Abstract
Since their discovery, at least 15 diatom viruses have been isolated and characterised using a culture method with two cycles of extinction dilution. However, the method is time consuming and laborious, and it isolates only the most dominant virus in a water sample. Recent studies have suggested inter-species host specificity of diatom viruses. Here, we describe a new protocol to estimate previously unrecognised host-virus relationships. Host cell cultures after inoculation of natural sediment pore water samples were obtained before complete lysis. The proliferated viral genomes in the host cells were amplified using degenerate primer pairs targeting protein replication regions of single-stranded RNA (ssRNA) and single-stranded DNA (ssDNA) viruses, and then sequenced. Diverse ssRNA virus types within known diatom virus group were detected from inoculated Chaetoceros tenuissimus and C. setoensis cells. A previously unknown ssDNA virus type was detected in inoculated C. tenuissimus cells, but not in C. setoensis cells. Despite the possible protocol biases, for example non-specific adsorptions of virions onto the host cells, the present method helps to estimate the viruses infectious to a single host species. Further improvements to this protocol targeting the proliferated viral genomes might reveal unexpected diatom–virus ecological relationships.
1. Introduction
Diatoms (Bacillariophyta) are unicellular, photosynthetic, eukaryotic algae found in aquatic environments worldwide. Their contribution to global biogeochemical cycling is significant [1], and they account for approximately 40% of marine primary production [2]. Complete diatom genome sequences reported to date [3,4] have revealed many aspects of diatom evolutionary history, ecology, metabolism, and influence on biogeochemical cycling [1,5,6,7].
Population dynamics of diatoms are affected by both abiotic factors [8] and biotic factors, and examples of the latter include viruses [9], zooplankton grazing, and bacterial and fungal parasites [10]. To date, at least 15 different diatom viruses have been reported [9]. Furthermore, recent metagenomic studies have predicted a high sequence diversity of diatom viruses in aquatic environments [11,12,13,14]. Metagenomic studies continue to provide diatom virus-like sequences owing to methodological progress. One of the problems in these studies is that the implemented methodologies can detect the virus-like sequences in the environment but not their host organisms. To understand the viral effects on the diatom dynamics, it is important to reveal the host-virus relationships. Therefore, many previous studies have reported isolations of viruses using specific diatom hosts [9].
The isolation of diatom viruses is essential to understand their basic biological features. The adopted method for virus isolation that employs two cycles of extinction dilution is time consuming and labour intensive. For example, establishment of an axenic diatom virus isolate using two cycles of extinction-dilution method generally required 2–3 months in our previous studies [9]. In the first step of the isolation procedure, samples of filtered water or sediment pore water are added to diatom cultures. After the lysis of cells, the viruses are cloned through two cycles of extinction dilution, which isolates only the most dominant virus but not the minor viruses. A recent study showed that infections with a single-stranded RNA (ssRNA) diatom virus CtenRNAV type II are not species specific, but the virus infects several species [15]. To understand the diverse relationship between the host and virus, efficient methods for identifying viruses other than the dominant ones are necessary.
To this end, we developed and tested a new protocol to estimate host-virus relationships and detected previously unrecognised viruses that specifically infect Chaetoceros species. In general, viral metagenomic studies directly analyse viral genomes in water. The new protocol we tested and present here aims to detect specific viral genomes amplified in the host algae. The host cell cultures after inoculation with natural sediment pore water samples were obtained before complete lysis. The proliferated viral genomes in the host cells were amplified using degenerate primer pairs, and then sequenced. We used two different primer pairs to amplify replication protein regions of single-stranded DNA (ssDNA) and ssRNA viruses; they include both conserved and highly diverse regions in general and are used for virus taxonomy [16,17]. One primer pair, Mplsc2, amplifies RNA-dependent RNA polymerase (RdRp) region of picornaviruses [11]. We designed the second primer pair that targeted the ssDNA diatom virus replication protein (Rep) region, based on several ssDNA diatom virus sequences. The goal of this method was not to isolate viruses, but to estimate potential relationships between host diatoms and their infectious viruses within the limit of the primer pair performance.
2. Materials and Methods
Sediment samples were collected in Hiroshima Bay using an Ekman–Birge type bottom sampler (0–1 cm depth) and preserved at −20 °C until analysis. Three grams of each sediment sample was shaken with 3 mL of modified SWM3 medium [18] at 400 rpm for 30 min, and centrifuged at 716× g for 10 min at 4 °C. The supernatant was filtered through a 0.2-μm pore-sized Dismic-25cs filter (Advantec, Tokyo, Japan) to remove bacteria. Aliquots (500 µL) of the filtered supernatants were added to 20 mL of exponentially growing C. setoensis (NIES-3712) and C. tenuissimus (NIES-3714 and NIES-3715) cultures. The cultures were incubated at 20 °C under a 12 h light and 12 h dark cycle and light intensity of 130 to 150 µmol photons m−2 s−1, which was produced by cool white fluorescent illumination. Two days post-inoculation, 2 mL of aliquots was centrifuged at 17,400× g for 10 min at 4 °C. These inoculation and culture procedures are necessary for the viruses to infect and amplify themselves in the specific microalgal host strain cells. The supernatants were removed and the host cell pellets were preserved at −80 °C until analysis.
