The Yeast Atlas of Appalachia: Species and Phenotypic Diversity of Herbicide Resistance in Wild Yeast
Abstract
1. Introduction
1.1. Diversity of Yeast
1.2. Glyphosate-Based Herbicides
1.3. Copper-Based Herbicides
2. Materials and Methods
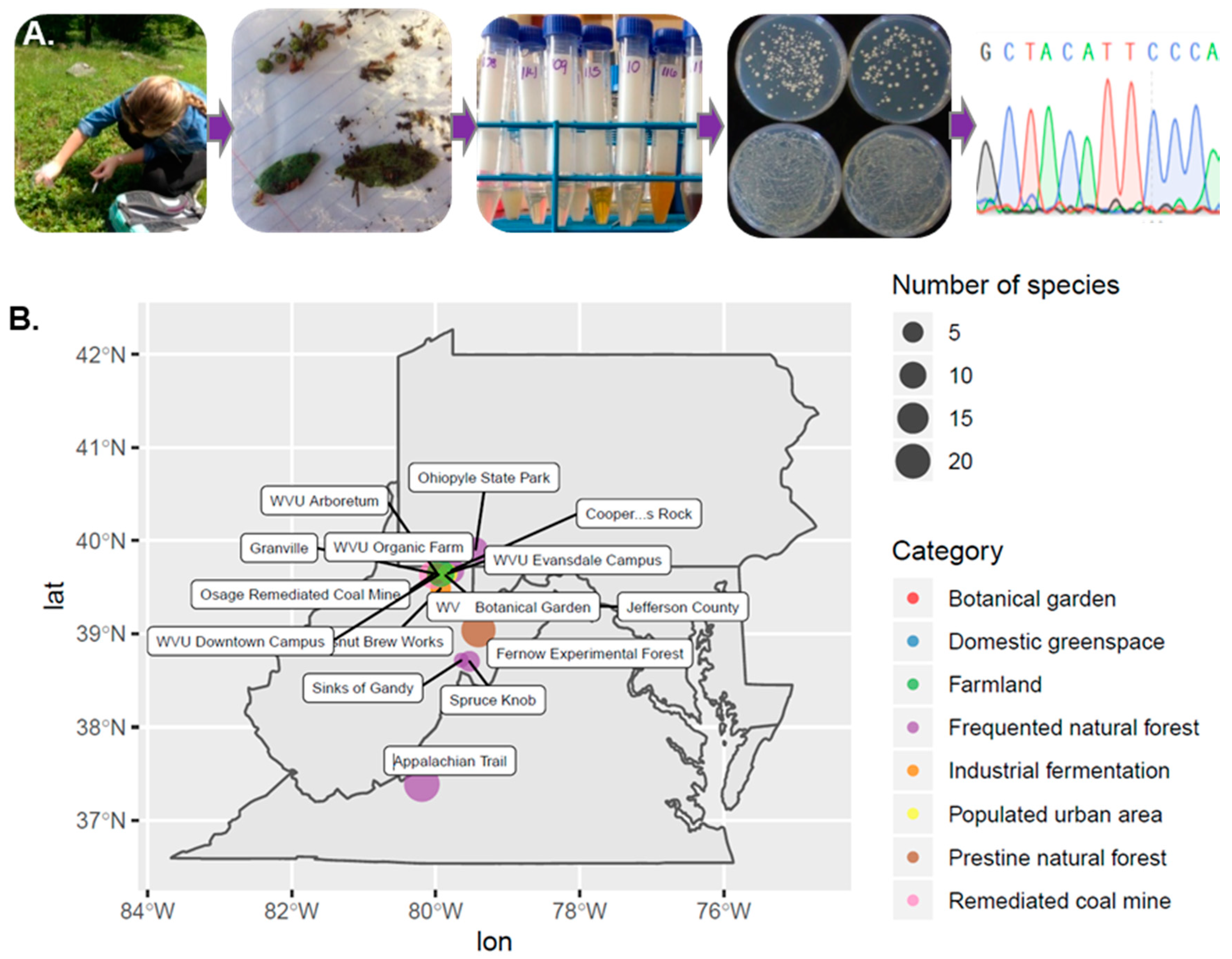
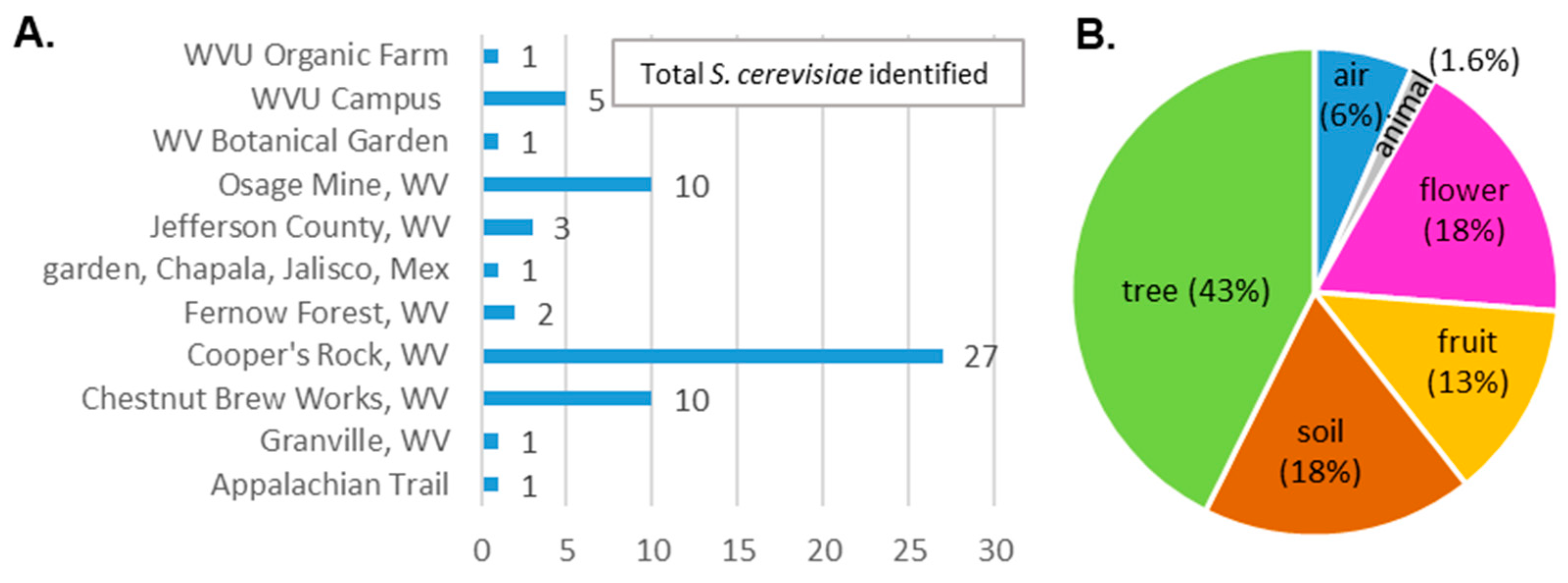
2.1. Collection of Samples
2.2. Isolation of Yeast-Like Colonies
2.3. Identification of Isolates
2.4. Phylogenetic Analysis
2.5. Phenotypic Characterization
3. Results/Discussion
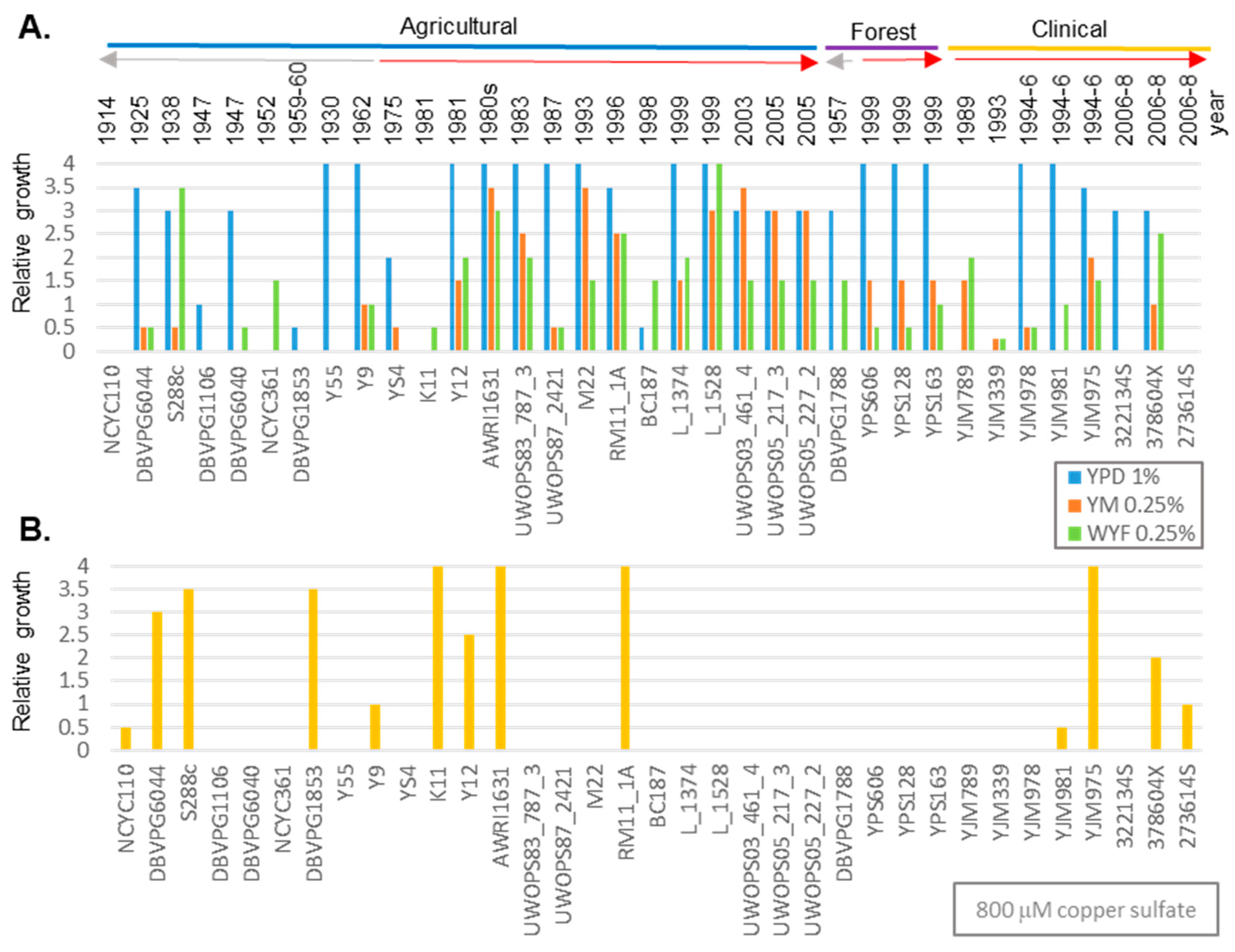
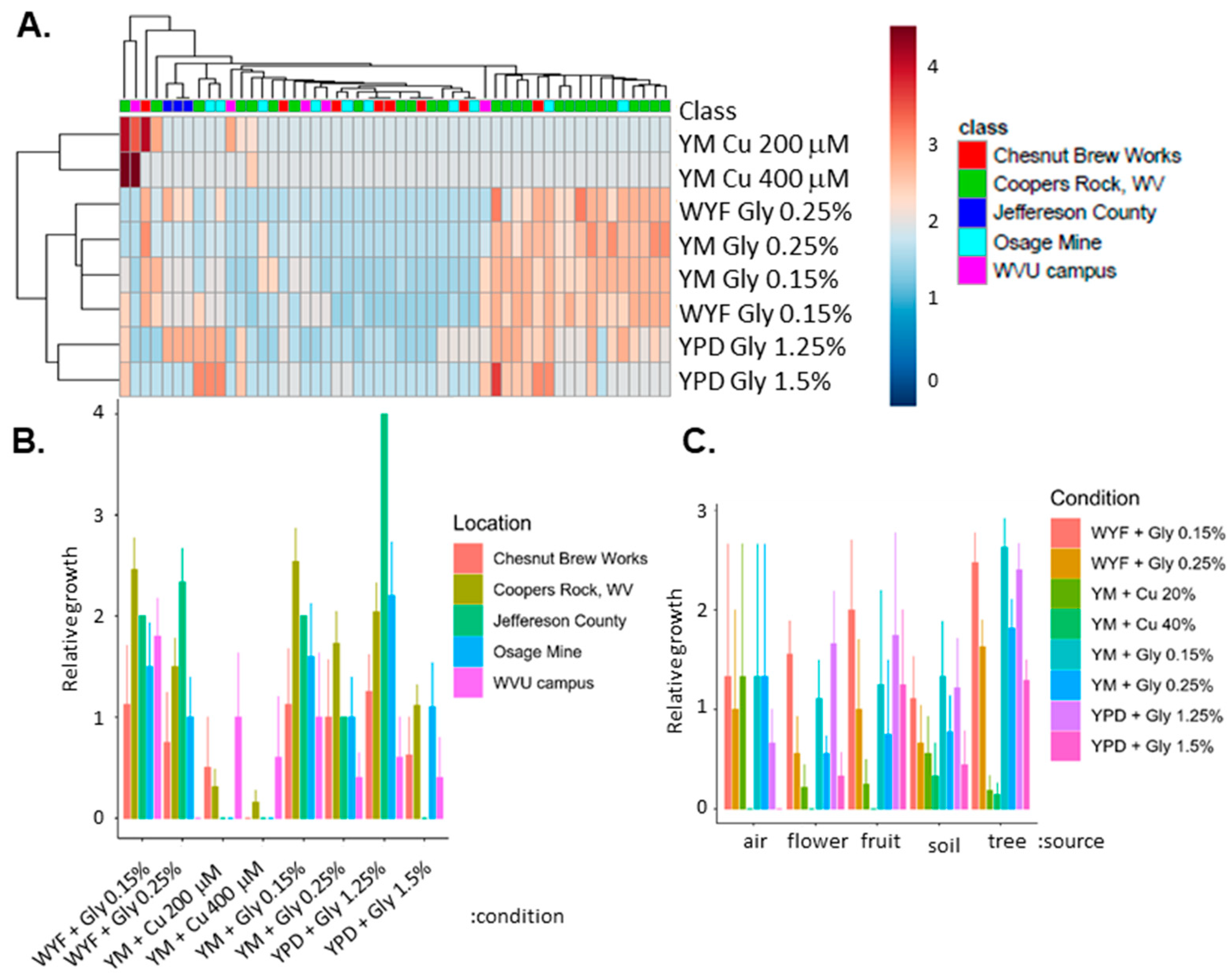
3.1. Genetic Variation of Fungicide in Geographically Diverse S. cerevisiae
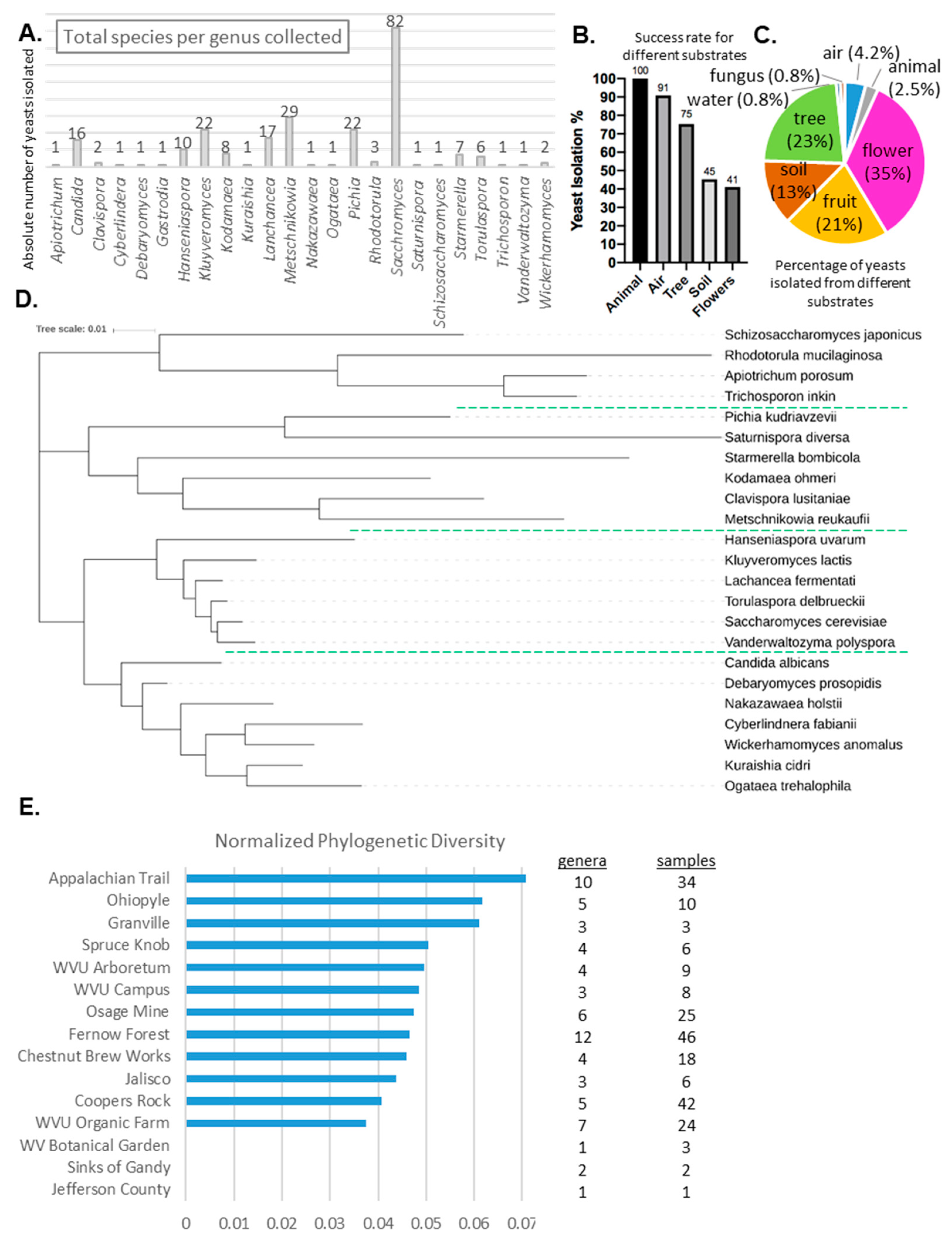
3.2. Isolate Identification
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Data Availability
References
- Kurtzman, C.; Fell, J.; Boekhout, T. The Yeasts: A Taxonomic Study; Elsevier: Amsterdam, The Netherlands, 2011. [Google Scholar]
- Shen, X.-X.; Opulente, D.A.; Kominek, J.; Zhou, X.; Steenwyk, J.L.; Buh, K.V.; Haase, M.A.B.; Wisecaver, J.H.; Wang, M.; Doering, D.T.; et al. Tempo and Mode of Genome Evolution in the Budding Yeast Subphylum. Cell 2018, 175, 1533–1545. [Google Scholar] [CrossRef]
- Libkind, D.; Hittinger, C.T.; Valério, E.; Gonçalves, C.; Dover, J.; Johnston, M.; Gonçalves, P.; Sampaio, J.P. Microbe domestication and the identification of the wild genetic stock of lager-brewing yeast. Proc. Natl. Acad. Sci. USA 2011, 108, 14539–14544. [Google Scholar] [CrossRef] [PubMed]
- Fay, J.C.; Liu, P.; Ong, G.T.; Dunham, M.J.; Cromie, G.A.; Jeffery, E.W.; Ludlow, C.L.; Dudley, A.M. A polyploid admixed origin of beer yeasts derived from European and Asian wine populations. PLoS Biol. 2019, 17, e3000147. [Google Scholar] [CrossRef] [PubMed]
- Liti, G.; Carter, D.M.; Moses, A.M.; Warringer, J.; Parts, L.; James, S.A.; Davey, R.P.; Roberts, I.N.; Burt, A.; Koufopanou, V.; et al. Population genomics of domestic and wild yeasts. Nature 2009, 458, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Peter, J.; De Chiara, M.; Friedrich, A.; Yue, J.