Abstract
Soil microorganisms are critical factors of plant productivity in terrestrial ecosystems. Coptis chinensis Franch is one of the most important medicinal plants in China. Soil types and cropping systems influence the diversity and composition of the rhizospheric microbial communities. In the current study, we provide detailed information regarding the diversity and composition of the rhizospheric bacterial communities of the C. chinensis plants in continuously cropped fields and fallow fields in two seasons (i.e., winter and summer) using next-generation sequencing. The alpha diversity was higher in the five-year cultivated C. chinensis field (CyS5) and lower in fallow fields (NCS). Significant differences analysis confirmed more biomarkers in the cultivated field soil than in fallow fields. Additionally, the principal coordinate analysis (PcoA) of the beta diversity indices revealed that samples associated with the cultivated fields and fallow fields in different seasons were separated. Besides, Proteobacteria, Actinobacteria, Chloroflexi, Acidobacteria, Bacteroidetes, Gemmatimonadetes were the top bacterial phyla. Among these phyla, Proteobacteria were found predominantly and showed a decreasing trend with the continuous cropping of C. chinensis. A phylogenetic investigation of communities by reconstruction of unobserved states (PICRUSt) revealed that the abundance of C and N functional genes had a significant difference between the soil samples from cultivated (CyS1, CyS3, and CyS5) and fallow (NCS) fields in two seasons (winter and summer). The principal coordinate analysis (PCoA) based on UniFrac distances (i.e., unweighted and weighted) revealed the variations in bacterial community structures in the soil samples. This study could provide a reference for solving the increasingly severe cropping obstacles and promote the sustainable development of the C. chinensis industry.
1. Introduction
Coptis chinensis Franch is one of the essential traditional medicinal plants in the family Ranunculaceae [1]. The C. chinensis rhizome is prescribed for many medicinal purposes such as diabetes treatment, distress, jaundice, acute fever and mild fever, seasonal fever disease, sore throat, fever reduction, and diarrhea [2,3]. Previous phytochemical studies on C. chinensis found more than 30 alkaloids. Among these alkaloids, berberine, epiberberine, palmatine, coptisine, and jatrorrhizine, which are predominantly active isoquinoline alkaloids, have been confirmed as significant constituents [4].
C. chinensis is distributed in areas such as Guizhou Province, Sichuan Province, Chongqing City, Shanxi Province, and Hunan Province in China. Currently, the high-quality and -quantity production of C. chinensis is more dependent on the cultivation method. Because of the increasing demands for this medicinal plant in the market, continuous cropping of C. chinensis has become common. Long-term continuous cropping of Chinese herbs often results in plant growth reduction, severe root rot disease, and significant loss of function. A continuous cropping of C. chinensis has resulted in a 70% to 80% yield reduction. Simultaneously, root rot disease has become a substantial threat to C. chinensis production [3,5].
Soil microorganisms are crucial for crop production in agriculture systems in order to maintain soil health [6,7]. Many previous studies showed that soil microbial communities are affected by many factors, including plant species, soil types, organic breeding, and agricultural management [8,9]. Bacteria are the most plentiful and diverse group of microorganisms in the soil, and it is estimated that there are 109–1010 bacteria and 6000 to 50,000 bacterial species per gram of soil [6].
It has been well documented that management practices (e.g., fertilizer application, periodic outflow, and crop rotation), plant species, soil type, geographical distance, and so forth, determine the structure of bacterial communities [6,10]. The choice of cultivation method is essential to increase crop yield and suppress disease, and it may cause changes in microbial communities as different plant species secrete different types of root exudates, which can alter the structure of the soil bacterial community [11]. Some molecular analytical technologies, such as 454 pyrosequencing analysis [5,12], denaturing gradient gel electrophoresis [13], terminal restriction fragment length polymorphism [14], and fluorescence in situ hybridization [15], are used to analyze the diversity of soil microbes in continuous cropping systems. They cannot overcome species deficiency and generally cannot provide sufficient data for analysis in a short time. Recently, high-throughput sequencing has often been used to detect the diversity of rhizosphere soil microbes as well as their relative abundance and evolution [16,17,18,19,20]. Despite that, most of the previous studies focused on the bacterial community structure and composition in the rhizospheric soil of crop and medicinal plants, but few studies on the metabolic activity and functional diversity of the bacterial community, which are crucial for C. chinesis production, exist. High-throughput sequencing could not directly present microbial function characteristics [7,11,12,16,21].
Recently, PICRUSt (phylogenetic investigation of communities by reconstruction of unobserved states) has been used to predict the functional characteristics of the bacterial community based on the 16s rRNA homology and conserved feature of functional contribution [22,23,24,25]. This cost-effective bioinformatics tool can use only a few hundred sequences to grasp most of the variations in gene content. The accuracy of PICRUSt prediction is above 85%. Therefore, PICRUSt is a powerful approach to understand the functional diversity of the bacterial community in the rhizosphere [26].
In this study, three replanted C. chinensis fields with continuous cropping histories for one, three, and five years and fallow fields in which C. chinensis was not cultivated for more than three years were selected to investigate the effect of continuous cropping on soil physicochemical properties, bacterial abundance, bacterial community structures, and bacterial functional activities. This study was conducted based on a large amount of data from next-generation sequencing (NGS) platforms such as Illumina MiSeq using 16S bacterial primers. The objectives of our study were 1) to explore the correlation between soil physicochemical properties and the bacterial community, 2) to find the bacterial community composition diversity and structure, and 3) to use PICRUSt to predict the metabolic properties of the bacterial community in rhizosphere soil.
2. Materials and Methods
2.1. Study Site and Soil Sampling
The soil samples were collected from the cropping fields of the C. chinensis plants at Jiannan town, Lichuan City, Hubei province, China (coordinates: 108°23′–108°35′ E, 30°18′–30°35′ N; altitude 1500 meters; soil types mostly sandy and clay; average annual rainfall between 1198 and 1650 mm and an average yearly temperature of 12.7 °C). The rhizospheric soil sampling (2 mm) was conducted on the continuously cropped C. chinensis fields for one year (CyS1), three years (CyS3), and five years (CyS5), and fallow fields (NCS) that were not cropped for more than three years, covering two seasons of winter and summer. Four C. chinensis fields were randomly selected for the experiment—three from the continuously cultivated areas and one from the fallow fields. One composite rhizosphere sample was taken per field from the roots of 10 randomly selected C. chinensis plants. The plant roots were shaken vigorously to separate the soil from tightly adhering to the roots. As there were no plants in the fallow fields, one composite soil sample consisting of five cores of about 15 cm of topsoil was taken. The soil was then transported to the laboratory in iceboxes. At the laboratory, each soil sample was properly sieved to remove debris and other stony materials using a (2 mm) sieve. Each sample was appropriately homogenized and 10 g of soils were put in the sterilized tubes and stored at −80 °C for DNA extraction; other soils were saved for an analysis of the physicochemical properties of the soil.