DNA and RNA of both host cell pellets with no inoculum and 500 µL of natural sediment pore water samples were isolated following the same method.
2.1. RNA Virus Detection by PCR
RNA was extracted from the cell pellets using the RNeasyPlus Mini Kit (Qiagen, Hilden, Germany) according to the manufacture’s recommendation. The extracted RNA was reverse transcribed to cDNA using the SuperScript III First-Strand Synthesis System with random hexamers (Thermo Fisher Scientific, Waltham, MA, USA). The partial RdRp region of ssRNA viruses was amplified using a degenerate primer pair, Mplsc2F: 3′-ITWGCIGGIGATTWCA-5′ and Mplsc2R: 3′-CKYTTCARRAAWTCAGCATC-5′ [11]. PCR amplification was conducted using 20 µL of mixtures containing 5 µL of cDNA, 1× BlendTaq buffer (Toyobo, Tokyo, Japan), 200 nM of dNTPs, 0.2 µM of each primer, and 1 U of BlendTaq DNA polymerase. The amplification was performed using GeneAmp PCR system 9700 (Thermo Fisher Scientific, USA), according to the following cycle parameters: 40 cycles of denaturation at 94 °C (30 s), annealing at 43 °C (30 s), and extension at 72 °C (30 s).
2.2. DNA Virus Detection by PCR
DNA was extracted from the cell pellets using the DNeasy Plant Mini Kit (Qiagen) according to the manufacture’s recommendation. The partial Rep region of diatom ssDNA viruses was amplified using a degenerate primer pair, BDVRepF1: 5′-GWCDTGGGAHCARATHSTTC-3′ and BDVRepR1: 5′-YKSMAHTCNTCVGTYTGRTC-3′, designed in this study (Figure S1). PCR amplification was conducted with 20 µL of reaction mixtures, which contained 5 µL of DNA, 1× BlendTaq buffer (Toyobo, Japan), 200 nM of dNTPs, 0.2 µM of each primer, and 1 U of BlendTaq DNA polymerase, using GeneAmp PCR system 9700 and the following cycle parameters: 40 cycles of denaturation at 95 °C (30 s), annealing at 55 °C or 50 °C (30 s), and extension at 72 °C (30 s). The annealing temperature of 55 °C was used for sediment pore water samples collected in July 2011 and August 2013 and inoculated to the two C. tenuissimus strains; 50 °C was used for all other samples.
2.3. Sequencing of PCR Products
The PCR products were then electrophoresed on 1% (w/v) agarose ME gels (WAKO Pure Chemical Industries, Osaka, Japan), and the nucleic acids were visualised using Midori green nucleic acid stain (NIPPON Genetics, Tokyo, Japan). PCR amplicons of approximately 0.7 kb were excised, and their nucleic acids were extracted (NucleoSpin® Gel and PCR Clean-up; Macherey-Nagel GmbH and Co., KG, Düren, Germany). The PCR products were ligated into the pGEM-T Easy vector (Promega, Madison, WI, USA) and transformed into Escherichia coli DH5α-competent cells (Toyobo, Japan). Sequencing was conducted using the dideoxy method with ABI PRISM 3100 Genetic Analyser (Thermo Fisher Scientific). The resultant sequences were treated with GENETYX v12 (GENETYX, Tokyo, Japan) and their homology to viral replication proteins was verified using BLASTP [19].
2.4. Phylogenetic Analysis
The sequenced PCR fragments of the partial RdRp gene in ssRNA viruses and the partial Rep gene in ssDNA viruses were translated to amino acid (AA) sequences. The obtained RdRp AA sequences of ssRNA viruses were compared with the diatom ssRNA virus RdRp sequence dataset. The merged ssRNA virus RdRp dataset was aligned using Clustal W [20], and then manually adjusted. Similarly, the AA sequences of the partial Rep were compared with those of the diatom ssDNA virus sequence dataset, aligned using Clustal W [20], and then manually adjusted. Phylogenetic trees based on ssRNA virus RdRp and ssDNA virus Rep were constructed using the maximum likelihood (ML) method with the Jones–Taylor–Thornton matrix (JTT model) package in MEGA X [21]. Bootstrap consensus trees were constructed using 1000× bootstrap replications in each tree. Accession numbers of the sequences used in this study are listed in Table A1.
