Molecular Diversity of Nematode Parasites in Afrotropical Reed Frogs (Hyperolius spp.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Morphological Identification of Nematode Vouchers
2.3. Nucleic Acid Extraction and Polymerase Chain Reactions
2.4. Sequencing and Phylogenetic Analysis
3. Results
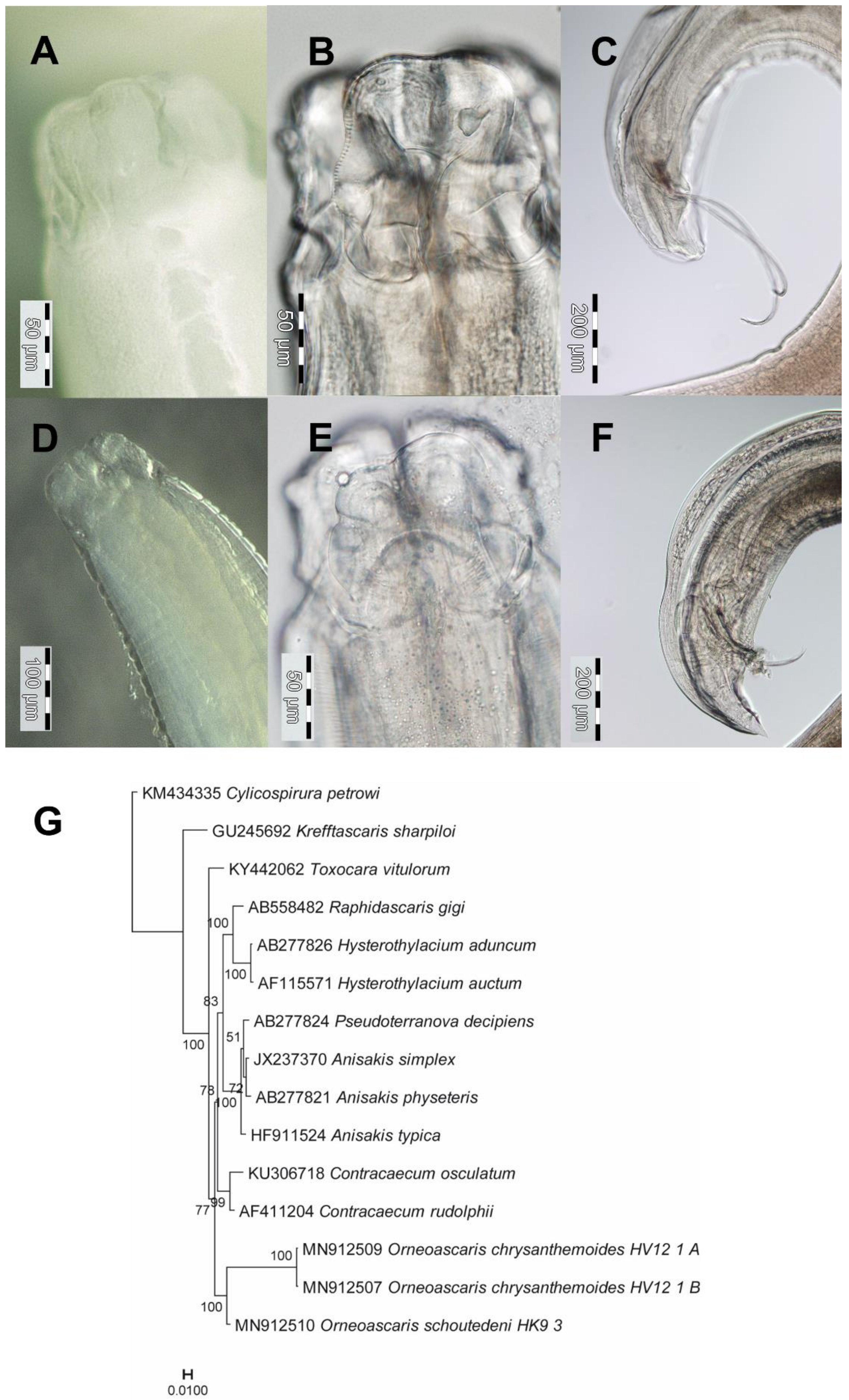
3.1. Orneoascaris chrysanthemoides (Ascaridoidea): Morphological, Ecological and Molecular Features
3.2. Orneoascaris schoutedeni (Ascaridoidea): Morphological, Ecological and Molecular Features
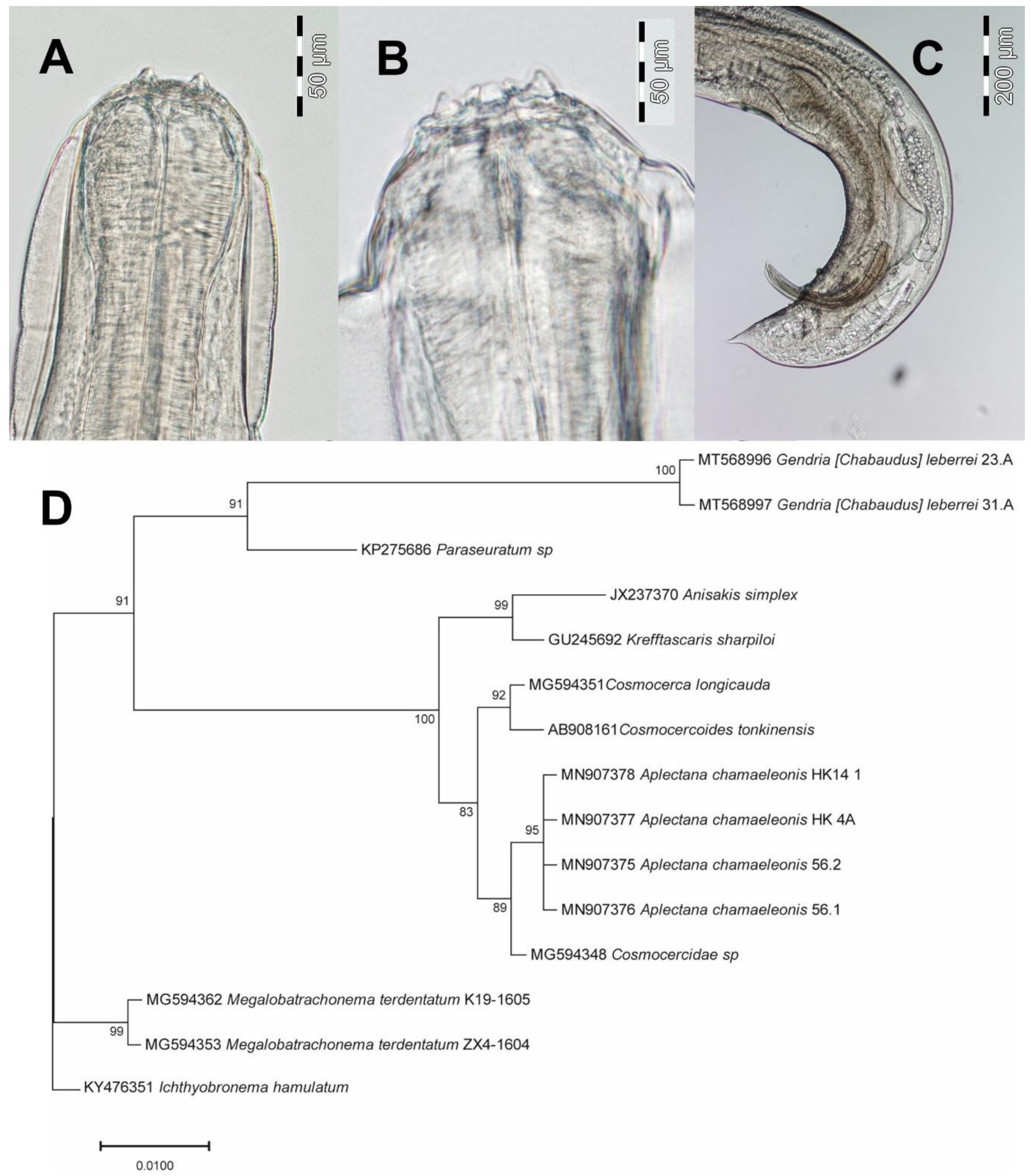
3.3. Gendria [Chabaudus] leberrei (Seuratoidea): Morphological, Ecological and Molecular Features
3.4. Aplectana Chamaeleonis (Cosmocercoidea): Morphological, Ecological and Molecular Features
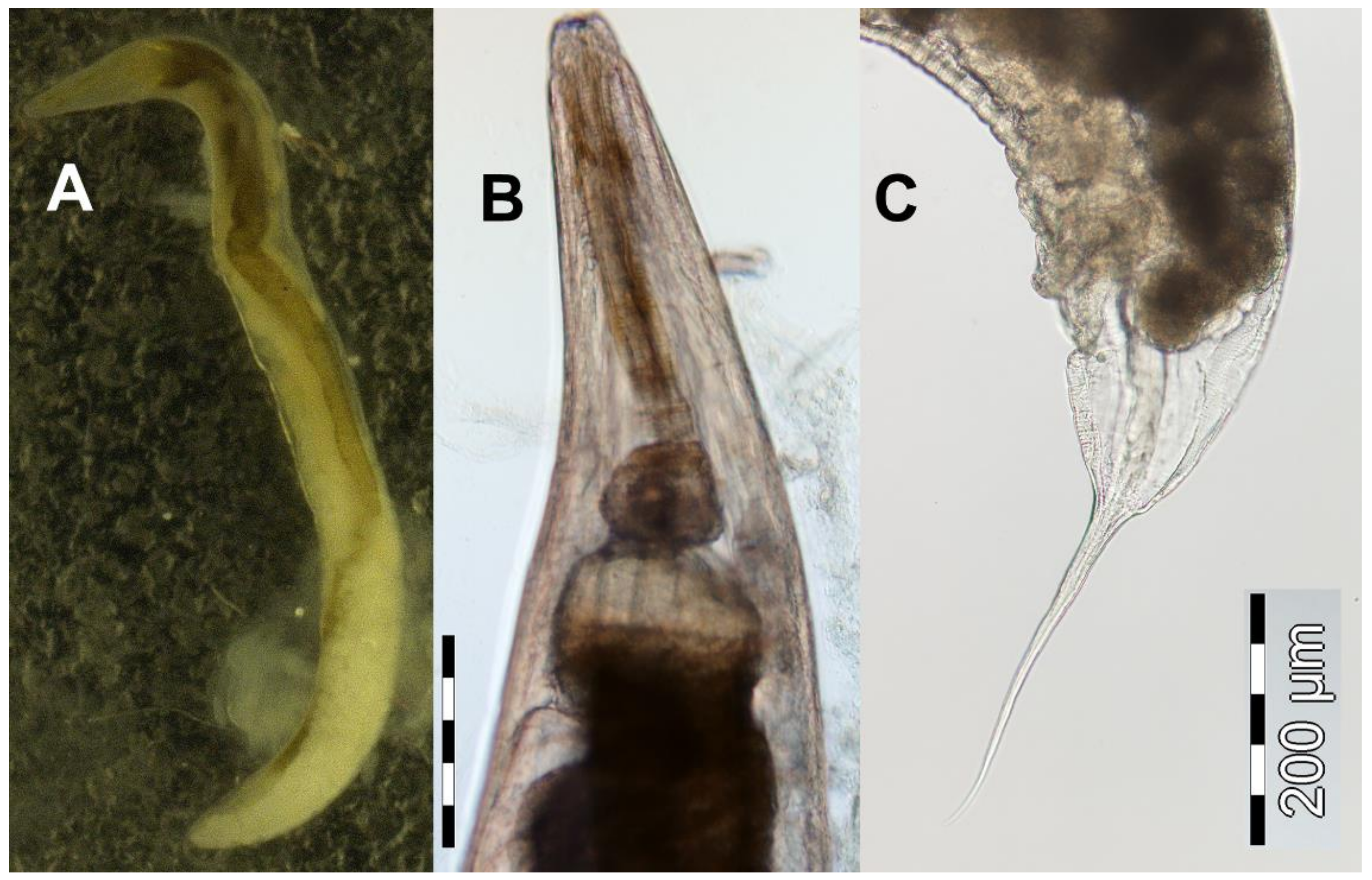
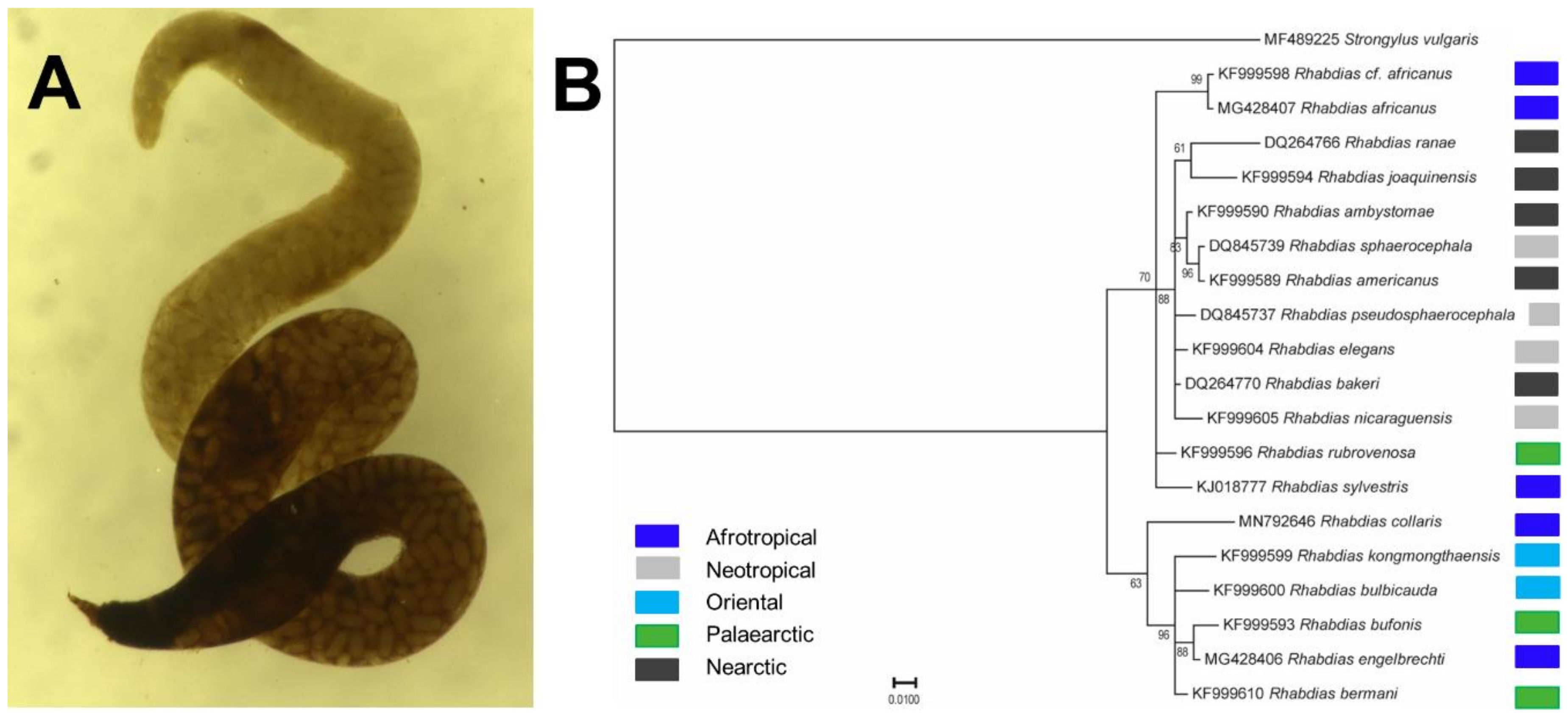
3.5. Rhabdias collaris (Rhabditoidea): Morphological, Ecological and Molecular Features
4. Discussion
4.1. Host–Parasite Ecology
4.2. Phylogenetic Implications
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Martinez-Arce, A.; De Jesus-Nayarrete, A.; Leasi, F. DNA Barcoding for Delimitation of Putative Mexican Marine Nematodes Species. Diversity 2020, 12, 107. [Google Scholar] [CrossRef]
- Hugot, J.P.; Baujard, P.; Morand, S. Biodiversity in helminths and nematodes as a field of study: An overview. Nematology 2001, 3, 199–208. [Google Scholar] [CrossRef]
- Dickson, D.W.; Chen, S.; Chen, Z. Nematology: Advances and Perspectives. Nematode Morphology, Physiology and Ecology; CABI: Wallingford, UK, 2004. [Google Scholar]
- Dorris, M.; De Ley, P.; Blaxter, M.L. Molecular analysis of nematode diversity and the evolution of parasitism. Parasitol. Today 1999, 15, 188–193. [Google Scholar] [CrossRef]
- Powers, T.; Harris, T.; Higgins, R.; Mullin, P.; Sutton, L.; Powers, K. MOTUs, morphology, and biodiversity estimation: A case study using nematodes of the suborder Criconematina and a conserved 18S DNA barcode. J. Nematol. 2011, 43, 35–48. [Google Scholar]
- Sinsch, U.; Heneberg, P.; Těšínský, M.; Balczun, C.; Scheid, P. Helminth endoparasites of the smooth newt Lissotriton vulgaris: Linking morphological identification and molecular data. J. Helminthol. 2019, 93, 332–341. [Google Scholar] [CrossRef] [PubMed]