2.2. Analysis of Soil Physicochemical Properties
The soil pH was measured with a Mettler-Toledo TE 20 (Mettler-Toledo, Columbus, OH USA) using soil suspension with deionized distilled water (1:20 w/v). The soil organic matter was tested using the potassium dichromate internal heating method. The total contents of N and P were measured using a SMARTCHEM 200 Discrete analyzer (Unity Scientific, Milford, MA USA), and the total content of K was measured using an FP series multielement flame photometer (Xiang Yi, Hunan, China).
2.3. DNA Extraction and MiSeq Sequencing
Genomic DNA was extracted from 0.5 g of soil samples (dry weight) using the PowerSoil kit (MO BIO Laboratories, Carlsbad, CA, USA). The concentration of the extracted DNA was determined using a Nanodrop 2000C Spectrophotometer (Thermo Scientific, Wilmington, DE, USA). The DNA extract from the soil samples was kept at −80 °C until it was used.
The V4–V5 regions of the bacterial 16S rRNA gene were amplified with primers 338F (5’-ACTCCTACGGGAGGCAGCAG-3’) and 806R (5’-GGACTACHVG GTWTCTAAT-3’). The following program was used to conduct the PCR reaction: 3 min of denaturation at 95 °C, 27 cycles of 30 s at 95 °C, 30 s for annealing at 55 °C, 45 s for elongation at 72 °C, and a final extension at 72 °C for 10 min.
The PCR reactions were performed in triplicate using a 20 μL mixture containing 4 μL of 5 × FastPfu Buffer, 2 μL of 2.5 mM dNTPs, 0.8 μL of each primer (5 μM), 0.4 μL of FastPfu Polymerase, 0.2 μL BSA, and 10 ng of template DNA.
The resultant PCR products were extracted from a 2% agarose gel and further purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) and quantified using QuantiFluor™-ST (Promega, Madison, WI, USA) according to the manufacturer’s protocol.
2.4. Bioinformatic Analyses
We pooled the purified amplicons in equimolar and paired-end sequenced (2 × 300) on an Illumina MiSeq platform (Illumina, San Diego, CA, USA) according to standard protocols by the company. Illumina sequencing was used to generate the raw gene sequences data and then the Trimmonmatic and FLASH were used to modify the data. Usearch 7.1 (http://qiime.org/) was used to cluster the sequences into operational taxonomic units (OTUs) after a 97% pairwise identity by QIIME. Ribosomal Database Project classifier (Release 11.1 http://rdp.cme.msu.edu/) was used for the taxonomic classification of the representative sequence for bacteria against the Greengenes 16S rRNA database (Release 13.5 http://greengenes.secondgenome.com/) and Silva (Release 115 http://www.arb-silva.de) [27]. The bacterial OTUs were imported into PICRUSt, which had demonstrated that the genome prediction accuracy was >0.80. The metabolic pathways were analyzed using the module KEGG after normalizing the OTU table [26].
2.5. Statistical Analyses
One-way analyses of variance (ANOVA) with Tukey’s HSD (honestly significant difference) multiple range tests were done for multiple comparisons and for comparison of N and C cycling genes’ relative abundance. Pearson correlation coefficients between the soil properties, and abundances of bacterial phyla were all calculated using SPSS v20.0 (SPSS Inc., Chicago, III, USA). For alpha diversity, all analyses were based on the OTU clusters with a cutoff of 3% dissimilarity. Chao, Shannon, Simpson, and Phylogenetic diversity (Pd) were calculated to estimate the richness and diversity of the bacterial community of each sample. Rarefaction curves with the average number of observed OTUs were generated using Mothur to compare relative levels of bacterial OTU diversity across the four different C. chinensis field soils. The weighted and unweighted UniFrac distance metrics (based on the phylogenetic structure) were used to generate PCoA plots to assess the similarities between the samples’ community memberships further. At the end, Venn diagrams were constructed to visualize shared and unique species between samples.
3. Results
3.1. Soil Chemical Properties
With the increasing years of C. chinensis cultivation, the total content of nitrogen, phosphorus, and organic matter significantly increased in both winter and summer seasons. The soil organic matter varied from 11.16 to 36.04 g kg−1 in winter and from 8.35 to 33.90 g kg−1 in summer. From this, the soil of the five-year (CyS5) fields was found to have the highest content of organic matter in winter and summer (36.04, 33.90 g kg−1, respectively) and the content of organic matter was lowest in the one-year (CyS1) fields (11.16, 8.35 g kg−1, respectively). The organic matter contents were significantly higher in the five-year cultivated fields (CyS5) and lower in the one-year cultivated fields (CyS1). The soil total nitrogen values varied from 0.99 to 3.27 g kg−1 in winter and from 1.02 to 2.68 g kg−1 in summer, among which the five-year (CyS5) soil had the highest content in winter and summer (3.27, 2.68 g kg−1, respectively) and the one-year (CyS1) soil had the lowest content in both winter and summer. The total soil potassium value was highest in the five-year (CyS5) fields in both winter and summer (4.18, 4.18 g kg−1, respectively) and had the lowest value in the fallow fields (NCS) (1.17, 1.47 g kg−1, respectively). Furthermore, the pH was slightly acidic and varied from 4.49 to 5.84 in winter and 5.59 to 6.48 in summer, the highest value in one-year (CyS1) soil and the lowest value in five-year (CyS5) soil in two seasons; see Table 1.

Table 1.
Physicochemical characteristics of the rhizospheric soil in two seasons (winter and summer).