2.5. Pairwise Sequence Identity Comparisons
The aligned AA database was rearranged according to an ML tree. Pairwise sequence identities for the AA sequences were calculated using the Sequence Identity Matrix implemented in the BioEdit Sequence Alignment Editor [22]. The resulting identity matrix was edited using Microsoft Excel.
3. Results
No virus-like sequence was detected in host diatom cultures with no inoculum. This suggested that the host genomes were harbouring no virus-like sequence targeted in this study, and therefore, did not affect the results.
The detected RdRp sequences were divided into 19 types (A–S) with pairwise identities of >0.9 (Figure 1, Figures S2 and S3). RdRp sequences were detected in 21 of the 28 samples of sediment pore water; all 19 types of RdRp sequences, except for types F, H, and P, were confirmed in these samples. Types F, H, and P were detected only in inoculated diatom cell pellets. The RdRp sequences detected in C. setoensis NIES-3712 belonged to types A, B, C, D, E, F, H, O, and P (Figure 1; Table 1). The RdRp sequences of C. tenuissimus NIES-3714 were detected in seven samples, those collected in July 2011 (type E), November 2011 (type D), July 2013 (type E), January 2014 (type N), April 2014 (type A), May 2014 (type A), and June 2014 (type C) (Table 1). RdRp types G, I, J, K, L, M, R, and S were detected only in sediment pore water samples, but not in inoculated diatom cells.
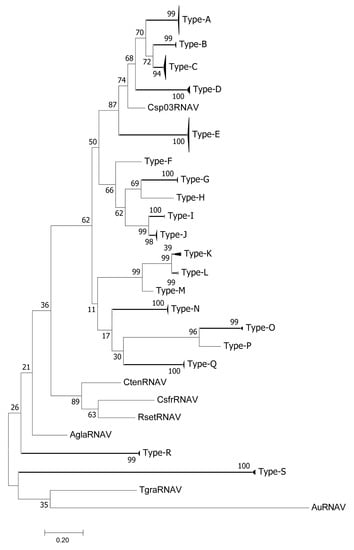
Figure 1.
Phylogenetic analysis of ssRNA viruses. Maximum likelihood (ML) trees of ssRNA viruses on the basis of RNA-dependent RNA polymerase sequences. Bootstrap values (percent) from 1000 replicates are shown at the nodes. ML distance scale is shown under the ML tree.

Table 1.
Viral RdRp and Rep types detected from sediment pore water in Hiroshima Bay, Japan, using diatom strains Chaetoceros setoensis and Chaetoceros tenuissimus during the study period. RdRp (A–S) and Rep (T–V) types were determined based on the phylogenetic trees in Figure 1 and Figure 2, respectively. Mplsc2F/R* and BDVrepF1/R1** are primer pairs used for the amplification of the RdRp and Rep regions, respectively. ‘-‘ indicates not detected.
The frequency of RdRp type E was only 3.4% of the sediment pore water samples (2/58, both samples collected in 2011), but it was dominant in >89% of C. tenuissimus NIES-3715 samples and was detected even when its presence was not confirmed in sediment pore water samples (Table 1). RdRp type E sequences of C. tenuissimus NIES-3714 or C. setoensis NIES-3712 were detected only in samples collected in July 2011 and July 2013.
The ssDNA virus Rep sequences detected in this study were grouped into three types (T, U, and V) at pairwise identities of >0.9 (Figure 2, Figures S4 and S5; Table 1). Most of the detected sequences were grouped into Rep type T, including CtenDNAV types I and II, which were detected only from the inoculated C. tenuissimus cells. Type U was detected only in sediment pore water samples collected in January 2014, February 2014, and May 2014. Type V was detected in inoculated C. tenuissimus NIES-3714, but not in NIES-3715 (Table 1). No Rep sequences were detected in C. setoensis.
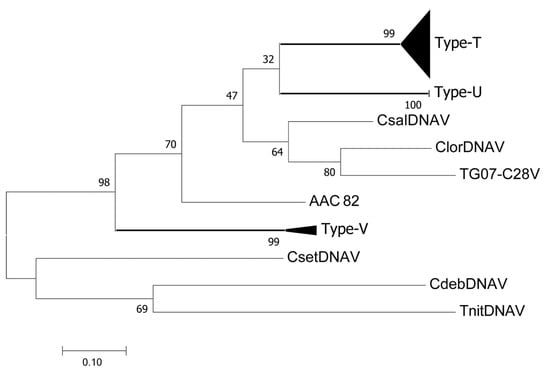
Figure 2.