- Anderson, R.C.; Chabaud, A.G.; Willmott, S. Keys to the Nematode Parasites of Vertebrates: Archival Volume; CABI: Wallingford, UK, 2009. [Google Scholar]
- Gibbons, L.M. Keys to the Nematode Parasites of Vertebrates: Supplementary Volume; CABI: Wallingford, UK, 2010; Volume 10. [Google Scholar]
- Aho, J.M. Helminth communities of amphibians and reptiles: Comparative approaches to understanding patterns and processes. In Parasite Communities: Patterns and Processes; Esch, G.W., Bush, A.O., Aho, J.M., Eds.; Springer Netherlands: Dordrecht, The Netherlands, 1990; pp. 157–195. [Google Scholar] [CrossRef]
- Baker, M.R. Nematode parasitism in amphibians and reptiles. Can. J. Zool. 1984, 62, 747–757. [Google Scholar] [CrossRef]
- Paredes-León, R.; Garcia-Prieto, L.; Guzman-Cornejo, C.; Leon-Regagnon, V.; Perez, T.M. Metazoan parasites of Mexican amphibians and reptiles. Zootaxa 2008, 1904, 1–166. [Google Scholar] [CrossRef]
- Halajian, A.; Bursey, C.R.; Goldberg, S.R.; Luus-Powell, W. Helminths of Six Species of Anurans from the Republic of South Africa: Amietophrynus garmani, Amietophrynus gutturalis, Amietophrynus maculatus, Schismaderma carens (Bufonidae), Amietia angolensis, and Strongylopus grayii (Pyxicephalidae), with a Review of South African Anuran Helminths. Comp. Parasitol. 2013, 80, 80–95. [Google Scholar] [CrossRef]
- Finnerty, P.B.; Shine, R.; Brown, G.P. The costs of parasite infection: Effects of removing lungworms on performance, growth and survival of free-ranging cane toads. Funct. Ecol. 2018, 32, 402–415. [Google Scholar] [CrossRef]
- Goater, C.P.; Semlitsch, R.D.; Bernasconi, M.V. Effects of body size and parasite infection on the locomotory performance of juvenile toads. Bufo bufo. Oikos 1993, 66, 129–136. [Google Scholar] [CrossRef]
- Pizzatto, L.; Shine, R. Lungworm infection modifies cardiac response to exercise in cane toads. J. Zool. 2012, 287, 150–155. [Google Scholar] [CrossRef]
- Kelehear, C.; Webb, J.K.; Shine, R. Rhabdias pseudosphaerocephala infection in Bufo marinus: Lung nematodes reduce viability of metamorph cane toads. Parasitology 2009, 136, 919–927. [Google Scholar] [CrossRef]