-X.; Pflieger, D.; Bergström, A.; Sigwalt, A.; Barre, B.; Freel, K.; Llored, A.; et al. Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature 2018, 556, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Hyma, K.E.; Fay, J.C. Mixing of vineyard and oak-tree ecotypes of Saccharomyces cerevisiae in North American vineyards. Mol. Ecol. 2013, 22, 2917–2930. [Google Scholar] [CrossRef]
- Chao, A.; Chiu, C.-H.; Jost, L. Phylogenetic diversity measures and their decomposition: A framework based on hill numbers. In Biodiversity Conservation and Phylogenetic Systematics; Pellens, R., Grandcolas, P., Eds.; Springer International Publishing: Basle, Switzerland, 2016; pp. 141–172. [Google Scholar]
- Botstein, D.; Chervitz, S.A.; Cherry, J.M. Yeast as a model organism. Science 1997, 277, 1259–1260. [Google Scholar] [CrossRef]
- Kachroo, A.H.; Laurent, J.M.; Yellman, C.M.; Meyer, A.G.; Wilke, C.O.; Marcotte, E.M. Systematic humanization of yeast genes reveals conserved functions and genetic modularity. Science 2015, 348, 921–925. [Google Scholar] [CrossRef]
- Belloch, C.; Pérez-Torrado, R.; González, S.S.; JoseE, P.-O.; García-Martínez, J.; Querol, A.; Barrio, E. Chimeric genomes of natural hybrids Of Saccharomyces cerevisiae and Saccharomyces kudriavzevii. Appl. Environ. Microbiol. 2009, 75, 2534–2544. [Google Scholar] [CrossRef]
- Winans, M.J.; Yamamoto, Y.; Fujimaru, Y.; Kusaba, Y.; Gallagher, J.E.G.; Kitagaki, H. Saccharomyces arboricola and Its Hybrids’ Propensity for Sake Production: Interspecific Hybrids Reveal Increased Fermentation Abilities and a Mosaic Metabolic Profile. Fermentation 2020, 6, 14. [Google Scholar] [CrossRef]
- Hittinger, C.T. Saccharomyces diversity and evolution: A budding model genus. Trends Genet. 2013, 29, 309–317. [Google Scholar] [CrossRef] [PubMed]
- Sniegowski, P.D.; Dombrowski, P.G.; Fingerman, E. Saccharomyces cerevisiae and Saccharomyces paradoxus coexist in a natural woodland site in North America and display different levels of reproductive isolation from European conspecifics. FEMS Yeast Res 2002, 1, 299–306. [Google Scholar] [PubMed]