Phylogenetic analysis of ssDNA viruses. Maximum likelihood (ML) trees of ssDNA viruses based on replication protein sequences. Bootstrap values (percent) from 1000 replicates are shown at the nodes. ML distance scale is shown under the ML tree.
4. Discussion
The results of the present study based on the new protocol suggested that the diverse RdRp type viruses have relationships with the specific diatom host strains. These results provide new insights into the diatom-virus ecology and evolution. For instance, no RNA virus infectious to C. setoensis NIES-3712 and C. tenuissimus NIES-3714 has been isolated to date (unpublished data), but RdRp sequences were detected in the inoculated cells of these two strains (Table 1). The diversity of RdRp types in the two diatom strains was higher than that of C. tenuissimus NIES-3715—nine and five types were detected using C. setoensis NIES-3712 and C. tenuissimus NIES-3714, respectively, vs. three in C. tenuissimus NIES-3715 (Table 1).
The major step in determining successful viral infection is dependent on the specific attachment of viruses to susceptible host cells; that is, viral tropism can be determined by cellular receptor-viral ligand interactions. One of the well-studied examples is poliovirus in the family Picornaviridae. The immunoglobulin superfamily molecule CD155 is regarded as the sole human cellular receptor for poliovirus [23]. Although rodents generally resist poliovirus infection because of the absence of CD155, transgenic mice expressing CD155 become susceptible to poliovirus infection [24]. Intra-species host specificity of HcRNAV infectious to the bloom-forming photosynthetic dinoflagellate Heterocapsa circularisquama is considered to be primarily determined in its upstream event—specific binding of a virus particle to its corresponding host cell surface receptor—as demonstrated in a transfection experiment [25]. Most of the substituted amino acid residues of the capsid proteins between two distinct HcRNAV isolates, which have different host specificities, are located on the exterior surface of the virion [26].
In the present study, RdRp type E was observed in all diatom strains. The currently isolated genomes of diatom RNA viruses are composed of two open reading frames, replication-related proteins including RdRp, and structural proteins [27]. Therefore, the RdRp type E viruses would share the same RdRp sequence but have diverse capsid proteins, for example, amino acid sequence and protein modification patterns, that determine viral infection specificities to these diatom species and/or strains. Similarly, type A was detected in all strains, whereas type C was detected in C. setoensis NIES-3712 and C. tenuissimus NIES-3714, but not in NIES-3715 (Table 1). These results suggest that diatom RNA virus genomes are diverse in terms of combinations of RdRp types and capsid proteins. The phylogenetic analysis of ssDNA virus Rep protein sequences placed CtenDNAV types I and II in the same group (Figure 2), although their structural protein similarity is only 27.6% [15]. Similar to ssRNA viruses, the combinations of Rep types and capsid proteins for diatom ssDNA viruses is expected to be diverse.
Another determinant of tropism is virus replication. Poxvirus tropism is regulated by intracellular events downstream of the virus binding and entry sites, rather than at the level of specific host receptors [28]. As for microalgal RNA viruses, the HcRNAV genome was successfully proliferated in cells of several H. circularisquama strains using transfection methods, but not in another species within the same genus, the marine dinoflagellate Heterocapsa triquetra [25]. Although both species are from the genus Heterocapsa, the intracellular condition of H. triquetra is not suitable for the replication of HcRNAV. The infections of CtenRNAV type II are inter-specific [15]. In addition to the original host strain C. tenuissimus NIES-3715, the virus infects and lyses at least four different Chaetoceros species including C. setoensis. However, the genome replication of CtenRNAV type II in the different host species seemed lower than that in the original host cells. These observations suggest that the cells permitting virus production may have different levels of permissiveness; that is, some isolates may produce large amounts of virus, whereas others may inhibit or permit only a low level of viral replication. The mechanisms determining the genome replication of microalgal RNA viruses are thus considered to be different at the intra-species and inter-species levels. The high frequency of RdRp type E in C. tenuissimus NIES-3715 reflects its higher replication efficiency in the cells. CtenRNAV type II was originally isolated in C. tenuissimus NIES-3715 [15] and was phylogenetically grouped within RdRp type E in this study. Considering the above discussion, the RdRp type E viruses and C. tenuissimus NIES-3715 are highly compatible and have successfully co-evolved.
Six different RdRp type viruses infectious to C. tenuissimus were present in the coastal sediments (Table 1). Of those, four types were also detected in the inoculated C. setoensis cells. The diatom virus densities are specifically increased during its host bloom periods as observed in other microalgae and virus systems [29]. In general, diatom blooms are usually composed of diverse species. Viruses produced by some diatom host species might not only affect host population dynamics, but also the dynamics of other species in diatom communities during bloom periods.