- Leary, C.J.; Ralicki, H.F.; Laurencio, D.; Crocker-Buta, S.; Malone, J.H. Assessing the links among environmental contaminants, endocrinology, and parasites to understand amphibian declines in montane regions of Costa Rica. PLoS ONE 2018, 13, e0191183. [Google Scholar] [CrossRef] [PubMed]
- Aisien, M.; Sampson, S.A.; Amuzie, C. Anuran parasites from three biotopes in Rivers State, Nigeria. Niger. J. Parasitol. 2017, 38, 129–135. [Google Scholar] [CrossRef]
- Aisien, M.S.O.; Ugbomeh, A.P.; Awharitoma, A.O. Parasitic infections of anurans from a freshwater creek community in Delta State, Niger Delta of Nigeria. Helminthologia 2017, 54, 132–144. [Google Scholar] [CrossRef][Green Version]
- McAllister, C.T.; Bursey, C.R.; Freed, P.S. Helminth parasites (Cestoidea, Nematoda, Pentastomida) of selected herpetofauna from Cameroon, West Africa. Acta Parasitol. 2010, 55, 90–93. [Google Scholar] [CrossRef]
- Skrjabin, K. Parasitic trematodes and nematodes collected by the expedition of Prof. V. Dogiel and I. Sokolov in British East Africa. In Scientific Results of the Zoological Expedition to British East Africa and Uganda Made by Prof. V. Dogiel and I. Sokolov in the Year 1914; Palala Press: Mishawaka, IN, USA, 1916; Volume 1, pp. 99–157. [Google Scholar]
- Junker, K.; Lhermitte-Vallarino, N.; Barbuto, M.; Ineich, I.; Wanji, S.; Bain, O. New species of Rhabdias (Nematoda: Rhabdiasidae) from afrotropical anurans, including molecular evidence and notes on biology. Folia Parasit. (Praha) 2010, 57, 47–61. [Google Scholar] [CrossRef] [PubMed]
- Kuzmin, Y.; Halajian, A.; Tavakol, S.; Luus-Powell, W.J.; Tkach, V.V. Description and phylogenetic position of a new species of Rhabdias Stiles et Hassall, 1905 (Nematoda: Rhabdiasidae) from the banded rubber frog, Phrynomantis bifasciatus (Smith) (Amphibia: Microhylidae), in South Africa. Folia Parasit. (Praha) 2017, 64, 35. [Google Scholar] [CrossRef]
- Sinsch, U.; Lümkemann, K.; Rosar, K.; Schwarz, C.; Dehling, J.M. Acoustic niche partitioning in an anuran community inhabiting an Afromontane wetland (Butare, Rwanda). Afr. Zool. 2012, 47, 60–73. [Google Scholar] [CrossRef]
- Tumushimire, L.; Mindje, M.; Sinsch, U.; Dehling, M.J. Anuran diversity of cultivated wetlands in Rwanda: Melting pot of generalists? Salamandra 2020, 56, 99–112. [Google Scholar]
