.886 | 1.377 | 0.068 | 0.015 | 0.000 | 1.011 | 6.39 × 105 | 3.05 × 105 | 1.50 × 104 | 0.022 | 0.000 | 8.96 × 105 |
| HPD95Hi | 3.998 | 2.152 | 1.327 | 0.135 | 0.137 | 1.704 | 8.86 × 105 | 4.77 × 105 | 2.94 × 105 | 0.270 | 0.147 | 1.51 × 106 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, S.; Zeng, W.; Fan, D.; Liang, H.; Yang, Y.; Kou, Y.; Zhang, Z. Subtle East–West Phylogeographic Break of Asteropyrum (Ranunculaceae) in Subtropical China and Adjacent Areas. Diversity 2021, 13, 627. https://doi.org/10.3390/d13120627
Cheng S, Zeng W, Fan D, Liang H, Yang Y, Kou Y, Zhang Z. Subtle East–West Phylogeographic Break of Asteropyrum (Ranunculaceae) in Subtropical China and Adjacent Areas. Diversity. 2021; 13(12):627. https://doi.org/10.3390/d13120627
Chicago/Turabian StyleCheng, Shanmei, Weidong Zeng, Dengmei Fan, Hua Liang, Yi Yang, Yixuan Kou, and Zhiyong Zhang. 2021. "Subtle East–West Phylogeographic Break of Asteropyrum (Ranunculaceae) in Subtropical China and Adjacent Areas" Diversity 13, no. 12: 627. https://doi.org/10.3390/d13120627
APA StyleCheng, S., Zeng, W., Fan, D., Liang, H., Yang, Y., Kou, Y., & Zhang, Z. (2021). Subtle East–West Phylogeographic Break of Asteropyrum (Ranunculaceae) in Subtropical China and Adjacent Areas. Diversity, 13(12), 627. https://doi.org/10.3390/d13120627