3.2. Description of Bacterial Community
Sequences were randomly selected from the modified sequences, and the number of sequences was plotted against the number of OTUs they represent in order to build the rarefaction curves (Figure S1). The curves tended to be flat as the number of sequences increased and with the upper limit of OTUs for one year. After the quality-filtering steps, 903,804 high-quality sequences were obtained with an average of 56,487.75 per sample in the most common length range (230–487 bp) in the winter and 940,104 high-quality sequences with an average of 58,756.5 sequences per sample in the most common length range (223–488 bp) in the summer (Table S1).
One-year (CyS1), three-year (CyS3), five-year (CyS5), and fallow (NCS) fields were 3135, 2388, 1900, and 1925, respectively, in the winter season, and 2089, 1745, 1410, and 1126, respectively, in the summer season. Thus, we concluded that our sequencing data were suitable for analysis and that more sequencing data would not give many more OTUs. The upper limits of the OTUs for both the three-year (CyS3) and five-year (CyS5) fields were lower than that of the one-year (CyS1) field, indicating that continuous cropping caused an increase in the community diversity; this was more evident after one year of constant cropping, which corresponded with the diversity indexes.
Venn diagrams can directly show the similarity and overlap of the species between different samples; 264, 13, 23, and 47 particular species were identified in the one-year (CyS1), three-year (CyS3), five-year (CyS5), and fallow (NCS) fields, respectively, in the winter, while 225, 32, 19, and 113 unique species were identified, respectively, in the summer. Among the samples, 730 (7.62%) species were shared in the winter and 584 (7.11%) species were divided in the summer while the one-year (CyS1) cultivated field had the most significant number of species in both winter and summer soil sampling. This result indicated that the continuous cropping of C. chinensis could change the species number in the rhizosphere (Figure 1).
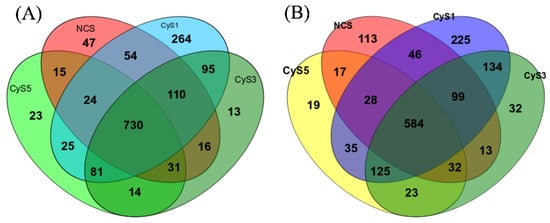
Figure 1.
Venn diagrams of unique species of bacteria in the soils from 4 time-series C. chinensis fields in winter (A) and summer (B). NCS, fallow field soil; CyS1, one-year C. chinensis cultivated soil; CyS3, three-year C. chinensis cultivated soil; CyS5, five-year C. chinensis cultivated soil.
3.3. Alpha Diversity
Alpha diversity indices (i.e., Chao, Shannon, Simpson, Observed OTUs, and Phylogenetic diversity (Pd)) were used to reveal the bacterial community diversity in the samples. The alpha diversity of the bacterial communities was considerably higher in the one-year (CyS1) fields as compared to the three-year (CyS3) and five-year (CyS5) fields. The diversity was lowest in the fallow fields (NCS) as compared to the cultivated fields (CyS1, CyS3, and CyS5). Among the cultivated fields, the observed OTUs were significantly higher in the one-year field (Table S2 and Figure 2).
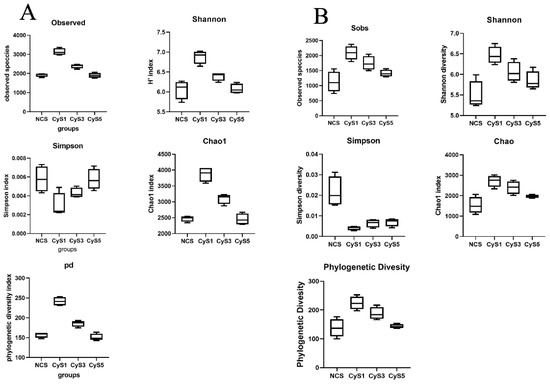
Figure 2.
Box plots showing alpha diversity (Sobs, Chao, Shannon, phylogenetic diversity, and Simpson Index) variation across samples in winter (A) and summer (B). NCS, fallow field soil; CyS1, one-year C. chinensis cultivated soil; CyS3, three-year C. chinensis cultivated soil; CyS5, five-year C. chinensis cultivated soil.
3.4. Composition of Bacterial Community
The OTUs present in these four samples and two seasons, including no-rank and unclassified, were divided into 40 phyla, 388 families, and 735 genera in the winter season and 34 phyla, 396 families, and 696 genera in the summer season. Among the 40 phyla, Proteobacteria, Actinobacteria, Chloroflexi, Acidobacteria, and Bacteroidetes with average relative abundances of 30.33%, 22.41%, 14.38%, 13.70%, and 4.34%, respectively, were the top five bacterial phyla in winter, whereas, among the 34 phyla, Proteobacteria, Acidobacteria, Chloroflexi, Actinobacteria, and Patescibacteria, with average relative abundances of 37.54%, 24.91%, 10.74%, 6.96%, and 4.77%, respectively, were the five most dominated bacterial phyla in the summer season (Table S3).
Additionally, in the winter, one-way ANOVA results showed that the relative abundances of Proteobacteria and Bacteroidetes phyla decreased with long-term continuous cropping of C. chinensis. Compared to the (NCS) fallow field, the cultivated fields (CyS1, CyS3, and CyS5) had higher relative abundances of Proteobacteria, Acidobacteria, and Gemmatimonadetes, and lower abundance of Actinobacteria, Chloroflexi, Bacteroidetes, and Cyanobacteria. However, in summer, the Acidobacteria phyla significantly increased with the increase in the number of cropping years, whereas the Proteobacteria, Patescibacteria, and Bacteroidetes phyla decreased significantly. Compared to the fallow field (NCS), the cultivated fields (CyS1, CyS3, and CyS5) had a higher relative abundance of Acidobacteria, Actinobacteria, and Gemmatimonadetes and lower relative abundance of Chloroflexi and Patescibacteria in summer (Figure 3).
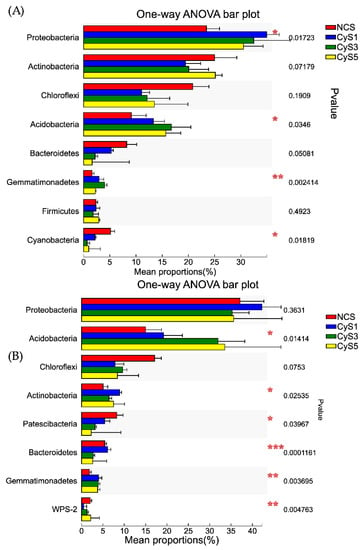
Figure 3.