- Bush, A.; Lafferty, K.; Lotz, J.; Shostak, A. Parasitology meets ecology on its own terms: Margolis et al. revisited. J. Parasitol. 1997, 83, 575–583. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef]
- Nicholas, K.B. GeneDoc: Analysis and visualization of genetic variation. Embnew. News 1997, 4, 14. [Google Scholar]
- Sinnappah, N.D.; Lim, L.-H.S.; Rohde, K.; Tinsley, R.; Combes, C.; Verneau, O. A paedomorphic parasite associated with a neotenic amphibian host: Phylogenetic evidence suggests a revised systematic position for Sphyranuridae within anuran and turtle Polystomatoineans. Mol. Phyl. Evol. 2001, 18, 189–201. [Google Scholar] [CrossRef] [PubMed]
- Kaci-Chaouch, T.; Verneau, O.; Desdevises, Y. Host specificity is linked to intraspecific variability in the genus Lamellodiscus (Monogenea). Parasitology 2008, 135, 607–616. [Google Scholar] [CrossRef]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Baylis, H.A. XXXVI.—On a further collection of parasitic worms from the Belgian Congo. Ann. Mag. Nat. Hist. 1940, 5, 401–417. [Google Scholar] [CrossRef]
- Bain, O.; Philippon, B. Recherche sur des larves de nematodes Ascaridida trouvees chez Simulium damnosum. Ann. Parasitol. Hum. Comp. 1969, 44, 147–156. [Google Scholar] [CrossRef]
- Baylis, H.A. XL.—Some parasitic Nematodes from the Uluguru and Usambara Mountains, Tanganyika Territory. Ann. Mag. Nat. Hist. 1929, 4, 372–381. [Google Scholar] [CrossRef]
- Baker, M.R. Rhabdias collaris n. sp. (Nematoda: Rhabdiasidae) from frogs of Tanzania. Syst. Parasitol. 1987, 9, 199–201. [Google Scholar] [CrossRef]
- Moravec, F.; Baruš, V. Some nematode parasites from amphibians and reptiles from Zambia and Uganda. Acta Soc. Zool. Bohem. 1990, 54, 177–192. [Google Scholar]
- Sprent, J. Ascaridoid nematodes of amphibians and reptiles: Orneoascaris. Ann. Parasitol. Hum. Comp. 1985, 60, 33–55. [Google Scholar] [CrossRef]
- Goldberg, S.R.; Bursey, C.R.; Greenbaum, E. Nematodes of Leptopelis christyi and Leptopelis karissimbensis (Anura: Arthroleptidae), from the Democratic Republic of Congo. Alytes 2016, 33, 21–24. [Google Scholar]
- Inglis, W.; Ogden, C. Chabaudus chabaudi gen. et sp. nov. from a fresh-water fish in Sierra Leone (Nematoda: Seuratoidea). Rev. Zool. Bot. Afr. 1965, 71, 171–176. [Google Scholar]