The protocol used in this study enables estimation of the host diatom and its infectious virus-type relationships, and is easier and quicker than the extinction dilution method. However, these results are affected by several technical problems and limitations. The detection efficiency of the target sequences, that is, viral replication proteins in this study, may be affected by higher concentrations of non-targeted nucleic acids derived from host organisms and bacteria, such as rRNA, during PCR amplifications. Further optimisation of the PCR protocols, for example, improving nucleic acid extraction and purification and PCR temperature settings, might eliminate these biases. The insufficient specificity of degenerate primer pairs for detecting diatom ssDNA viruses would also limit detection. Our preliminary tests showed that the degenerate primer pair for amplifying the ssDNA virus replication protein region failed to detect the previously known CtenDNAV type II. Hence, further improvements to the primers and protocols would be necessary to elucidate the host-virus systems, especially for ssDNA viruses.
We could not completely exclude the possibility of non-specific adsorptions of virions onto the host cells, which will overestimate the diversity of infectious viruses. However, the present results demonstrated that the viral RdRp types detected in cell pellets were often not detected in sediment pore water samples used for inoculation, and vice versa. For example, no RdRp sequence was detected in sediment pore water collected in September 2013, but type E was detected in the inoculated C. tenuissimus NIES-3715 cells. Furthermore, six different RdRp types were detected in the sediment pore water sampled in December 2013, but not in inoculated cells. The direct detection of ssDNA virus Rep proteins from sediment pore water samples was mostly unsuccessful, and only Rep type U was detected three times in water samples but never confirmed in the inoculated C. tenuissimus cells (Table 1). Rep types T and V were detected at higher frequencies in inoculated C. tenuissimus cells (Table 1). Therefore, the present method may not have enough resolution to detect non-specific virions adsorbed onto the host cell surface. For an improved understanding of the host-virus system, development of efficient protocols is necessary to detect the proliferated target sequences in the host cell. For example, detection of negative-stranded genome RNA for ssRNA viruses and mRNA sequences for ssDNA viruses might be more effective in determining the host organisms.
The lytic effects of microalgal viruses are affected by abiotic environmental factors and their effect on host cells, especially water temperature [30,31]. Viral proliferation in C. tenuissimus cells changes with respect to water temperature [27,32]. In this study, water temperature was set at 20 °C, but different experimental conditions may affect the results. In addition to water temperature, salinity and nutrient conditions potentially affect viral infection [33].
Nevertheless, although the present protocol has several technical problems, its methodology can be utilised to understand diatom and virus relationships. For example, our results indicated that a diatom host permits the proliferation of various RdRp type viruses. Furthermore, an efficient proliferation of the viral genome is determined by the compatibility of the RdRp types to the host intracellular machinery. Our study lays the foundation for further studies on virus–diatom interactions.
Supplementary Materials
The following are available online at https://www.mdpi.com/1424-2818/12/6/225/s1, Figure S1: A primer pair, BDVRepF1/R1, used for the amplification of the partial replication protein (Rep) region of diatom ssDNA viruses. The primers were designed based on the alignment result of diatom ssDNA virus Rep sequences.; Figure S2: Full data set of Figure 1; Figure S3: Pairwise sequence identity comparisons (%) of the RNA-dependent RNA polymerase (RdRp) region among all ssRNA virus samples; Figure S4: Full data set of Figure 2; Figure S5: Pairwise sequence identity comparisons (%) of the replication protein (Rep) region among all ssDNA virus samples.
Author Contributions
Conceptualisation, Y.T.; methodology, Y.T.; validation, Y.T. and K.K.; formal analysis, K.K.; resources, Y.T. and K.K.; writing—original draft preparation, Y.T.; writing—review and editing, Y.T. and K.K.; visualisation, K.K.; supervision, Y.T. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by KAKENHI (19H00956) and grants for Scientific Research on Innovative Areas (16H06429, 16K21723, and 16H06437) from the Ministry of Education, Culture, Science, Sports and Technology (MEXT) of Japan.
Acknowledgments
We are grateful to T.M. for technical assistance.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A

Table A1.
List of accession numbers of the sequences determined in this study.
Table A1.
List of accession numbers of the sequences determined in this study.