Multiple comparisons between the relative abundance of 8 top bacterial phyla in winter (A) and summer (B). NCS, fallow field soil; CyS1, one-year C. chinensis cultivated soil; CyS3, three-year C. chinensis cultivated soil; CyS5, five-year C. chinensis cultivated soil.
3.5. Beta Diversity
The principal coordinate analysis (PCoA) based on UniFrac distances (i.e., unweighted and weighted) revealed that the rhizospheric bacterial communities of C. chinensis in the different years clustered separately in the summer season of the cropping years. However, the rhizospheric bacterial communities of the three-year (CyS3) and five-year (CyS5) fields were grouped in the winter season. The result indicates that the bacterial communities of the three-year (CyS3) and five-year (CyS5) fields are more similar to each other than the bacterial communities of the one-year (CyS1) field. When compared to the cultivated fields, the bacterial communities in the fallow fields (NCS) clustered in different axes: the first principal component (35.81% contribution) and the second principal component (32.78% contribution) axes in winter and 37.36% and 20.03% contribution in the first and second principal component axes, respectively, in the summer. In order to distinguish the bacterial communities in soils used for cultivating three-year (CyS3) and five-year (CyS5) C. chinensis, the cultivated C. chinensis fields were separated using PC1 and the cultivated C. chinensis fields (CyS1, CyS3, CyS5) were separated from the fallow field by PC2. The unweighted UniFrac algorithm showed the same results. For clarification, in this study, only the weighted UniFrac PCoA plots were shown (Figure 4).
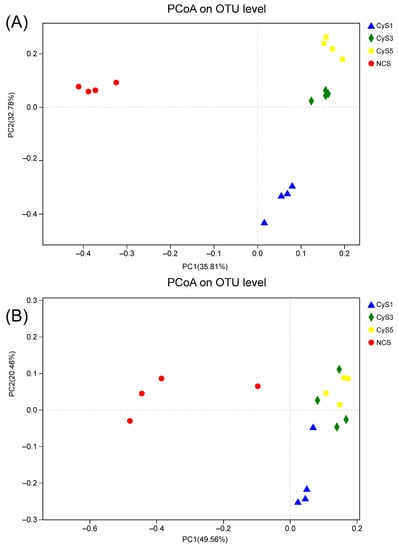
Figure 4.
Principal coordinate analysis (PcoA) in winter (A) and summer (B). NCS, fallow field soil; CyS1, one-year C. chinensis cultivated soil; CyS3, three-year C. chinensis cultivated soil; CyS5, five-year C. chinensis cultivated soil.
3.6. Functional Prediction of the Bacterial Community
The 16S rRNA gene profiling information PICRUSt [26] to predict the abundance of C and N functional genes [27] found a significant difference between the soil samples from cultivated (CyS1, CyS3, and CyS5) and fallow (NCS) fields in two seasons (winter and summer). The abundance of the C and N functional genes found significant differences and higher abundance in the fallow field in summer. In contrast, there was lower abundance in winter. One-way ANOVA results show that the continuous cropping of C. chinensis significantly altered the abundance of the N2-fixing functional gene, nifD (nitrogenase molybdenum-iron protein alpha chain [EC 1.18.6.1]) (p-value = 0.016), in summer and showed an increasing trend. The relative abundance of the functional genes involved in denitrification, nosZ (nitrous oxide reductase [EC 1.7.2.4]), showed a significant variation among the cultivated and fallow fields (p-value = 0.048); it was higher in fallow fields and at the same time showed significant variation with the cultivated fields (p-value = 0.020), in which it was higher in five-year cultivated fields (CyS5) in winter.
The relative abundance of predicted carbon-degrading functional genes, alpha-amylase [EC 3.2.1.1], was significantly altered (p-value = <0.001) in summer among the cultivated and fallow fields; these genes showed higher abundance in the fallow field (NCS). The relative abundance of the chitinase [EC 3.2.1.14]-degrading genes was significantly higher in the fallow fields (NCS) (p-value = <0.001) in summer. The lignin-degrading catalase [EC 1.11.1.6] had a significant higher abundance in fallow fields (NCS) (p-value = <0.001) in winter. No meaningful difference was found in the relative abundance of the predicted N and C cycling genes ALAS (5-aminolevulinate synthase [EC: 2.3.1.37]) and glucoamylase [EC: 3.2.1.3] among the cultivated fields and between cultivated and fallow fields in both seasons.
The abundance of the napA (periplasmic nitrate reductase NapA [EC: 1.7.99.4]) for dissimilatory N reduction to ammonium (DNRA) was significantly (p < 0.05) higher in cultivated fields than in fallow fields in both winter and summer seasons (Table 2 and Figures S3 and S4).

Table 2.
The relative abundance of predicted nitrogen and carbon cycling genes in two seasons (winter and summer). Significant P-values indicated in bold. NCS, fallow field soil; CyS1, one-year C. chinensis cultivated soil; CyS3, three-year C. chinensis cultivated soil; CyS5, five-year C. chinensis cultivated soil.
Spearman’s rank correlation was calculated between the most abundant bacterial phyla and environmental factors (Table S4, Figure 5). In winter, the relative abundance of Actinobacteria was significantly positively correlated with TCN and significantly negatively correlated with pH. The relative abundance of Acidobacteria and Saccharibacteria had a significant positive correlation with TCP, while Bacteroidetes and Cyanobacteria had a meaningful negative relationship. However, in summer, Patescibacteria, Bacteroidetes, and Cyanobacteria significantly negatively correlated with the TCP. On the other hand, the abundance of Acidobacteria significantly positively correlated with TCP.
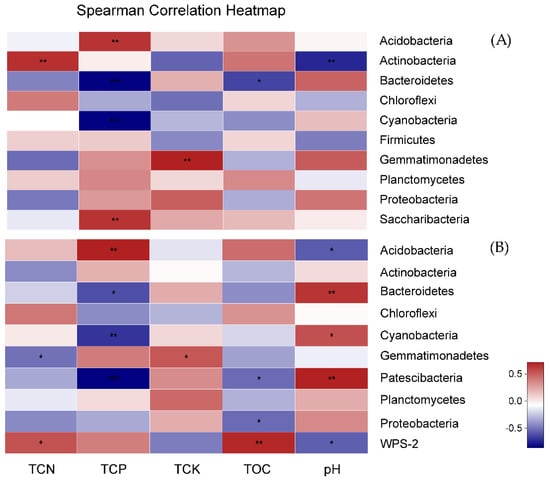
Figure 5.