- Moravec, F.; Jirků, M. Some nematodes from freshwater fishes in central Africa. Folia Parasit. 2017, 64, 33. [Google Scholar] [CrossRef] [PubMed]
- Jackson, J.A.; Tinsley, R.C.; Du Preez, L.H.; Henderson, A.C. A redescription of Chabaudus leberrei (Bain & Philippon, 1969) (Nematoda: Seuratoidea) from Xenopus spp. in Swaziland. Syst. Parasitol. 2001, 50, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Durette-Desset, M.-C.; Batcharov, G. Deux nematodes parasites d’amphibiens du Togo. Ann. Parasitol. Hum. Comp. 1974, 49, 567–576. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pike, A.W. Helminth parasites of the amphibians Dicroglossus occipitalis (Günther) and Bufo regularis Reuss, in Khartoum, Republic of Sudan. J. Nat. Hist. 1979, 13, 337–376. [Google Scholar] [CrossRef]
- Baker, M.R. Revision of Old World species of the genus Aplectana Railliet & Henry, 1916 (Nematoda, Cosmocercidae). Bull. Mus. Natl. Hist. Nat. 1980, 2, 955–998. [Google Scholar]
- Chen, P. Aplectana chamaeleonis (Baylis, 1929) from a frog and a freshwater fish in Ethiopia. Ann. Parasitol. Hum. Comp. 1966, 9, 333–336. [Google Scholar] [CrossRef]
- Goldberg, S.R.; Bursey, C.R. Helminths from eight species of African skinks (Trachylepis: Scincidae). Com. Parasitol. 2010, 77, 236–241. [Google Scholar] [CrossRef]
- Rasheed, S. Some parasitic nematodes from the Cameroons (W. Africa). J. Helminthol. 1965, 39, 67–100. [Google Scholar] [CrossRef] [PubMed]
- Aisien, S.O.; Ugbo, A.D.; Ilavbare, A.N.; Ogunbor, O. Endoparasites of amphibians from South-Western Nigeria. Acta Parasitol. 2001, 46, 299–305. [Google Scholar]
- Aisien, M.S.O.; Ogoannah, S.O.; Imasuen, A.A. Helminth parasites of amphibians from a rainforest reserve in southwestern Nigeria. Afr. Zool. 2009, 44, 1–7. [Google Scholar] [CrossRef]
- Amuzie, C.C.; Ekerette, I.B. Variations in parasitic helminths of Amietophrynus species collected from similar habitats, Rivers State, Nigeria. Int. J. Biosci. 2017, 10, 76–79. [Google Scholar]
- Amuzie, C.C.; Aisien, M. Parasitic helminths of some Ptychadena species from Ogoniland, Niger Delta, Nigeria. NISEB J. 2018, 18, 68–71. [Google Scholar]
- Okere, S.; Joseph, F.; Amuzie, C. Endo-parasitic helminths of amphibians, Ptychadena mascareniensis and Ptychadena pumilio at Rumuji-emohua, Rivers state, Nigeria. Afr. J. Environ. Sci. 2019, 2, 71–76. [Google Scholar]