- Sylvester, K.; Wang, Q.-M.; James, B.; Mendez, R.; Hulfachor, A.B.; Hittinger, C.T. Temperature and host preferences drive the diversification of Saccharomyces and other yeasts: A survey and the discovery of eight new yeast species. FEMS Yeast Res. 2015, 15, fov002. [Google Scholar] [CrossRef]
- Maganti, H.; Bartfai, D.; Xu, J. Ecological structuring of yeasts associated with trees around Hamilton, Ontario, Canada. FEMS Yeast Res. 2012, 12, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Stefanini, I.; Dapporto, L.; Berná, L.; Polsinelli, M.; Turillazzi, S.; Cavalieri, D. Social wasps are a Saccharomyces mating nest. Proc. Natl. Acad. Sci. USA 2015, 113, 2–6. [Google Scholar] [CrossRef] [PubMed]
- Cromie, G.A.; Hyma, K.E.; Ludlow, C.L.; Garmendia-Torres, C.; Gilbert, T.L.; May, P.; Huang, A.A.; Dudley, A.M.; Fay, J.C. Genomic Sequence Diversity and Population Structure of Saccharomyces cerevisiae Assessed by RAD-seq. G3 Genes Genomes Genet. 2013, 3, 2163–2171. [Google Scholar] [CrossRef]
- Baucom, R.S.; Mauricio, R. Constraints of the evolution of toleracne to herbicide in the common morning glory: Resistance and tolerance are mutually exclusive. Evolution 2008, 62, 2842–2854. [Google Scholar] [CrossRef]
- Chaney, L.; Baucom, R.S. The costs and benefits of tolerance to competition in Ipomoea purpurea, the common morning glory. Evolution 2014, 68, 1698–1709. [Google Scholar] [CrossRef]
- Powles, S.B.; Preston, C. Evolved Glyphosate Resistance in Plants: Biochemical and Genetic Basis of Resistance. Weed Technol. 2006, 20, 282–289. [Google Scholar] [CrossRef]
- Schönbrunn, E.; Eschenburg, S.; Shuttleworth, W.A.; Schloss, J.V.; Amrhein, N.; Evans, J.N.S.; Kabsch, W. Interaction of the herbicide glyphosate with its target enzyme 5-enolpyruvylshikimate 3-phosphate synthase in atomic detail. Proc. Natl. Acad. Sci. USA 2001, 98, 1376–1380. [Google Scholar] [CrossRef]
- Healy-Fried, M.L.; Funke, T.; Priestman, M.A.; Han, H.; Schönbrunn, E. Structural Basis of Glyphosate Tolerance Resulting from Mutations of Pro101 in Escherichia coli 5-Enolpyruvylshikimate-3-phosphate Synthase. J. Biol. Chem. 2007, 282, 32949–32955. [Google Scholar] [CrossRef] [PubMed]