The heatmap of the correlation between bacterial phyla and physicochemical characteristics of rhizospheric soil in winter (A) and summer (B). NCS, fallow field soil; CyS1, one-year C. chinensis cultivated soil; CyS3, three-year C. chinensis cultivated soil; CyS5, five-year C. chinensis cultivated soil. TCN, total content of nitrogen; TCP, total content of phosphorus; TCK, total content of potassium; TOC, total organic carbon; and pH. The heatmap was created according to the results from the Spearman’s correlation analysis. Positive correlations are represented in red, while negative correlations are represented in blue. The significant correlations are presented as asterisks (*, P < 0.05; **, P < 0.01, ***; P < 0.001).
4. Discussion
Changing the diversity and composition of the rhizosphere soil microorganisms have considerable influence on the soil health and fertility [28,29,30,31,32]. In the meantime, soil microorganisms play an essential role in maintaining plant health, and the associated microbial community is also introduced as the second genome of the plant [33]. Understanding the bacterial community structure and diversity in the rhizosphere soil could provide novel insights into potential hostile microorganisms that might hinder plant pathogens [33]. In this study, we found that the continuous cropping of C. chinensis resulted in a significant decline in bacterial richness (Sobs, Chao) and diversity (Shannon, PD) (Table S2 and Figure 2). This result is in agreement with several previous studies in which the bacterial diversity in black pepper was found to decrease significantly after 55 years of continuous cropping [12]. Fierer et al. [34] reported that the soil microbial diversity had been almost completely eradicated with decades of intense agricultural practices in the tall grass prairie soils of the United States. Similar results have also been observed in cucumber [35] and potato [36]; it was found that the soil bacterial community richness and diversity indices have been reduced by continuous cropping. Soil microbial richness and diversity have an essential role in soil quality and health, and soil ecosystem sustainability [37]. Decreasing soil microbial diversity was responsible for the development of soil-borne plant diseases [38]. Hence, the reduction of microbial community richness and diversity might contribute to sick plant performance and insufficient resistance against disease and pests in C. chinensis continuous cropping.
In this study, after the quality-filtering steps, 903,804 high-quality sequences in winter and 940,104 high-quality sequences in summer were obtained. Moreover, Proteobacteria, Acidobacteria, Chloroflexi, and Actinobacteria were the four top bacterial phyla in both seasons (i.e., winter and summer). This result has also been reported in black pepper by Xiong et al. [12], in German forest and grassland soils by Nacke et al. [21], and banana by Shen et al. [39]. This could be attributed to the pH decrease caused by a continuous cropping system [40]. Among the other phyla, the relative abundance of Bacteroidetes was significantly decreased with the continuous cropping of C. chinensis. This finding was consistent with those of previous research [7]. One of the essential results of this study was that Bacteroidetes is very important for soil health, which was found by Xiong et al. in vanilla-grown soil. Moreover, the relative abundance of Firmicutes corresponding with soil-borne disease suppression [41] made no significant difference in the soil samples, but higher abundance was observed in CyS5 in winter and summer. A similar result was also reported by Xiong et al. [12]. In the genus, the level of the relative abundance of Bradyrhizobium, which is essential for the promotion of plant growth [42], significantly increased in winter but significantly decreased in summer (data not shown). Similarly, Xiong et al. [7] have observed a declining result in Bradyrhizobium in vanilla-grown soil. This result indicates that the continuous cropping of C. chinensis decreased the relative abundance of plant-beneficial bacteria, which is critical for C. chinensis production.
The UniFrac-weighted principal coordinate analysis PcoA (Figure 4) showed evident variations in the bacterial community structure across the two time-series soil sampling from the three time-scales replanted C. chinensis fields and fallow fields as a control. These results were consistent with the findings of Xiong et al. [12] that long-term continuous cropping of black pepper showed noticeable variations in the bacterial community structure, and Li et al. [43] found that the soil microbial community composition and structures among three peanut fields altered significantly with different monoculture histories using 454-pyrosequencing analysis. Hence, we speculated that a continuous cropping of C. chinensis greatly affected the soil bacterial community. Venn diagrams showed that the number of unique bacterial species decreased along the years of C. chinensis cultivation (Figure 1). Moreover, following our results, the previous study reported that the number of unique bacterial OTUs declined along the years of vanilla monoculture [7].
Understanding the characteristics of the soil physicochemical properties in the C. chinensis continuous cropping system could help provide an essential consideration of soil productivity in C. chinensis replanted fields. Additionally, changes in the soil physicochemical properties are likely to lead to changes in the microbial community composition [44]. In the present study, the total content of nitrogen, phosphorus, and organic matter showed an increasing trend with an increase in the number of cultivation years in winter and summer. On the other hand, lower soil pH was observed with a continuous cropping of C. chinensis in two time-series soil sampling. It may be attributed to the following factors: the application of nitrogen fertilizer [45] and accumulation of allelochemicals [46]. A similar result was reported by Tan et al. [18] in continuous P. notoginseng cropping. Another possible reason for acidification in C. chinensis continuous cropping systems may be the different bioactive catechins produced by roots, leaves, and other residues that are used in the fields.
Moreover, determining the correlation between the diversity of the soil bacterial community and the soil environmental factors could provide direct insights into the mechanisms of the continuous cropping obstacle [12]. In the present study, we observed that the relative abundance of Actinobacteria was significantly positively correlated with TCN and significantly negatively correlated with pH, and the relative abundance of Acidobacteria and Saccharibacteria had a significant positive correlation with TCP, while Bacteroidetes and Cyanobacteria had a significant negative relationship in winter. Moreover, the relative abundance of Patescibacteria, Bacteroidetes, and Cyanobacteria significantly negatively correlated with TCP, and the relative abundance of Acidobacteria significantly positively correlated with TCP in summer (Table S4). A change in the microbial community structure in response to continuous cropping of C. chinensis could not only refer to deviations in the soil chemical properties but may also be due to long-term influences of C. chinensis plant root exudates or residues [47], which could not be determined in the range of this study.