- Pessier, A.P.; Baitchman, E.J.; Crump, P.; Wilson, B.; Griffith, E.; Ross, H. Causes of mortality in anuran amphibians from an ex situ survival assurance colony in Panama. Zoo Biol. 2014, 33, 516–526. [Google Scholar] [CrossRef]
- Wiley, M.; Wu, J.; Dawson, B. A comparison of two treatments for nematode infections in the Túngara Frog, Engystomops pustulosus. J. Herpetol. Med. Surg. 2009, 19, 21–22. [Google Scholar] [CrossRef]
- Moretti, E.H.; Madelaire, C.B.; Silva, R.J.; Mendonça, M.T.; Gomes, F.R. The relationships between parasite intensity, locomotor performance, and body condition in adult toads (Rhinella icterica) from the wild. J. Herpetol. 2014, 48, 277–283. [Google Scholar] [CrossRef]
- Koprivnikar, J.; Marcogliese, D.J.; Rohr, J.R.; Orlofske, S.A.; Raffel, T.R.; Johnson, P.T.J. Macroparasite infections of amphibians: What can they tell us? Ecohealth 2012, 9, 342–360. [Google Scholar] [CrossRef] [PubMed]
- McElwain, A.; Warren, M.B.; Pereira, F.B.; Ksepka, S.P.; Bullard, S.A. Pathobiology and first report of larval nematodes (Ascaridomorpha sp.) infecting freshwater mussels (Villosa nebulosa, Unionidae), including an inventory of nematode infections in freshwater and marine bivalves. Int. J. Parasitol. 2019, 10, 41–58. [Google Scholar] [CrossRef] [PubMed]
- Sokolov, S.; Malysheva, S. Molecular characterization of Ichtyobronema hamulatum (Moulton, 1931) (Nematoda: Quimperiidae), a common parasite of burbot Lota lota (Linnaeus) (Actinopterygii: Lotidae). Helminthologia 2017, 54, 183–188. [Google Scholar] [CrossRef]
- Bursey, C.R.; Goldberg, S.R.; Kraus, A.F. New species of Aplectana (Nematoda: Cosmocercidae) in Sphenomorphus pratti from Papua New Guinea. J. Parasitol. 2011, 97, 654–660. [Google Scholar] [CrossRef]
- Bursey, C.R.; Goldberg, S.R.; Grismer, L.L. A new species of Aplectana (Nematoda, Cosmocercidae) in Goniurosaurus bawanglingensis (Squamata, Eublepharidae), from Hainan Province, China. Acta Parasitol. 2018, 63, 190–197. [Google Scholar] [CrossRef]
- Sou, S.K.; Sow, K.K.; Nandi, A.P. Aplectana hoplobatrachusia sp nov (Nematoda: Cosmocercidae) in Hoplobatrachus crassus (Jerdon, 1853) (Anura: Dicroglossidae) from Birbhum District, West Bengal, India. Zootaxa 2018, 4472, 194–200. [Google Scholar] [CrossRef]