- Rogers, S.G.; Brand, L.A.; Holder, S.B.; Sharps, E.S.; Brackin, M.J. Amplification of the aroA gene from Escherichia coli results in tolerance to the herbicide glyphosate. Appl. Environ. Microbiol. 1983, 46, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.-C.; Chen, Y.-C.; Tian, Z.-X.; Li, F.-M.; Wang, X.-Y.; Zhang, J.; Xiao, Z.-L.; Lin, M.; Gilmartin, N.; Dowling, D.N.; et al. Novel AroA with High Tolerance to Glyphosate, Encoded by a Gene of Pseudomonas putida 4G-1 Isolated from an Extremely Polluted Environment in China. Appl. Environ. Microbiol. 2005, 71, 4771–4776. [Google Scholar] [CrossRef]
- Rong-Mullins, X.; Ravishankar, A.; McNeal, K.A.; Lonergan, Z.R.; Biega, A.C.; Creamer, J.P.; Gallagher, J.E.G.; Louis, E.J.; Ravishankar, A.; McNeal, K.A.; et al. Genetic variation in Dip5, an amino acid permease, and Pdr5, a multiple drug transporter, regulates glyphosate resistance in S. cerevisiae. PLoS ONE 2017, 12, e0187522. [Google Scholar] [CrossRef] [PubMed]
- Ravishankar, A.; Pupo, A.; Gallagher, J.E.G. Resistance mechanisms of Saccharomyces cerevisiae to commercial formulations of glyphosate involve DNA damage repair, the cell cycle and the cell wall structure. G3 Genes Genomes Genet. 2020. accepted. [Google Scholar]
- Ravishankar, A.; Cumming, J.R.; Gallagher, J.E.G. Mitochondrial metabolism is central for response and resistance of Saccharomyces cerevisiae to exposure to a glyphosate-based herbicide. Environ. Pollut. 2020, 262, 114359. [Google Scholar] [CrossRef]
- Song, G.; Dickins, B.J.A.; Demeter, J.; Engel, S.; Gallagher, J.; Choe, K.; Dunn, B.; Snyder, M.; Cherry, J.M.; Choe Barbara, K.D.; et al. AGAPE (Automated Genome Analysis PipelinE) for pan-genome analysis of Saccharomyces cerevisiae. PLoS ONE 2015, 10, e0120671. [Google Scholar] [CrossRef]
- Dixon, B. Pushing Bordeaux mixture. Lancet Infect. Dis. 2004, 4, 594. [Google Scholar] [CrossRef]
- Komárek, M.; Čadková, E.; Chrastný, V.; Bordas, F.; Bollinger, J.-C. Contamination of vineyard soils with fungicides: A review of environmental and toxicological aspects. Environ. Int. 2009, 36, 138–151. [Google Scholar] [CrossRef]