Wide ranges of bacteria have the ability to convert atmospheric N2 into ammonia through the process of biological N fixation. To benefit from this function, various plants establish mutualistic interactions by releasing signals in the form of secondary metabolites to free living N-fixing bacteria so that they can enter roots. In the present study, we used PICRUSt to predict the potential to alter the N and C metabolism with different soil samples from cultivated and fallow fields. PICRUSt could produce accurate metagenome predictions with a mean NSTI lower than 0.17 [26]. The abundance of the N and C metabolism genes were both directly influenced by the cropping system. This study was consistent with many previous studies such as that of Mickan et al., conducted in 2017 and 2018. The abundance of the N fixation, ammonification, denitrification, and N reduction was higher in the C. chinensis rhizosphere than in the fallow field’s soil. It possibly leads to more active microbially mediated N transformation in the rhizosphere. This result was consistent with the findings of Li et al. (2014) [48] that N fixation, ammonification, denitrification, and N reduction were significantly higher in the maize rhizosphere when compared to the bulk soil.
5. Conclusion
In conclusion, in the present study, we found that the continuous cropping of C. chinensis resulted in a significant decline in bacterial richness (Sobs, Chao) and diversity (Shannon, PD). Moreover, Proteobacteria, Acidobacteria, Chloroflexi, and Actinobacteria were the four top bacterial phyla in winter and summer. Hence, we speculated that continuous cropping of C. chinensis greatly affected the soil bacterial community. At the same time, the bacterial communities in different functional groups were affected by the continuous cropping of C. chinensis.
Supplementary Materials
The following are available online at https://www.mdpi.com/1424-2818/12/2/57/s1; Figure S1. Rarefaction curves of soil bacterial communities based on observations at 97% sequence similarity level in four times soil samples from C. chinensis fields. The X-axis represents the number of randomly selected sequences; Y-axis represents the number of species. NCS, CyS1, CyS3, and CyS5 represent the fallow field, one-year cultivated field, three-year cultivated field, and five-year cultivated field, respectively; (A) winter, (B) summer. Figure S2. Relative abundance of N and C cycling genes in summer season. NCS, CyS1, CyS3, and CyS5 represent the fallow field, one-year cultivated field, three-year cultivated field, and five-year cultivated field, respectively. Figure S3. Relative abundance of N and C cycling genes in winter season. NCS, CyS1, CyS3, and CyS5 represent the fallow field, one-year cultivated field, three-year cultivated field, and five-year cultivated field, respectively. Table S1. Bacterial numbers of sequences, base number, and average length in each sample in two seasons (winter and summer). NCS; fallow field soil, CyS1; one-year C. chinensis cultivated soil; CyS3, three-year C. chinensis cultivated soil; CyS5, five-year C. chinensis cultivated soil. Table S2. Bacterial Alpha diversity in two seasons (winter and summer). TCN, total content of nitrogen; TCP, total content of phosphorus; TCK, total content of potassium; TOC, total organic carbon; and pH. NCS, fallow field soil; CyS1, one-year C. chinensis cultivated soil; CyS3, three-year C. chinensis cultivated soil; CyS5, five-year C. chinensis cultivated soil. Values are means ± standard deviation (n = 4). Means followed by the same letter for a given factor are not significantly different (P < 0.05; Tukey’s HSD test). Table S3. The relative abundance (<0.01%) of bacterial groups of each sample at the phylum level in two seasons (winter and summer). NCS, fallow field soil; CyS1, one-year C. chinensis cultivated soil; CyS3, three-year C. chinensis cultivated soil; CyS5, five-year C. chinensis cultivated soil. Table S4. Spearman’s rank correlation coefficients (r) between the abundant bacterial phyla and soil properties. In two seasons (i.e., winter and summer). TCN, total content of nitrogen; TCP, total content of phosphorus; TCK, total content of potassium; TOC, total organic carbon; and pH. * indicates significant correlations (P < 0.05), ** indicates significant correlations (P < 0.01), *** indicates significant correlations (P < 0.001).
Author Contributions
Conceptualization, X.W.; Data curation, X.W.; Formal analysis, M.M.A.; Investigation, Y.M. and X.W.; Methodology, M.M.A.; Project administration, X.W.; Resources, J.X., Y.M., D.Z., Z.G. and S.S.; Software, M.M.A.; Visualization, M.M.A.; Writing—original draft, M.M.A.; Writing—review & editing, M.M.A. All authors have read and agreed to the published version of the manuscript.
Funding
The National Key R&D Program financially supported this research. Specialized Research on Modernization of Traditional Chinese Medicine. Large-scale cultivation of Huanglian (Coptis chinensis Franch) and two other Chinese medicinal plants with high values and their roles in the precise alleviation of poverty (2017YFC1701000).
Acknowledgments
The authors thank Xu Zegang from the Lichuan city Zhuanzhuxi Huanglian Professional Cooperative Company for the preparation of fields and their cooperation to collect the soil samples. Moreover, we thank the Chinese Scholarship Council (CSC) for providing scholarships for our studies.
