Enough Is Enough? Searching for the Optimal Sample Size to Monitor European Habitats: A Case Study from Coastal Sand Dunes
Abstract
1. Introduction
2. Methods
2.1. Study Area and Sampling Design
2.2. Workflow of the Analysis
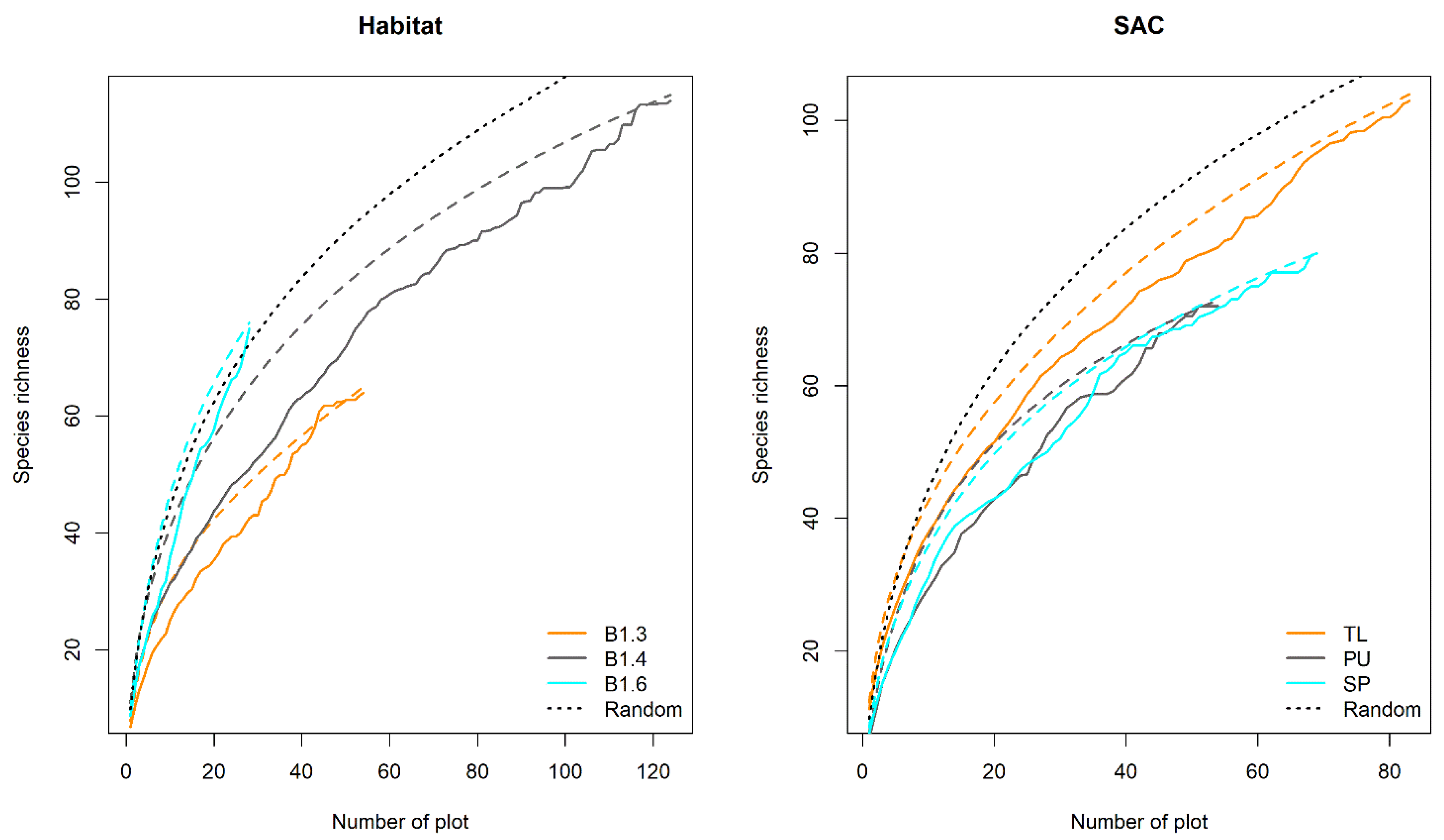
2.2.1. Description of Diversity Patterns
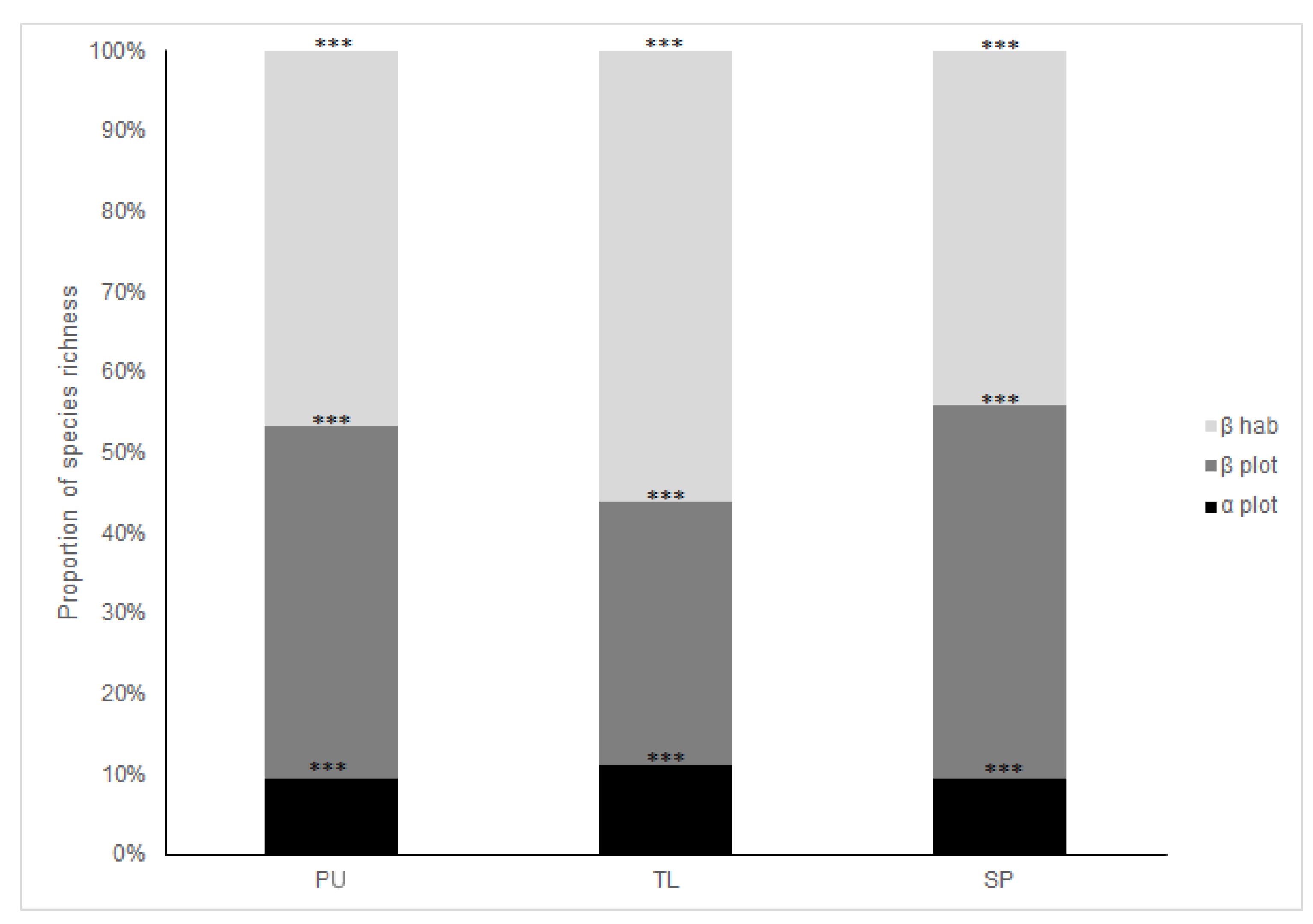
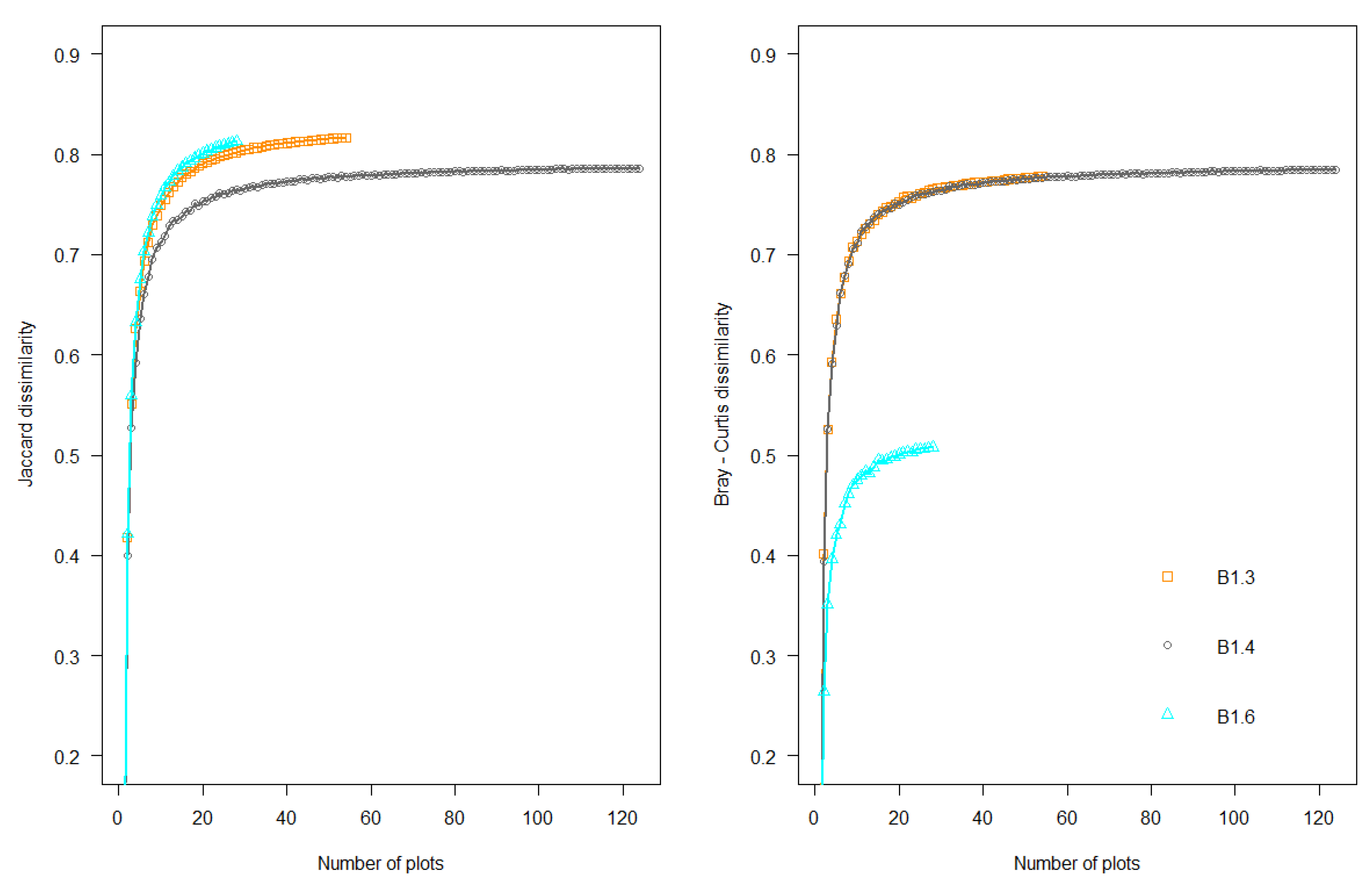