| Gene | Primer | Sequence | Reference |
|---|---|---|---|
| 18S-ITS1-5.8S | S1 | 5′-ATTCCGATAACGAACGAGACT-3′ | [30] |
| IR8 | 5′-GCTAGCTGCGTTCTTCATCGA-3′ | [31] | |
| COI | LCO1490 | 5′-GGTCAACAAATCATAAAGATATTGG-3′ | [32] |
| HCO2198 | 5′-TGATTTTTTGGTCACCCTGAAGTTTA-3′ | [32] |
| Species | Locus | |
|---|---|---|
| Nuclear Ribosomal Sequence | COI | |
| Rhabdias collaris | MN792646 | MN927222 |
| Orneoascaris chrysanthemoides HV12_1_A | MN912509 | - |
| Orneoascaris chrysanthemoides HV12_1_B | MN912507 | - |
| Orneoascaris schoutedeni | MN912510 | - |
| Gendria [Chabaudus] leberrei | MT568996 | - |
| Gendria [Chabaudus] leberrei | MT568997 | - |
| Aplectana chamaeleonis 56.2 | MN907375 | - |
| Aplectana chamaeleonis 56.1 | MN907376 | - |
| Aplectana chamaeleonis HK_4A | MN907377 | - |
| Aplectana chamaeleonis HK14_1 | MN907378 | - |
| Orneoascaris chrysanthemoides | Orneoascaris schoutedeni | Gendria [Chabaudus] leberrei | Aplectana chamaeleonis | Rhabdias collaris | |
|---|---|---|---|---|---|
| Locality | Huye | Musanze | Cyamudongo | Huye | Huye |
| Hyperolius kivuensis (n = 115) Infection site N specimens Prevalence [%] Infection intensity Range | intestines (s, l) Ntot = 24 3.5 6.0 ± 7.1 1–16 | intestines (s, l) Ntot = 3 0.9 3 - | - | intestines (l) Ntot = 23 8.7 2.3 ± 2.0 1–7 | - |
| Hyperolius parallelus (n = 45) Infection site N specimens Prevalence [%] Infection intensity Range | - | - | intestines (s, l) Ntot = 6 6.7 2.0 ± 1.7 1–4 | - | - |
| Hyperolius viridi-flavus (n = 100) Infection site N specimens Prevalence [%] Infection intensity Range | Intestines (s, l) Ntot = 3 2.0 1.5 ± 0.7 1–2 | - | - | - | lungs Ntot = 1 0.7 1 1 |
| Species | R. collaris 927222 * | R. engelbrechti MG428410 * | R. picardiae FN434095 * | R. tanyai FN434107 * | R. vencesi FN434104 * | R. africanus MG428411 * |
|---|---|---|---|---|---|---|
| R. collaris | 0 | 89 | 89 | 88 | 87 | 86 |
| R. engelbrechti | 532 | 0 | 91 | 88 | 90 | 89 |
| R. picardiae | 536 | 546 | 0 | 87 | 90 | 87 |
| R. tanyai | 526 | 530 | 523 | 0 | 85 | 87 |
| R. vencesi | 523 | 537 | 540 | 509 | 0 | 87 |
| R. africanus | 515 | 533 | 524 | 522 | 524 | 0 |
| Orneoascaris chrysanthemoides | Orneoascaris schoutedeni | Gendria [Chabaudus] leberrei | Aplectana chamaeleonis | Rhabdias collaris | |