Conflicts of Interest
The authors have no conflict of interest in this work
Abbreviations
| NGS | Next-Generation Sequencing |
| RDA | Redundancy Analysis |
| PcoA | Principal Coordinate Analysis |
| PICRUSt | Phylogenetic Investigation of Communities by Reconstruction of Unobserved States |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| OTUs | Operational Taxonomic Units |
References
- Xiang, K.-L.; Wu, S.-D.; Yu, S.-X.; Liu, Y.; Jabbour, F.; Erst, A.S.; Zhao, L.; Wang, W.; Chen, Z.-D. The First Comprehensive Phylogeny of Coptis (Ranunculaceae) and Its Implications for Character Evolution and Classification. PLoS ONE 2016, 11, e0153127. [Google Scholar] [CrossRef] [PubMed]
- Li, C.B.; Li, X.X.; Chen, Y.G.; Gao, H.Q.; Bu, P.L.; Zhang, Y.; Ji, X.P. Huang-Lian-Jie-Du-Tang Protects Rats from Cardiac Damages Induced by Metabolic Disorder by Improving Inflammation-Mediated Insulin Resistance. PLoS ONE 2013, 8, e67530. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Pan, Y.; Li, L.; Wu, X.; Wang, Y. Composition and diversity of rhizosphere fungal community in Coptis chinensis Franch. continuous cropping fields. PLoS ONE 2018, 13, e0193811. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, S.Y.; Chen, L.; Huang, X.J.; Zhang, Q.W.; Jiang, R.W.; Yao, F.; Ye, W.C. New enantiomeric isoquinoline alkaloids from Coptis chinensis. Phytochem. Lett. 2014, 7, 89–92. [Google Scholar] [CrossRef]
- Wu, L.; Chen, J.; Wu, H.; Wang, J.; Wu, Y.; Lin, S.; Khan, M.U.; Zhang, Z.; Lin, W. Effects of consecutive monoculture of Pseudostellaria heterophylla on soil fungal community as determined by pyrosequencing. Sci. Rep. 2016, 6, 26601. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhang, R.; Xue, C.; Xun, W.; Sun, L.; Xu, Y.; Shen, Q. Pyrosequencing Reveals Contrasting Soil Bacterial Diversity and Community Structure of Two Main Winter Wheat Cropping Systems in China. Microb. Ecol. 2014, 67, 443–453. [Google Scholar] [CrossRef]
- Zhao, J.; Ni, T.; Li, Y.; Xiong, W.; Ran, W.; Shen, B.; Shen, Q.; Zhang, R. Responses of bacterial communities in arable soils in a rice-wheat cropping system to different fertilizer regimes and sampling times. PLoS ONE 2014, 9, e85301. [Google Scholar] [CrossRef]
- Klein, E.; Katan, J.; Gamliel, A. Soil suppressiveness by organic amendment to Fusarium disease in cucumber: Effect on pathogen and host. Phytoparasitica 2016, 44, 239–249. [Google Scholar] [CrossRef]
- Qiu, M.; Zhang, R.; Xue, C.; Zhang, S.; Li, S.; Zhang, N.; Shen, Q. Application of bio-organic fertilizer can control Fusarium wilt of cucumber plants by regulating microbial community of rhizosphere soil. Biol. Fertil. Soils 2012, 48, 807–816. [Google Scholar] [CrossRef]
- Sun, B.; Dong, Z.-X.; Zhang, X.-X.; Li, Y.; Cao, H.; Cui, Z.-L. Rice to Vegetables: Short- Versus Long-Term Impact of Land-Use Change on the Indigenous Soil Microbial Community. Microb. Ecol. 2011, 62, 474–485. [Google Scholar] [CrossRef]
- Xuan, D.T.; Guong, V.T.; Rosling, A.; Alström, S.; Chai, B.; Högberg, N. Different crop rotation systems as drivers of change in soil bacterial community structure and yield of rice, Oryza sativa. Biol. Fertil. Soils 2012, 48, 217–225. [Google Scholar] [CrossRef]
- Xiong, W.; Li, Z.; Liu, H.; Xue, C.; Zhang, R.; Wu, H.; Li, R.; Shen, Q. The Effect of Long-Term Continuous Cropping of Black Pepper on Soil Bacterial Communities as Determined by 454 Pyrosequencing. PLoS ONE 2015, 10, e0136946. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Long, X.Q.; Huo, X.D.; Chen, Y.F.; Lou, K. 16S rRNA-Based PCR-DGGE Analysis of Actinomycete Communities in Fields with Continuous Cotton Cropping in Xinjiang, China. Microb. Ecol. 2013, 66, 385–393. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Y.; Lin, W.X.; Yang, Y.H.; Chen, H.; Chen, X.J. Effects of consecutively monocultured Rehmannia glutinosa L. on diversity of fungal community in rhizospheric soil. Agric. Sci. China 2011, 10, 1374–1384. [Google Scholar] [CrossRef]
- Eickhorst, T.; Tippkötter, R. Improved detection of soil microorganisms using fluorescence in situ hybridization (FISH) and catalyzed reporter deposition (CARD-FISH). Soil Biol. Biochem. 2008, 40, 1883–1891. [Google Scholar] [CrossRef]
- Han, Y.; Xu, L.; Liu, L.; Yi, M.; Guo, E.; Zhang, A.; Yi, H. Illumina sequencing reveals a rhizosphere bacterial community associated with foxtail millet smut disease suppression. Plant Soil 2017, 410, 411–421. [Google Scholar] [CrossRef]
- Ashworth, A.J.; DeBruyn, J.M.; Allen, F.L.; Radosevich, M.; Owens, P.R. Microbial community structure is affected by cropping sequences and poultry litter under long-term no-tillage. Soil Biol. Biochem. 2017, 114, 210–219. [Google Scholar] [CrossRef]
- Tan, Y.; Cui, Y.; Li, H.; Kuang, A.; Li, X.; Wei, Y.; Ji, X. Rhizospheric soil and root endogenous fungal diversity and composition in response to continuous Panax notoginseng cropping practices. Microbiol. Res. 2017, 194, 10–19. [Google Scholar] [CrossRef]
- Degnan, P.H.; Ochman, H. Illumina-based analysis of microbial community diversity. ISME J. 2012, 6, 183–194. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef]
- Nacke, H.; Thürmer, A.; Wollherr, A.; Will, C.; Hodac, L.; Herold, N.; Schöning, I.; Schrumpf, M.; Daniel, R. Pyrosequencing-Based Assessment of Bacterial Community Structure Along Different Management Types in German Forest and Grassland Soils. PLoS ONE 2011, 6, e17000. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Mao, H.; Li, X. Functional characteristics and influence factors of microbial community in sewage sludge composting with inorganic bulking agent. Bioresour. Technol. 2018, 249, 527–535. [Google Scholar] [CrossRef] [PubMed]