2.2.2. Species Composition Variation across Spatial Scales
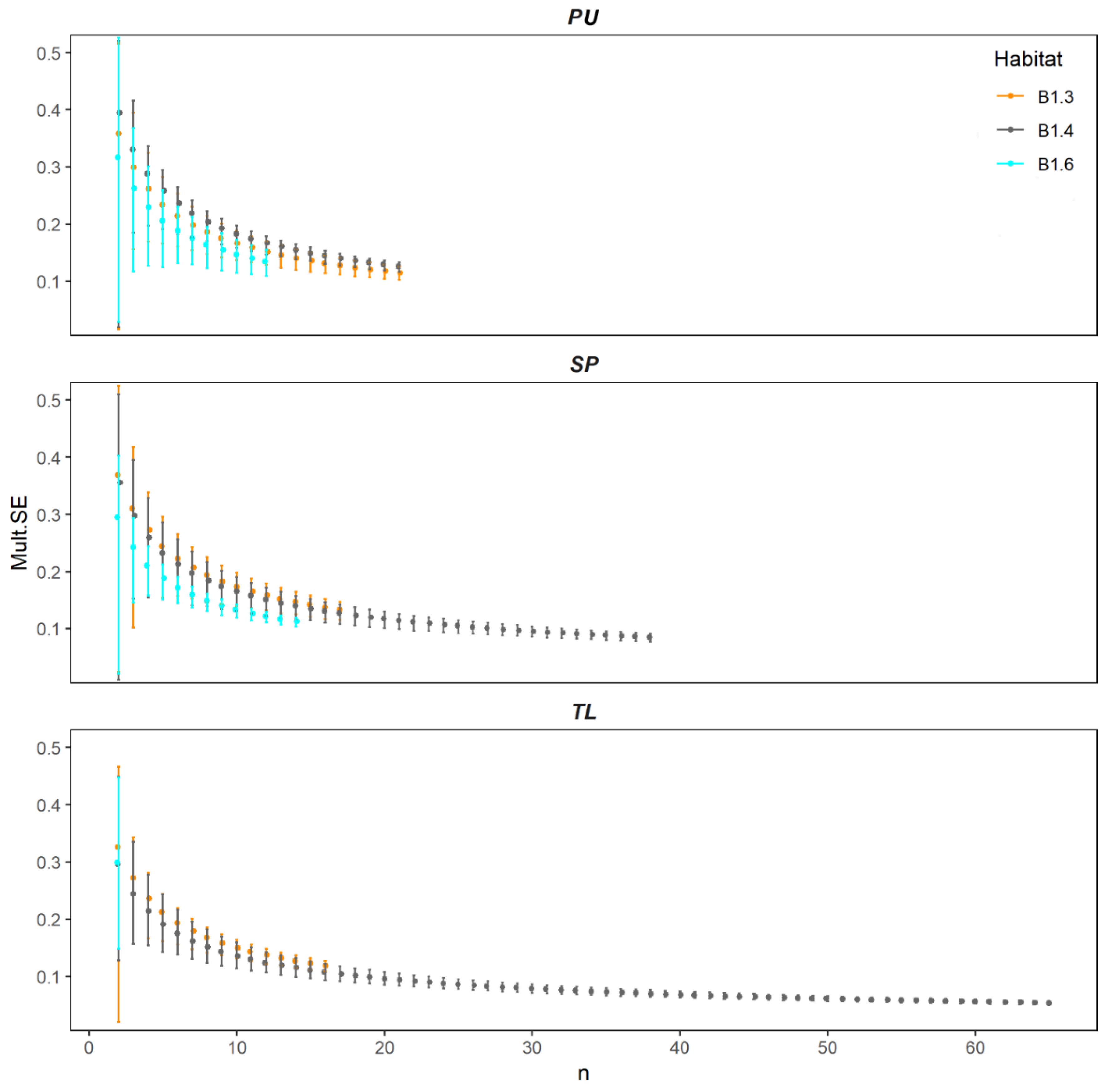
2.2.3. Measurement of Multivariate Precision and Associated Dissimilarities (MultSE)
2.2.4. Effect of Sampling Size Reduction
3. Results
4. Discussion
4.1. Diversity Patterns Across Habitats and SACs
4.2. Spatial Variation of Reduced Dataset
4.3. The Lesson We Learned
5. Conclusions
- conduct pilot studies testing different sampling probabilistic methods;
- wisely plan sampling efforts taking into account resource availability (i.e., time and costs);
- approaches based on plant functional traits and remote sensing may provide novel insights on ecosystem functioning, the latter revealed to be also a cost-effective way to handle biodiversity measurements and to predict species changes through time.