- Dollwet, H.H.A.; Sorenson, J.R.J. Historic uses of copper compounds in medicine. Trace Elem. Med. 1985, 2, 80–87. [Google Scholar]
- Lloyd, R.V.R.V.; Hanna, P.M.P.M.; Mason, R.P.R.P. The origin of the hydroxyl radical oxygen in the Fenton reaction. Free Radic. Biol. Med. 1997, 22, 885–888. [Google Scholar] [CrossRef]
- Avery, S.S.V.; Howlett, N.N.G.; Radice, S. Copper toxicity towards Saccharomyces cerevisiae: Dependence on plasma membrane fatty acid composition. Appl. Environ. Microbiol. 1996, 62, 3960–3966. [Google Scholar] [CrossRef] [PubMed]
- Winans, M.J.; Gallagher, J.E.G. Metallomic and Lipidomic Analysis of S. cerevisiae Response to Cellulosic Copper Nanoparticles Uncover Drivers of Toxicity. Metallomics 2020, in press. [Google Scholar] [CrossRef]
- Quaranta, D.; Krans, T.; Santo, C.E.; Elowsky, C.G.; Domaille, D.W.; Chang, C.J.; Grass, G. Mechanisms of Contact-Mediated Killing of Yeast Cells on Dry Metallic Copper Surfaces. Appl. Environ. Microbiol. 2011, 77, 416–426. [Google Scholar] [CrossRef]
- Rong-Mullins, X.; Winans, M.J.; Lee, J.B.; Lonergan, Z.R.; Pilolli, V.A.; Weatherly, L.M.; Carmenzind, T.W.; Jiang, L.; Cumming, J.R.; Oporto, G.S.; et al. Proteomic and genetic analysis of the response of S. cerevisiae to soluble copper leads to improvement of the antimicrobial function of cellulosic copper nanoparticles. Metallomics 2017, 9, 1304–1315. [Google Scholar] [CrossRef]
- Samanovic, M.I.; Ding, C.; Thiele, D.J.; Darwin, K.H. Copper in microbial pathogenesis: Meddling with the metal. Cell Host Microbe 2012, 11, 106–115. [Google Scholar] [CrossRef]
- Fay, J.C.; McCullough, H.L.; Sniegowski, P.D.; Eisen, M.B. Population genetic variation in gene expression is associated with phenotypic variation in Saccharomyces cerevisiae. Genome Biol 2004, 5, R26. [Google Scholar] [CrossRef]
- Tohoyama, H.; Inouhe, M.; Joho, M.; Murayama, T. Production of metallothionein in copper- and cadmium-resistant strains of Saccharomyces cerevisiae. J. Ind. Microbiol. 1995, 14, 126–131. [Google Scholar] [CrossRef]