| Accession Number | Sequence | Accession Number | Sequence |
|---|---|---|---|
| LC534420 | 2011_Aug_Sd-a | LC534488 | 2013_Jul_3715 |
| LC534421 | 2011_Aug_Sd-b | LC534489 | 2013_Aug_3715 |
| LC534422 | 2011_Aug_Sd-c | LC534490 | 2013_Sep_3715 |
| LC534423 | 2011_Aug_Sd-d | LC534491 | 2013_Oct_3715 |
| LC534424 | 2011_Dec_Sd-a | LC534492 | 2014_Jan_3714 |
| LC534425 | 2011_Dec_Sd-b | LC534493 | 2014_Jan_3715 |
| LC534426 | 2011_Dec_Sd-c | LC534494 | 2014_Apr_3714 |
| LC534427 | 2011_Dec_Sd-d | LC534495 | 2014_May_3714 |
| LC534428 | 2011_Dec_Sd-e | LC534496 | 2014_Jun_3714 |
| LC534429 | 2011_Jul_Sd-a | LC534497 | 2014_Jun_3715 |
| LC534430 | 2011_Jul_Sd-d | LC534498 | 2014_Jul_3715 |
| LC534431 | 2011_Jul_Sd-e | LC534499 | 2014_Aug_3715 |
| LC534432 | 2011_Jul_Sd-f | LC534500 | 2014_Sep_3715 |
| LC534433 | 2011_Nov_Sd-a | LC534501 | 2014_Oct_3715 |
| LC534434 | 2011_Nov_Sd-b | LC534502 | 2014_Nov_3715 |
| LC534435 | 2011_Oct_Sd-a | LC534503 | 2011_Jul_3712 |
| LC534436 | 2011_Oct_Sd-b | LC534504 | 2011_Nov_3712 |
| LC534437 | 2011_Oct_Sd-c | LC534505 | 2011_Dec_3712-A |
| LC534438 | 2011_Sep_Sd-a | LC534506 | 2011_Dec_3712-B |
| LC534439 | 2011_Sep_Sd-b | LC534507 | 2013_May_3712 |
| LC534440 | 2013_Aug_Sd | LC534508 | 2014_Feb_3712-A |
| LC534441 | 2013_Dec_Sd-a | LC534509 | 2014_Feb_3712-B |
| LC534442 | 2013_Dec_Sd-b | LC534510 | 2014_Apr_3712 |
| LC534443 | 2013_Dec_Sd-c | LC534511 | 2014_Jun_3712 |
| LC534444 | 2013_Dec_Sd-d | LC534512 | 2014_Jul_3712 |
| LC534445 | 2013_Dec_Sd-e | LC534513 | 2014_Sep_3712 |
| LC534446 | 2013_Dec_Sd-f | LC534514 | 2014_Oct_3712 |
| LC534447 | 2013_Jul_Sd | LC534515 | 2014_Nov_3712 |
| LC534448 | 2013_Jun_Sd-a | LC534516 | 2015_Jan_3712 |
| LC534449 | 2013_Jun_Sd-b | LC534517 | 2015_Feb_3712-A |
| LC534450 | 2013_May_Sd-a | LC534518 | 2015_Feb_3712-B |
| LC534451 | 2013_May_Sd-b | LC534519 | 2015_Mar_3712-A |
| LC534452 | 2013_Nov_Sd-a | LC534520 | 2015_Mar_3712-B |
| LC534453 | 2013_Nov_Sd-b | LC534521 | 2014_Jan_Sd |
| LC534454 | 2013_Nov_Sd-c | LC534522 | 2014_Feb_Sd |
| LC534455 | 2013_Nov_Sd-d | LC534523 | 2014_May_Sd |
| LC534456 | 2013_Oct_Sd | LC534524 | 2011_Sep_3715 |
| LC534457 | 2014_Apr_Sd | LC534525 | 2011_Sep_3714 |
| LC534458 | 2014_Feb_Sd-a | LC534526 | 2011_Aug_3714 |
| LC534459 | 2014_Feb_Sd-b | LC534527 | 2011_Jul_3714 |
| LC534460 | 2014_Feb_Sd-d | LC534528 | 2015_Mar_3715 |
| LC534461 | 2014_Feb_Sd-e | LC534529 | 2015_Feb_3714 |
| LC534462 | 2014_Feb_Sd-g | LC534530 | 2014_Mar_3714 |
| LC534463 | 2014_Feb_Sd-h | LC534531 | 2014_Mar_3715 |
| LC534464 | 2014_Feb_Sd-i | LC534532 | 2014_Feb_3714-a |
| LC534465 | 2014_Feb_Sd-j | LC534533 | 2014_Feb_3714-b |
| LC534466 | 2014_Jan_Sd-a | LC534534 | 2014_Feb_3715 |
| LC534467 | 2014_Jan_Sd-b | LC534535 | 2011_Dec_3714 |
| LC534468 | 2014_Jun_Sd | LC534536 | 2013_May_3714 |
| LC534469 | 2014_May_Sd-a | LC534537 | 2013_May_3715 |
| LC534470 | 2014_May_Sd-b | LC534538 | 2013_Jun_3714 |
| LC534471 | 2014_Nov_Sd-a | LC534539 | 2011_Aug_3715 |
| LC534472 | 2014_Nov_Sd-c | LC534540 | 2013_Sep_3714 |
| LC534473 | 2014_Nov_Sd-d | LC534541 | 2013_Oct_3714 |
| LC534474 | 2014_Nov_Sd-e | LC534542 | 2013_Nov_3715 |
| LC534475 | 2015_Jan_Sd | LC534543 | 2013_Dec_3715 |
| LC534476 | 2015_Mar_Sd-a | LC534544 | 2013_Nov_3714 |
| LC534477 | 2015_Mar_Sd-b | LC534545 | 2014_Apr_3715 |
| LC534478 | 2011_Jul_3714 | LC534546 | 2014_Jun_3714 |