- Koo, H.; Mojib, N.; Hakim, J.A.; Hawes, I.; Tanabe, Y.; Andersen, D.T.; Bej, A.K. Microbial communities and their predicted metabolic functions in growth laminae of a unique large conical mat from Lake Untersee, East Antarctica. Front. Microbiol. 2017, 8, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Wei, M.; Xu, C.; Chen, J.; Zhu, C.; Li, J.; Lv, G. Characteristics of bacterial community in cloud water at Mt Tai: Similarity and disparity under polluted and non-polluted cloud episodes. Atmos. Chem. Phys. 2017, 17, 5253–5270. [Google Scholar] [CrossRef]
- Hariharan, J.; Sengupta, A.; Grewal, P.; Dick, W.A. Functional Predictions of Microbial Communities in Soil as Affected by Long-term Tillage Practices. Agric. Environ. Lett. 2017, 2, 170031. [Google Scholar] [CrossRef]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef]
- del Carmen Portillo, M.; Franquès, J.; Araque, I.; Reguant, C.; Bordons, A. Bacterial diversity of Grenache and Carignan grape surface from different vineyards at Priorat wine region (Catalonia, Spain). Int. J. Food Microbiol. 2016, 219, 56–63. [Google Scholar] [CrossRef]
- Wang, R.; Xiao, Y.; Lv, F.; Hu, L.; Wei, L.; Yuan, Z.; Lin, H. Bacterial community structure and functional potential of rhizosphere soils as influenced by nitrogen addition and bacterial wilt disease under continuous sesame cropping. Appl. Soil Ecol. 2018, 125, 117–127. [Google Scholar] [CrossRef]
- Enwall, K.; Nyberg, K.; Bertilsson, S.; Cederlund, H.; Stenström, J.; Hallin, S. Long-term impact of fertilization on activity and composition of bacterial communities and metabolic guilds in agricultural soil. Soil Biol. Biochem. 2007, 39, 106–115. [Google Scholar] [CrossRef]
- Janvier, C.; Villeneuve, F.; Alabouvette, C.; Edel-Hermann, V.; Mateille, T.; Steinberg, C. Soil health through soil disease suppression: Which strategy from descriptors to indicators? Soil Biol. Biochem. 2007, 39, 1–23. [Google Scholar] [CrossRef]
- De Quadros, P.D.; Zhalnina, K.; Davis-Richardson, A.; Fagen, J.R.; Drew, J.; Bayer, C.; Camargo, F.A.O.; Triplett, E.W. The effect of tillage system and crop rotation on soil microbial diversity and composition in a subtropical acrisol. Diversity 2012, 4, 375–395. [Google Scholar] [CrossRef]
- Cardinale, B.J.; Srivastava, D.S.; Emmett Duffy, J.; Wright, J.P.; Downing, A.L.; Sankaran, M.; Jouseau, C. Effects of biodiversity on the functioning of trophic groups and ecosystems. Nature 2006, 443, 989–992. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, R.L.; Pieterse, C.M.J.; Bakker, P.A.H.M. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Ladau, J.; Clemente, J.C.; Leff, J.W.; Owens, S.M.; Pollard, K.S.; Knight, R.; Gilbert, J.A.; McCulley, R.L. Reconstructing the microbial diversity and function of pre-agricultural tallgrass prairie soils in the United States. Science 2013, 342, 621–624. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.H.; Huang, X.Q.; Zhang, F.G.; Zhao, D.K.; Yang, X.M.; Shen, Q.R. Application of Trichoderma harzianum SQR-T037 bio-organic fertiliser significantly controls Fusarium wilt and affects the microbial communities of continuously cropped soil of cucumber. J. Sci. Food Agric. 2012, 92, 2465–2470. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, J.; Gu, T.; Zhang, W.; Shen, Q.; Yin, S.; Qiu, H. Microbial community diversities and taxa abundances in soils along a seven-year gradient of potato monoculture using high throughput pyrosequencing approach. PLoS ONE 2014, 9, 1–10. [Google Scholar] [CrossRef]
- Kennedy, A.C.; Smith, K.L. Soil microbial diversity and the sustainability of agricultural soils. Plant Soil 1995, 170, 75–86. [Google Scholar] [CrossRef]
- Mazzola, M. Assessment and Management of Soil Microbial Community Structure for Disease Suppression. Annu. Rev. Phytopathol. 2004, 42, 35–59. [Google Scholar] [CrossRef]
- Shen, Z.; Wang, D.; Ruan, Y.; Xue, C.; Zhang, J.; Li, R.; Shen, Q. Deep 16S rRNA pyrosequencing reveals a bacterial community associated with banana Fusarium wilt disease suppression induced by bio-organic fertilizer application. PLoS ONE 2014, 9, 1–10. [Google Scholar] [CrossRef]
- Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 2009, 75, 5111–5120. [Google Scholar] [CrossRef]
- Mendes, R.; Kruijt, M.; De Bruijn, I.; Dekkers, E.; Van Der Voort, M.; Schneider, J.H.M.; Piceno, Y.M.; DeSantis, T.Z.; Andersen, G.L.; Bakker, P.A.H.M.; et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 2011, 332, 1097–1100. [Google Scholar] [CrossRef] [PubMed]
- Antoun, H.; Beauchamp, C.J.; Goussard, N.; Chabot, R.; Lalande, R. Potential of Rhizobium and Bradyrhizobium species as plant growth promoting rhizobacteria on non-legumes: Effect on radishes (Raphanus sativus L.). Plant Soil 1998, 204, 57–67. [Google Scholar] [CrossRef]
- Li, X.g.; Ding, C.f.; Zhang, T.l.; Wang, X. xiang Fungal pathogen accumulation at the expense of plant-beneficial fungi as a consequence of consecutive peanut monoculturing. Soil Biol. Biochem. 2014, 72, 11–18. [Google Scholar] [CrossRef]
- Dong, L.; Xu, J.; Zhang, L.; Yang, J.; Liao, B.; Li, X.; Chen, S. High-throughput sequencing technology reveals that continuous cropping of American ginseng results in changes in the microbial community in arable soil. Chin. Med. 2017, 12, 1–11. [Google Scholar] [CrossRef]
- Barak, P.; Jobe, B.O.; Krueger, A.R.; Peterson, L.A.; Laird, D.A. Effects of long-term soil acidification due to nitrogen fertilizer inputs in Wisconsin. Plant Soil 1997, 197, 61–69. [Google Scholar] [CrossRef]
- Ehrenfeld, J.G.; Ravit, B.; Elgersma, K. FEEDBACK IN THE PLANT-SOIL SYSTEM. Annu. Rev. Environ. Resour. 2005, 30, 75–115. [Google Scholar] [CrossRef]
- LIU, J.; WU, F.; YANG, Y. Effects of Cinnamic Acid on Bacterial Community Diversity in Rhizosphere Soil of Cucumber Seedlings Under Salt Stress. Agric. Sci. China 2010, 9, 266–274. [Google Scholar] [CrossRef]
- Li, X.; Rui, J.; Xiong, J.; Li, J.; He, Z.; Zhou, J.; Yannarell, A.C.; Mackie, R.I. Functional Potential of Soil Microbial Communities in the Maize Rhizosphere. PLoS ONE 2014, 9, e112609. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).