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Evans, D.; Arvela, M. Assessment and Reporting under Article 17 of the Habitats Directive. Explanatory Notes & Guidelines for the Period 2007–2012; Final Version; European Commission: Brussels, Belgium, 2011. [Google Scholar]
- Alberdi, I.; Nunes, L.; Kovac, M.; Bonheme, I.; Cañellas, I.; Rego, F.C.; Dias, S.; Duarte, I.; Notarangelo, M.; Rizzo, M.; et al. The conservation status assessment of Natura 2000 forest habitats in Europe: Capabilities, potentials and challenges of national forest inventories data. Ann. For. Sci. 2019, 76, 34. [Google Scholar] [CrossRef]
- Ellwanger, G.; Runge, S.; Wagner, M.; Ackermann, W.; Neukirchen, M.; Frederking, W.; Müller, C.; Ssymank, A.; Sukopp, U. Current status of habitat monitoring in the European Union according to Article 17 of the Habitats Directive, with an emphasis on habitat structure and functions and on Germany. Nat. Conserv. 2018, 29, 57–78. [Google Scholar] [CrossRef]
- Del Vecchio, S.; Fantinato, E.; Silan, G.; Buffa, G. Trade-offs between sampling effort and data quality in habitat monitoring. Biodivers. Conserv. 2019, 28, 55–73. [Google Scholar] [CrossRef]
- Yoccoz, N.G.; Nichols, J.D.; Boulinier, T. Monitoring of biological diversity in space and time. Trends Ecol. Evol. 2001, 16, 446–453. [Google Scholar] [CrossRef]
- Balmford, A.; Green, R.E.; Jenkins, M. Measuring the changing state of nature. Trends Ecol. Evol. 2003, 18, 326–330. [Google Scholar] [CrossRef]
- Legg, C.J.; Nagy, L. Why most conservation monitoring is, but need not be, a waste of time. J. Environ. Manag. 2006, 78, 194–199. [Google Scholar] [CrossRef]
- Cao, Y.; Larsen, D.P.; Hughes, R.M.; Angermeier, P.L.; Patton, T.M. Sampling effort affects multivariate comparisons of stream assemblages. J. N. Am. Benthol. Soc. 2002, 21, 701–714. [Google Scholar] [CrossRef]
- Marignani, M.; Del Vico, E.; Maccherini, S. Spatial scale and sampling size affect the concordance between remotely sensed information and plant community discrimination in restoration monitoring. Biodivers. Conserv. 2007, 16, 3851–3861. [Google Scholar] [CrossRef]
- Schmera, D.; Erős, T. The role of sampling effort, taxonomical resolution and abundance weight in multivariate comparison of stream dwelling caddisfly assemblages collected from riffle and pool habitats. Ecol. Ind. 2011, 11, 230–239. [Google Scholar] [CrossRef]
- Anderson, M.J.; Santana-Garcon, J. Measures of precision for dissimilarity-based multivariate analysis of ecological communities. Ecol. Lett. 2015, 18, 66–73. [Google Scholar] [CrossRef]
- Tordoni, E.; Napolitano, R.; Maccherini, S.; Da Re, D.; Bacaro, G. Ecological drivers of plant diversity patterns in remnants coastal sand dune ecosystems along the northern Adriatic coastline. Ecol. Res. 2018, 33, 1157–1168. [Google Scholar] [CrossRef]
- Maun, M.A. The Biology of Coastal Sand Dunes; Oxford University Press: New York, NY, USA, 2009. [Google Scholar]
- Dolan, A.H.; Walker, I.J. Understanding vulnerability of coastal communities to climate change related risks. J. Coast. Res. 2006, 39, 1316–1323. [Google Scholar]
- Coombes, E.G.; Jones, A.P.; Sutherland, W.J. The biodiversity implications of changes in coastal tourism due to climate change. Environ. Conserv. 2008, 35, 319–330. [Google Scholar] [CrossRef]
- Miller, T.E.; Gornish, E.S.; Buckley, H. Weather and coastal vegetation: Effects of storms and drought. Plant Ecol. 2010, 206, 97–104. [Google Scholar] [CrossRef]
- Ciccarelli, D. Mediterranean coastal dune vegetation: Are disturbance and stress the key selective forces that drive the psammophilous succession. Estuar. Coast. Shelf Sci. 2015, 165, 247–253. [Google Scholar] [CrossRef]
- Ciccarelli, D.; Pinna, M.S.; Alquini, F.; Cogoni, D.; Ruocco, M.; Bacchetta, G.; Sarti, G.; Fenu, G. Development of a coastal dune vulnerability index for Mediterranean ecosystems: A useful tool for coastal managers? Estuar. Coast. Shelf Sci. 2017, 187, 84–95. [Google Scholar] [CrossRef]
- Bertacchi, A. Dune habitats of the Migliarino—San Rossore—Massaciuccoli Regional Park (Tuscany—Italy). J. Maps 2017, 13, 322–331. [Google Scholar] [CrossRef]
- Carboni, M.; Santoro, R.; Acosta, A.T.R. Are some communities of the coastal dune zonation more susceptible to alien plant invasion? J. Plant Ecol. 2010, 3, 139–147. [Google Scholar] [CrossRef]
- Novoa, A.; González, L.; Moravcová, L.; Pyšek, P. Constraints to native plant species establishment in coastal dune communities invaded by Carpobrotus edulis: Implications for restoration. Biol. Conserv. 2013, 164, 1–9. [Google Scholar] [CrossRef]
- Tordoni, E.; Petruzzellis, F.; Nardini, A.; Savi, T.; Bacaro, G. Make it simpler: Alien species decrease functional diversity of coastal plant communities. J. Veg. Sci. 2019, 30, 498–509. [Google Scholar] [CrossRef]
- Carmignani, L.; Lazzarotto, A.; Brogi, A.; Conti, P.; Cornamusini, G.; Costantini, A.; Sandrelli, P. Carta geologica della Toscana (1: 250 000). Regione Toscana, Direzione Generale delle Politiche Territoriali e Ambientali, Servizio Geologico; Litografia Artistica Cartografica: Firenze, Italy, 2004. [Google Scholar]
- Pesaresi, S.; Galdenzi, D.; Biondi, E.; Casavecchia, S. Bioclimate of Italy: Application of the worldwide bioclimatic classification system. J. Maps 2014, 10, 538–553. [Google Scholar] [CrossRef]
- Medvecká, J.; Jarolímek, I.; Senko, D.; Svitok, M. Fifty years of plant invasion dynamics in Slovakia along a 2,500 m altitudinal gradient. Biol. Invasions 2014, 16, 1627. [Google Scholar] [CrossRef]
- Sperandii, M.G.; Bazzichetto, M.; Acosta, A.T.R.; Barták, V.; Malavasi, M. Multiple drivers of plant diversity in coastal dunes: A Mediterranean experience. Sci. Total Environ. 2019, 652, 1435–1444. [Google Scholar] [CrossRef]
- Davies, C.E.; Moss, D.; Hill, M.O. EUNIS Habitat Classification Revised; Report to European Environment Agency; European Topic Centre on Nature Protection and Biodiversity: Paris, France, 2004. [Google Scholar]
- Biondi, E.; Blasi, C.; Burrascano, S.; Casavecchia, S.; Copiz, R.; Del Vico, E.; Galdenzi, D.; Gigante, D.; Lasen, C.; Spampinato, G.; et al. Manuale Italiano di interpretazione degli habitat della Direttiva 92/43/CEE (Italian Interpretation Manual of the 92/43/EEC Directive habitats). Available online: http://vnr.unipg.it/habitat/index.jsp (accessed on 15 September 2019).