- Jensen, L.T.; Howard, W.R.; Strain, J.J.; Winge, D.R.; Culotta, V.C. Enhanced effectiveness of copper ion buffering by CUP1 metallothionein compared with CRS5 metallothionein in Saccharomyces cerevisiae. J. Biol. Chem. 1996, 271, 18514–18519. [Google Scholar] [CrossRef]
- Karin, M.; Najarian, R.; Haslinger, A.; Valenzuela, P.; Welch, J.; Fogel, S.; Najariant, R.; Haslinger, A.; Valenzuelat, P.; Welcht, J. Primary structure and transcription of an amplified genetic locus: The CUP1 locus of yeast. Proc. Natl. Acad. Sci. USA 1984, 81, 337–341. [Google Scholar] [CrossRef]
- Rees, E.M.M.; Lee, J.; Thiele, D.J.J. Mobilization of intracellular copper stores by the ctr2 vacuolar copper transporter. J. Biol. Chem. 2004, 279, 54221–54229. [Google Scholar] [CrossRef] [PubMed]
- Butler, G.; Thiele, D.J. ACE2, an activator of yeast metallothionein expression which is homologous to SWI5. Mol. Cell. Biol. 1991, 11, 476–485. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gross, C.; Kelleher, M.; Iyer, V.R.; Brown, P.O.; Winge, D.R. Identification of the Copper Regulon in Saccharomyces cerevisiae by DNA Microarrays. J. Biol. Chem. 2000, 275, 32310–32316. [Google Scholar] [CrossRef] [PubMed]
- Hodgins-Davis, A.; Adomas, A.B.; Warringer, J.; Townsend, J.P. Abundant gene-by-environment interactions in gene expression reaction norms to copper within Saccharomyces cerevisiae. Genome Biol. Evol. 2012, 4, 1061–1079. [Google Scholar] [CrossRef]
- Welch, J.; Fogel, S.; Buchman, C.; Karin, M. The CUP2 gene product regulates the expression of the CUP1 gene, coding for yeast metallothionein. EMBO J. 1989, 8, 255–260. [Google Scholar] [CrossRef]
- Zhao, Y.; Strope, P.K.K.; Kozmin, S.G.G.; McCusker, J.H.H.; Dietrich, F.S.S.; Kokoska, R.J.J.; Petes, T.D.D. Structures of naturally evolved CUP1 tandem arrays in yeast indicate that these arrays are generated by unequal nonhomologous recombination. G3 Genes Genomes Genet. 2014, 4, 2259–2269. [Google Scholar] [CrossRef]