|---|---|---|---|---|---|
| Type host | Sclerophrys spec. (Toad) | Varanusniloticus (Nile monitor) | Sclerophrysregularis (Toad) | Kinyongia matschiei (Chamaeleon) | Leptopelisvermiculatus (Frog) |
| Amphibian hosts | Hyperolius kivuensis H. viridiflavus Amietia nutti Arthroleptis stenodactylus Breviceps macrodactylus Leptopelis christyi L. karissimbensis Sclerophrys regularis S. maculatus S. superciliaris Trichobatrachus robustus Xenopus laevis | Hyperolius kivuensis | Hyperolius parallelus Aubria subsigillata Hoplobatrachus occipitalis Ptychadena mascareniensis P. pumilio Sclerophrys camerunensis S. maculatus Xenopus laevis X. muelleri | Hyperolius kivuensis Amietia angolensis Bufotes viridis Cacosternum capense Epidalea calamita Hylarana spec. Leptopelis christyi, L. karissimbensis Ptychadena oxyrhynchus Scotobleps gabonicus Sclerophrys rangeri Sclerophrys spp. Schismadera carens Trichobatrachus robustus Vandijkophrynus angusticeps | Hyperolius viridiflavus Hyperolius spec. |
| Reptile hosts | Varanus niloticus Chameleo dilepis Trachylepis affinis Kinixys erosa Natrix olivaceus Bitis cornuta Dispholidus typus Causus rhombeatus Crocodylus Niloticus | Varanus exanthematicus | - | - | - |
| References | [21,37,38,39,47,48] | [33,38] | [18,34,42,49,50,51,52,53] | [12,35,39,45,46] | [36] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sinsch, U.; Dehling, J.M.; Scheid, P.; Balczun, C. Molecular Diversity of Nematode Parasites in Afrotropical Reed Frogs (Hyperolius spp.). Diversity 2020, 12, 265. https://doi.org/10.3390/d12070265
Sinsch U, Dehling JM, Scheid P, Balczun C. Molecular Diversity of Nematode Parasites in Afrotropical Reed Frogs (Hyperolius spp.). Diversity. 2020; 12(7):265. https://doi.org/10.3390/d12070265
Chicago/Turabian StyleSinsch, Ulrich, J. Maximilian Dehling, Patrick Scheid, and Carsten Balczun. 2020. "Molecular Diversity of Nematode Parasites in Afrotropical Reed Frogs (Hyperolius spp.)" Diversity 12, no. 7: 265. https://doi.org/10.3390/d12070265
APA StyleSinsch, U., Dehling, J. M., Scheid, P., & Balczun, C. (2020). Molecular Diversity of Nematode Parasites in Afrotropical Reed Frogs (Hyperolius spp.). Diversity, 12(7), 265. https://doi.org/10.3390/d12070265