- Chiarucci, A.; Bacaro, G.; Rocchini, D.; Ricotta, C.; Palmer, M.; Scheiner, S. Spatially constrained rarefaction: Incorporating the autocorrelated structure of biological communities into sample-based rarefaction. Commun. Ecol. 2009, 10, 209–214. [Google Scholar] [CrossRef]
- Bacaro, G.; Rocchini, D.; Ghisla, A.; Marcantonio, M.; Neteler, M.; Chiarucci, A. The spatial domain matters: Spatially constrained species rarefaction in a Free and Open Source environment. Ecol. Complex. 2012, 12, 63–69. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. R package Version 2.5-6; Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 15 September 2019).
- Bacaro, G.; Altobelli, A.; Cameletti, M.; Ciccarelli, D.; Martellos, S.; Palmer, M.W.; Ricotta, C.; Rocchini, D.; Scheiner, S.M.; Tordoni, E.; et al. Incorporating spatial autocorrelation in rarefaction methods: Implications for ecologists and conservation biologists. Ecol. Indic. 2016, 69, 233–238. [Google Scholar] [CrossRef]
- Lande, R. Statistics and partitioning of species diversity, and similarity among multiple communities. Oikos 1996, 76, 5–13. [Google Scholar] [CrossRef]
- Crist, T.O.; Veech, J.A.; Gering, J.C.; Summerville, K.S. Partitioning species diversity across landscapes and regions: A hierarchical analysis of α, β, and γ-diversity. Am. Nat. 2003, 162, 734–743. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001, 26, 32–46. [Google Scholar]
- Anderson, M.J.; TerBraak, C.J.F. Permutation tests for multi-factorial analysis of variance. J. Stat. Comput. Sim. 2003, 73, 85–113. [Google Scholar] [CrossRef]
- Clarke, K.R.; Gorley, R.N. PRIMER v6: Users Manual/Tutorial; PRIMER-E: Plymouth, UK, 2006. [Google Scholar]
- Anderson, M.J.; Gorley, R.N.; Clarke, K.R. PERMANOVA+ for PRIMER: Guide to Software and Statistical Methods; PRIMER-E: Plymouth, UK, 2008; p. 214. [Google Scholar]
- Clarke, K.R. Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 1993, 18, 117–143. [Google Scholar] [CrossRef]
- Muggeo, V.M.R. Estimating regression models with unknown break-points. Stat. Med. 2003, 22, 3055–3071. [Google Scholar] [CrossRef] [PubMed]
- Muggeo, V.M.R. Segmented: An R Package to Fit Regression Models with Broken-Line Relationships. R. News 2008, 8/1, 20–25. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria, 2020. Available online: https://www.R-project.org (accessed on 15 September 2019).
- Foster, B.L.; Tilman, D. Dynamic and static views of succession: Testing the descriptive power of the chronosequence approach. Plant Ecol. 2000, 146, 1–10. [Google Scholar] [CrossRef]
- Acosta, A.; Carranza, M.L.; Izzi, C.F. Are there habitats that contribute best to plant species diversity in coastal dunes? Biodivers. Conserv. 2009, 18, 1087–1098. [Google Scholar] [CrossRef]
- Kuiters, A.; Kramer, K.V.; Van der Hagen, H.G.J.M.; Schaminée, J.H.J. Plant diversity, species turnover and shifts in functional traits in coastal dune vegetation: Results from permanent plots over a 52-year period. J. Veg. Sci. 2009, 20, 1053–1063. [Google Scholar] [CrossRef]
- Ciccarelli, D.; Bacaro, G. Quantifying plant species diversity in coastal dunes: A piece of help from spatially constrained rarefaction. Folia Geobot. 2016, 51, 129–141. [Google Scholar] [CrossRef]
- Carranza, M.L.; Acosta, A.T.R.; Stanisci, A.; Pirone, G.; Ciaschetti, G. Ecosystem classification for EU habitat distribution assessment in sandy coastal environments: An application in central Italy. Environ. Monit. Assess. 2008, 140, 99–107. [Google Scholar] [CrossRef]
- Calvão, T.; Pessoa, M.F.; Lidon, F.C. Impact of human activities on coastal vegetation: A review. Emir. J. Food Agric. 2013, 25, 926. [Google Scholar]