- Kwiatkowski, N.P.; Babiker, W.M.; Merz, W.G.; Carroll, K.C.; Zhang, S.X. Evaluation of nucleic acid sequencing of the D1/D2 region of the large subunit of the 28S rDNA and the internal transcribed spacer region using smartgene idn software for identification of filamentous fungi in a clinical laboratory. J. Mol. Diagn. 2012, 14, 393–401. [Google Scholar] [CrossRef]
- Hirt, B. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 1967, 26, 365–369. [Google Scholar] [CrossRef]
- White, T.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR Protocols: A guide to methods and applications. Acad. Press 1990, 64, 315–322. [Google Scholar]
- Taverna, C.G.; Bosco-Borgeat, M.E.; Murisengo, O.A.; Davel, G.; Boité, M.C.; Cupolillo, E.; Canteros, C.E. Comparative analyses of classical phenotypic method and ribosomal RNA gene sequencing for identification of medically relevant Candida species. Mem. Inst. Oswaldo Cruz 2013, 108, 178–185. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, J.E.G.; Zheng, W.; Rong, X.; Miranda, N.; Lin, Z.; Dunn, B.; Zhao, H.; Snyder, M.P. Divergence in a master variator generates distinct phenotypes and transcriptional responses. Genes Dev. 2014, 28, 409–421. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Forlani, G.; Mangiagalli, A.; Nielsen, E.; Suardi, C. Degradation of the phosphonate herbicide glyphosate in soil: Evidence for a possible involvement of unculturable microorganisms. Soil Biol. Biochem. 1999, 31, 991–997. [Google Scholar] [CrossRef]
- Wang, Q.-Y.; Zhou, D.-M.; Cang, L. Microbial and enzyme properties of apple orchard soil as affected by long-term application of copper fungicide. Soil Biol. Biochem. 2009, 41, 1504–1509. [Google Scholar] [CrossRef]
- Battaglin, W.A.; Meyer, M.T.; Kuivila, K.M.; Dietze, J.E. Glyphosate and Its Degradation Product AMPA Occur Frequently and Widely in U.S. Soils, Surface Water, Groundwater, and Precipitation. JAWRA J. Am. Water Resour. Assoc. 2014, 50, 275–290. [Google Scholar] [CrossRef]
- Adamo, G.M.; Lotti, M.; Tamas, M.J.; Brocca, S. Amplification of the CUP1 gene is associated with evolution of copper tolerance in Saccharomyces cerevisiae. Microbiology 2012, 158, 2325–2335. [Google Scholar] [CrossRef]