| LC534479 | 2011_Jul_3715 | LC534547 | 2014_Jun_3715 |
| LC534480 | 2011_Aug_3715 | LC534548 | 2014_Aug_3714 |
| LC534481 | 2011_Sep_3715 | LC534549 | 2014_Nov_3714 |
| LC534482 | 2011_Oct_3715 | LC534550 | 2014_Oct_3715 |
| LC534483 | 2011_Nov_3714 | LC534551 | 2014_Oct_3714 |
| LC534484 | 2011_Nov_3715 | LC534552 | 2014_Sep_3714 |
| LC534485 | 2011_Dec_3715 | LC534553 | 2014_Nov_3715 |
| LC534486 | 2013_Jun_3715 | LC534554 | 2015_Jan_3714 |
| LC534487 | 2013_Jul_3714 | LC534555 | 2015_Jan_3715 |
References
- Tréguer, P.; Bowler, C.; Moriceau, B.; Dutkiewicz, S.; Gehlen, M.; Aumont, O.; Bittner, L.; Dugdale, R.; Finkel, Z.; Iudicone, D.; et al. Influence of diatom diversity on the ocean biological carbon pump. Nat. Geosci. 2018, 11, 27–37. [Google Scholar]
- Field, C.B.; Behrenfeld, M.J.; Randerson, J.T.; Falkowski, P.G. Primary production of the biosphere: Integrating terrestrial and oceanic components. Science 1998, 281, 237–240. [Google Scholar] [CrossRef]
- Armbrust, E.V.; Berges, J.A.; Bowler, C.; Green, B.R.; Martinez, D.; Putnam, N.H.; Zhou, S.; Allen, A.E.; Apt, K.E.; Bechner, M.; et al. The genome of the diatom Thalassiosira pseudonana: Ecology, evolution, and metabolism. Science 2004, 306, 79–86. [Google Scholar] [CrossRef]
- Bowler, C.; Allen, A.E.; Badger, J.H.; Grimwood, J.; Jabbari, K.; Kuo, A.; Maheswari, U.; Martens, C.; Maumus, F.; Otillar, R.P.; et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 2008, 456, 239–244. [Google Scholar] [CrossRef]
- Armbrust, E.V. The life of diatoms in the world’s oceans. Nature 2009, 459, 185–192. [Google Scholar] [CrossRef]
- Bowler, C.; Vardi, A.; Allen, A.E. Oceanographic and biogeochemical insights from diatom genomes. Annu. Rev. Mar. Sci. 2010, 2, 333–365. [Google Scholar] [CrossRef]
- Benoiston, A.-S.; Ibarbalz, F.M.; Bittner, L.; Guidi, L.; Jahn, O.; Dutkiewicz, S.; Bowler, C. The evolution of diatoms and their biogeochemical functions. Philos. Trans. R. Soc. Lond., B Biol. Sci. 2017, 372, 20160397. [Google Scholar] [CrossRef]
- Sarthou, G.; Timmermans, K.R.; Blain, S.; Tréguer, P. Growth physiology and fate of diatoms in the ocean: A review. J. Sea Res. 2005, 53, 25–42. [Google Scholar] [CrossRef]
- Tomaru, Y.; Toyoda, K.; Kimura, K. Marine diatom viruses and their hosts: Resistance mechanisms and population dynamics. Perspect. Phycol. 2015, 2, 69–81. [Google Scholar] [CrossRef]
- Raven, J.A.; Waite, A.M. The evolution of silicification in diatoms: Inescapable sinking and sinking as escape? New Phytol. 2004, 162, 45–61. [Google Scholar] [CrossRef]
- Culley, A.I.; Mueller, J.A.; Belcaid, M.; Wood-Charlson, E.M.; Poisson, G.; Steward, G.F. The characterization of RNA viruses in tropical seawater using targeted PCR and metagenomics. mBio 2014, 5, e01210–01214. [Google Scholar] [CrossRef]
- McDaniel, L.D.; Rosario, K.; Breitbart, M.; Paul, J.H. Comparative metagenomics: Natural populations of induced prophages demonstrate highly unique, lower diversity viral sequences. Environ. Microbiol. 2014, 16, 570–585. [Google Scholar] [CrossRef]
- Dayaram, A.; Goldstien, S.; Argüello-Astorga, G.R.; Zawar-Reza, P.; Gomez, C.; Harding, J.S.; Varsani, A. Diverse small circular DNA viruses circulating amongst estuarine molluscs. Infect. Genet. Evol. 2015, 31, 284–295. [Google Scholar] [CrossRef]