- Angiolini, C.; Landi, M.; Pieroni, G.; Frignani, F.; Finoia, M.G.; Gaggi, C. Soil chemical features as key predictors of plant community occurrence in a Mediterranean coastal ecosystem. Estuar. Coast. Shelf Sci. 2013, 119, 91–100. [Google Scholar] [CrossRef]
- Angiolini, C.; Bonari, G.; Landi, M. Focal plant species and soil factors in Mediterranean coastal dunes: An undisclosed liaison? Estuar. Coast. Shelf Sci. 2018, 211, 248–258. [Google Scholar] [CrossRef]
- Del Vecchio, S.; Fantinato, E.; Janssen, J.A.M.; Bioret, F.; Acosta, A.; Prisco, I.; Tzonev, R.; Marcenò, R.; Rodwell, J.; Buffa, G. Biogeographic variability of coastal perennial grasslands at the European scale. Appl. Veg. Sci. 2018, 21, 312–321. [Google Scholar] [CrossRef]
- Maccherini, S.; Marignani, M.; Gioria, M.; Renzi, M.; Rocchini, D.; Santi, E.; Torri, D.; Tundo, J.; Honnay, O. Determinants of plant community composition of remnant biancane badlands: A hierarchical approach to quantify species-environment relationships. Appl. Veg. Sci. 2011, 14, 378–387. [Google Scholar] [CrossRef]
- Torri, D.; Rossi, M.; Brogi, F.; Marignani, M.; Bacaro, G.; Santi, E.; Tordoni, E.; Amici, V.; Maccherini, S. Badlands and the dynamics of human history, land use, and vegetation through centuries—Chapter IV. In Badlands Dynamics in a Context of Global Change; Nadal-Romero, E., Martínez-Murillo, J.F., Khun, N., Eds.; Elsevier: Amsterdam, The Netherlands, 2018; pp. 111–153. [Google Scholar]
- De l Vecchio, S.; Slaviero, A.; Fantinato, E.; Buffa, G. The use of plant community attributes to detect habitat quality in coastal environments. AoB Plants 2016, 8, plw040. [Google Scholar] [CrossRef]
- Wiedemann, A.M.; Pickart, A.J. Temperate zone coastal dunes. In Coastal Dunes: Ecology and Conservation. Ecological Studies: Analysis and Synthesis; Martínez, M.L., Psuty, N.P., Eds.; Springer: Heidelberg, Germany, 2004; pp. 53–65. [Google Scholar]
- Chelli, S.; Simonetti, E.; Campetella, G.; Chiarucci, A.; Cervellini, M.; Tardella, F.M.; Tomasella, M.; Canullo, R. Plant diversity changes in a nature reserve: A probabilistic sampling method for quantitative assessments. Nat. Conserv. 2019, 34, 145–161. [Google Scholar] [CrossRef]
- Diekmann, M.; Kühne, A.; Isermann, M. Random vs non-random sampling: Effects on patterns of species abundance, species richness and vegetation-environment relationships. Folia Geobot. 2007, 42, 179. [Google Scholar] [CrossRef]
- Lájer, K. Statistical tests as inappropriate tools for data analysis performed on non-random samples of plant communities. Folia Geobot. 2007, 42, 115–122. [Google Scholar] [CrossRef]
- Swacha, G.; Botta-Dukát, Z.; Kącki, Z.; Pruchniewicz, D.; Żołnierz, L.A. Performance comparison of sampling methods in the assessment of species composition patterns and environment–vegetation relationships in species-rich grasslands. Acta Soc. Bot. Pol. 2017, 86, 3561. [Google Scholar] [CrossRef]
- Stohlgren, T.J.; Chong, G.W.; Kalkhan, M.A.; Schell, L.D. Rapid assessment of plant diversity patterns: A methodology for landscapes. Environ. Monit. Assess. 1997, 48, 25–43. [Google Scholar] [CrossRef]
- Loos, J.; Hanspach, J.; von Wehrden, H.; Beldean, M.; Moga, C.I.; Fischer, J. Developing robust field survey protocols in landscape ecology: A case study on birds, plants and butterflies. Biodivers. Conserv. 2015, 24, 33–46. [Google Scholar] [CrossRef][Green Version]
- Bried, J.T.; Pellet, J. Optimal design of butterfly occupancy surveys and testing if occupancy converts to abundance for sparse populations. J. Insect. Conserv. 2012, 16, 489–499. [Google Scholar] [CrossRef]
- Maccherini, S.; Bacaro, G.; Giovannetti, G.; Angiolini, C.; Chiarucci, A. Analysing methodological issues in short-term monitoring of rare European beech forests restoration. Plant Biosyst. 2019, 153, 60–67. [Google Scholar] [CrossRef]