- Jang, J.H.; Lee, J.H.; Ki, C.S.; Lee, N.Y. Identification of clinical mold isolates by sequence analysis of the internal transcribed spacer region, ribosomal large-subunit D1/D2, and β-tubulin. Ann. Lab. Med. 2012, 32, 126–132. [Google Scholar] [CrossRef]
- Hinrikson, H.P.; Hurst, S.F.; Lott, T.J.; Warnock, D.W.; Morrison, C.J. Assessment of ribosomal large-subunit D1-D2, internal transcribed spacer 1, and internal transcribed spacer 2 regions as targets for molecular identification of medically important Aspergillus species. J. Clin. Microbiol. 2005, 43, 2092–2103. [Google Scholar] [CrossRef]
- Quan, A.S.; Eisen, M.B. The ecology of the Drosophila-yeast mutualism in wineries. PLoS ONE 2018, 13, e0196440. [Google Scholar] [CrossRef]
- West Virginia Division of Forestry. Coopers Rock State Forest, Forest Resources Management Plan. 2006. Available online: https://www.wvforestry.com/pdf/CRSF%20final%20draft%201-19-06.pdf (accessed on 13 May 2016).
- Difazio, S. West Virginia University: Morgantown, WV, USA, Unpublished work. 2014.

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barney, J.B.; Winans, M.J.; Blackwood, C.B.; Pupo, A.; Gallagher, J.E.G. The Yeast Atlas of Appalachia: Species and Phenotypic Diversity of Herbicide Resistance in Wild Yeast. Diversity 2020, 12, 139. https://doi.org/10.3390/d12040139
Barney JB, Winans MJ, Blackwood CB, Pupo A, Gallagher JEG. The Yeast Atlas of Appalachia: Species and Phenotypic Diversity of Herbicide Resistance in Wild Yeast. Diversity. 2020; 12(4):139. https://doi.org/10.3390/d12040139
Chicago/Turabian StyleBarney, Jordan B., Matthew J. Winans, Catherine B. Blackwood, Amaury Pupo, and Jennifer E.G. Gallagher. 2020. "The Yeast Atlas of Appalachia: Species and Phenotypic Diversity of Herbicide Resistance in Wild Yeast" Diversity 12, no. 4: 139. https://doi.org/10.3390/d12040139
APA StyleBarney, J. B., Winans, M. J., Blackwood, C. B., Pupo, A., & Gallagher, J. E. G. (2020). The Yeast Atlas of Appalachia: Species and Phenotypic Diversity of Herbicide Resistance in Wild Yeast. Diversity, 12(4), 139. https://doi.org/10.3390/d12040139