- Miranda, J.A.; Culley, A.I.; Schvarcz, C.R.; Steward, G.F. RNA viruses as major contributors to Antarctic virioplankton. Environ. Microbiol. 2016, 18, 3714–3727. [Google Scholar] [CrossRef]
- Kimura, K.; Tomaru, Y. Discovery of two novel viruses expands the diversity of ssDNA and ssRNA viruses infecting a cosmopolitan marine diatom. Appl. Environ. Microbiol. 2015, 81, 1120–1131. [Google Scholar] [CrossRef]
- Varsani, A.; Krupovic, M. Sequence-based taxonomic framework for the classification of uncultured single-stranded DNA viruses of the family Genomoviridae. Virus Evol. 2017, 3, vew037. [Google Scholar] [CrossRef]
- Vlok, M.; Lang, A.S.; Suttle, C.A. Application of a sequence-based taxonomic classification method to uncultured and unclassified marine single-stranded RNA viruses in the order Picornavirales. Virus Evol. 2019, 5, vez056. [Google Scholar] [CrossRef]
- Imai, I.; Itakura, S.; Matsuyama, Y.; Yamaguchi, M. Selenium requirement for growth of a novel red tide flagellate Chattonella verruculosa (Raphidophyceae) in culture. Fish. Sci. 1996, 62, 834–835. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Mendelsohn, C.L.; Wimmer, E.; Racaniello, V.R. Cellular receptor for poliovirus: Molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell 1989, 56, 855–865. [Google Scholar] [CrossRef]
- Ren, R.; Costantini, F.; Gorgacz, E.J.; Lee, J.J.; Racaniello, V.R. Transgenic mice expressing a human poliovirus receptor: A new model for poliomyelitis. Cell 1990, 63, 353–362. [Google Scholar] [CrossRef]
- Mizumoto, H.; Tomaru, Y.; Takao, Y.; Shirai, Y.; Nagasaki, K. Intraspecies host specificity of a single-stranded RNA virus infecting a marine photosynthetic protist is determined at the early steps of infection. J. Virol. 2007, 81, 1372–1378. [Google Scholar] [CrossRef]
- Nagasaki, K.; Shirai, Y.; Takao, Y.; Mizumoto, H.; Nishida, K.; Tomaru, Y. Comparison of genome sequences of single-stranded RNA virus infecting the bivalve-killing dinoflagellate Heterocapsa circularisquama. Appl. Environ. Microbiol. 2005, 71, 8888–8894. [Google Scholar] [CrossRef]
- Tomaru, Y.; Kimura, K.; Yamaguchi, H. Temperature alters algicidal activity of DNA and RNA viruses infecting Chaetoceros tenuissimus. Aquat. Microb. Ecol. 2014, 73, 171–183. [Google Scholar] [CrossRef]
- McFadden, G. Poxvirus tropism. Nat. Rev. Microbiol. 2005, 3, 201–213. [Google Scholar] [CrossRef]
- Tomaru, Y.; Fujii, N.; Oda, S.; Toyoda, K.; Nagasaki, K. Dynamics of diatom viruses on the western coast of Japan. Aquat. Microb. Ecol. 2011, 63, 223–230. [Google Scholar] [CrossRef]
- Demory, D.; Arsenieff, L.; Simon, N.; Six, C.; Rigaut-Jalabert, F.; Marie, D.; Ge, P.; Bigeard, E.; Jacquet, S.; Sciandra, A.; et al. Temperature is a key factor in Micromonas–virus interactions. ISME J. 2017, 11, 601–612. [Google Scholar] [CrossRef]
- Maat, D.S.; Biggs, T.; Evans, C.; Van Bleijswijk, J.D.; Van Der Wel, N.N.; Dutilh, B.E.; Brussaard, C.P. Characterization and temperature dependence of arctic Micromonas polaris viruses. Viruses 2017, 9, 134. [Google Scholar] [CrossRef] [PubMed]
- Kimura, K.; Tomaru, Y. Effects of temperature and salinity on diatom cell lysis by DNA and RNA viruses. Aquat. Microb. Ecol. 2017, 79, 79–83. [Google Scholar] [CrossRef]
- Mojica, K.D.; Brussaard, C.P. Factors affecting virus dynamics and microbial host-virus interactions in marine environments. FEMS Microbiol. Ecol. 2014, 89, 495–515. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).