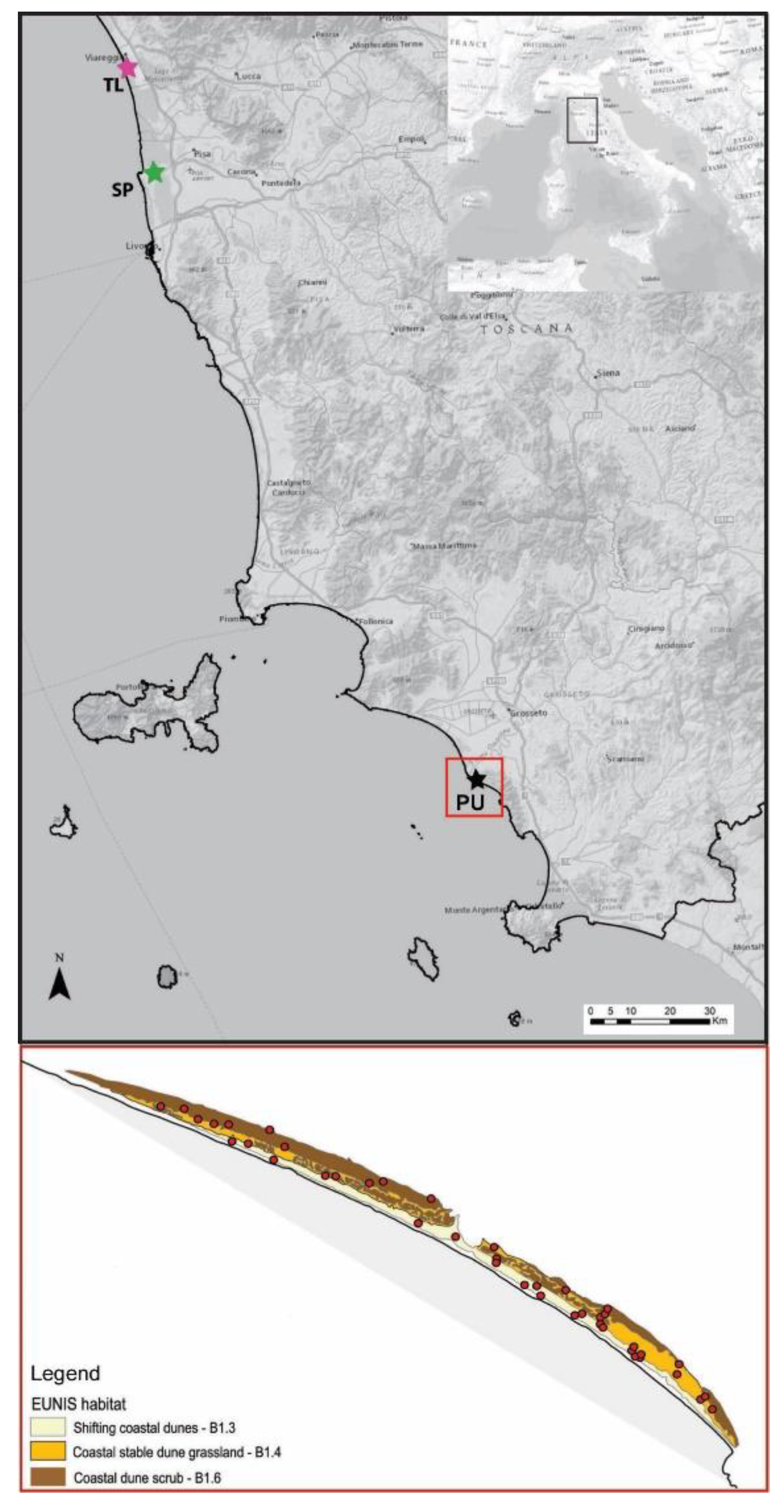
| EUNIS Habitat | EU Habitat (Directive 92/43/EEC) | Target Species |
|---|---|---|
| Shifting coastal dunes (B1.3) | 2110- Embryonic shifting dunes | Ammophila arenaria, Anthemis maritima, Calystegia soldanella, Cyperus capitatus, Echinophora spinosa, Elymus farctus, Eryngium maritimum, Euphorbia peplis, Euphorbia paralias, Lotus creticus, L. cytisoides, Medicago marina, Othantus maritimus, Pancratium maritimum, Polygonum maritimum, Solidago litoralis, Sporobolus arenarius, Stachys maritima |
| 2120 -Shifting dunes along the shoreline with Ammophila arenaria (white dunes) | ||
| Coastal stable dune grassland (B1.4) | 2210 -Fixed coastal dunes with herbaceous vegetation (grey dunes) | Anthemis mixta, Bromus diandrus, Corrigiola telephifolia, Corynephorus divaricatus, Crucianella maritima, Cutandia maritima, Daucus pumilus, Eva xpygmaea, Helichrysum stoechas, Lagurus ovatus, Lupinus angustifolius, Malcolmia ramosissima, Maresia nana, Matthiola tricuspidata, Medicago littoralis, Ononis variegata, Pancratium maritimum, Phleum arenarium, Polycarpon diphyllum, Pseudorlaya pumila, Pycnocomon rutifolium, Seseli tortuosum, Silene canescens, S. gallica, S. niceensis, Sonchus bulbosus, Thesium humile, Vulpia fasciculata |
| 2230 - Malcolmietalia dune grasslands | ||
| Coastal dune scrub (B1.6) | 2230 -Malcolmietalia dune grasslands 2240 -Brachypodietalia dune grasslands with annuals 2250* - Coastal dunes with Juniperus spp. | Aetheorhiza bulbosa, Aira elegans, Andryala integrifolia, Anthyllis barba-jovis, Asparagus acutifolius, Brachypodium distachyum, Briza maxima, Clematis flammula, Corynephorus divaricatus, Corrigiola telephifolia, Cutandia maritima, Evax pygmaea, Galium divaricatum, Juniperus communis, Juniperus macrocarpa, J. turbinata, Lagurus ovatus, Lonicera implexa, Lotus angustissimus, Lupinus angustifolius, Malcolmia ramosissima, Maresia nana, Matthiola tricuspidata, Medicago littoralis, Myrtus communis, Ononis variegata, Ornithopus compressus, Phillyrea angustifolia, P. latifolia, Phleum arenarium, Pistacia lentiscus, Polycarpon diphyllum, Plantago lagopus, P. bellardii, Prasium majus, Pseudorlaya pumila, Rhamnus alaternus, Rubia peregrina, Rumex bucephalophorus, Ruscus aculeatus, Silene canescens, S. nicaensis, S. gallica, Smilax aspera, Tuberaria guttata, Vulpia membranacea |
| Term | Levels | Name | Area (Ha) | N° Plots | Average Richness (Min-Max) |
|---|---|---|---|---|---|
| HABITAT | B1.3 | Shifting coastal dunes | 54.70 | 54 | 7 (2–12) |
| B1.4 | Stable dune grasslands | 121.94 | 124 | 10 (3–21) | |
| B1.6 | Coastal dune scrubs | 42.33 | 28 | 9 (2–17) | |
| SAC | TL | Torre del Lago | 81.26 | 83 | 11 (4–21) |
| PU | Parco dell’Uccellina | 73.33 | 54 | 7 (3–12) | |
| SP | SelvaPisana | 64.39 | 69 | 8 (2–17) | |
| HABITAT:SAC | B1.3:PU | Shifting coastal dunes: Parco dell’Uccellina | 20.11 | 21 | 7 (3–12) |
| B1.3:SP | Shifting coastal dunes: SelvaPisana | 19.26 | 17 | 6 (2–11) | |
| B1.3:TL | Shifting coastal dunes: Torre del Lago | 15.32 | 16 | 8 (4–11) | |
| B1.4:PU | Stable dune grasslands: Parco dell’Uccellina | 20.08 | 21 | 6 (3–11) | |
| B1.4:SP | Stable dune grasslands: SelvaPisana | 40.96 | 38 | 8 (3–16) | |
| B1.4:TL | Stable dune grasslands: Torre del Lago | 60.91 | 65 | 12 (6–21) | |
| B1.6:PU | Coastal dune scrub: Parco dell’Uccellina | 33.14 | 12 | 8 (4–12) | |
| B1.6:SP | Coastal dune scrub: Selva Pisana | 41.62 | 14 | 9 (2–17) | |
| B1.6:TL | Coastal dune scrub: Torre del Lago | 5.03 | 2 | 12 (11–12) |
| Source of Variation | df | MS | F | Variance Components (%) |
|---|---|---|---|---|
| Abundance | ||||
| SAC | 2 | 14,817 | 6.17 *** | 15.58 |
| Habitat | 2 | 42,946 | 17.86 *** | 25.68 |
| SAC × Habitat | 4 | 8988 | 3.74 *** | 16.97 |
| Residual | 197 | 2405 | 41.77 | |
| Total | 205 | |||
| presence/absence | ||||
| SAC | 2 | 19,916 | 8.99 *** | 18.53 |
| Habitat | 2 | 40,571 | 18.31 *** | 24.86 |
| SAC × Habitat | 4 | 8653 | 3.91 *** | 16.70 |
| Residual | 197 | 2215 | 39.91 | |
| Total | 205 |
| Levels | Name | Abundance | Presence/Absence |
|---|---|---|---|
| B1.3 | Shifting coastal dunes | 9 ± 0.3 | 10 ± 0.4 |
| B1.4 | Stable dune grasslands | 25 ± 2.2 | 14 ± 0.4 |
| B1.6 | Coastal dune scrubs | 7 ± 0.3 | 7 ± 0.3 |
| TL | Torre del Lago | 12 ± 0.4 | 11 ± 0.4 |
| PU | Parco dell’Uccellina | 9 ± 0.3 | 9 ± 0.4 |
| SP | SelvaPisana | 10 ± 0.4 | 10 ± 0.4 |
| B1.3:PU | Shifting coastal dunes: Parco dell’Uccellina | 6 ± 0.3 | 6 ± 0.3 |
| B1.3:SP | Shifting coastal dunes: SelvaPisana | 5 ± 0.3 | 5 ± 0.3 |
| B1.3:TL | Shifting coastal dunes: Torre del Lago | 5 ± 0.3 | 5 ± 0.3 |
| B1.4:PU | Stable dune grasslands: Parco dell’Uccellina | 6 ± 0.3 | 6 ± 0.3 |
| B1.4:SP | Stable dune grasslands: SelvaPisana | 8 ± 0.3 | 8 ± 0.3 |
| B1.4:TL | Stable dune grasslands: Torre del Lago | 10 ± 0.4 | 10 ± 0.4 |
| B1.6:PU | Coastal dune scrub: Parco dell’Uccellina | 4 ± 0.2 | 4 ± 0.2 |
| B1.6:SP | Coastal dune scrub: Selva Pisana | 5 ± 0.3 | 5 ± 0.2 |
| B1.6:TL | Coastal dune scrub: Torre del Lago | - | - |
| Abundance | Presence/Absence | ||||||
|---|---|---|---|---|---|---|---|
| Term | Statistic | F | R2 | Rate of Significance (p < 0.05) | F | R2 | Rate of Significance (p < 0.05) |
| SAC | Min. | 2.62 | 0.06 | 100% | 2.31 | 0.08 | 100% |
| 1st quart. | 3.86 | 0.09 | 3.11 | 0.10 | |||
| Median | 4.31 | 0.01 | 3.38 | 0.10 | |||
| 3rd quart. | 4.77 | 0.11 | 3.65 | 0.11 | |||
| Max. | 6.84 | 0.15 | 5.22 | 0.15 | |||
| Habitat | Min. | 4.67 | 0.13 | 100% | 2.74 | 0.09 | 100% |
| 1st quart. | 7.20 | 0.19 | 4.03 | 0.13 | |||
| Median | 8.11 | 0.21 | 4.47 | 0.14 | |||
| 3rd quart. | 9.06 | 0.23 | 4.93 | 0.15 | |||
| Max. | 12.97 | 0.30 | 7.31 | 0.20 | |||
| SAC × Habitat | Min. | 1.11 | 0.06 | 94.3% | 1.15 | 0.08 | 98.7% |
| 1st quart. | 1.67 | 0.09 | 1.53 | 0.10 | |||
| Median | 1.88 | 0.01 | 1.67 | 0.10 | |||
| 3rd quart. | 2.13 | 0.11 | 1.83 | 0.11 | |||
| Max. | 3.38 | 0.15 | 2.70 | 0.14 | |||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maccherini, S.; Bacaro, G.; Tordoni, E.; Bertacchi, A.; Castagnini, P.; Foggi, B.; Gennai, M.; Mugnai, M.; Sarmati, S.; Angiolini, C. Enough Is Enough? Searching for the Optimal Sample Size to Monitor European Habitats: A Case Study from Coastal Sand Dunes. Diversity 2020, 12, 138. https://doi.org/10.3390/d12040138
Maccherini S, Bacaro G, Tordoni E, Bertacchi A, Castagnini P, Foggi B, Gennai M, Mugnai M, Sarmati S, Angiolini C. Enough Is Enough? Searching for the Optimal Sample Size to Monitor European Habitats: A Case Study from Coastal Sand Dunes. Diversity. 2020; 12(4):138. https://doi.org/10.3390/d12040138
Chicago/Turabian StyleMaccherini, Simona, Giovanni Bacaro, Enrico Tordoni, Andrea Bertacchi, Paolo Castagnini, Bruno Foggi, Matilde Gennai, Michele Mugnai, Simona Sarmati, and Claudia Angiolini. 2020. "Enough Is Enough? Searching for the Optimal Sample Size to Monitor European Habitats: A Case Study from Coastal Sand Dunes" Diversity 12, no. 4: 138. https://doi.org/10.3390/d12040138
APA StyleMaccherini, S., Bacaro, G., Tordoni, E., Bertacchi, A., Castagnini, P., Foggi, B., Gennai, M., Mugnai, M., Sarmati, S., & Angiolini, C. (2020). Enough Is Enough? Searching for the Optimal Sample Size to Monitor European Habitats: A Case Study from Coastal Sand Dunes. Diversity, 12(4), 138. https://doi.org/10.3390/d12040138








