Genomic Screening Reveals That the Endangered Eucalyptus paludicola (Myrtaceae) Is a Hybrid
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Species
2.2. Sample Collection and DNA Extraction
2.3. Marker Development and Sequencing
2.4. Data Processing
2.5. Data Analyses
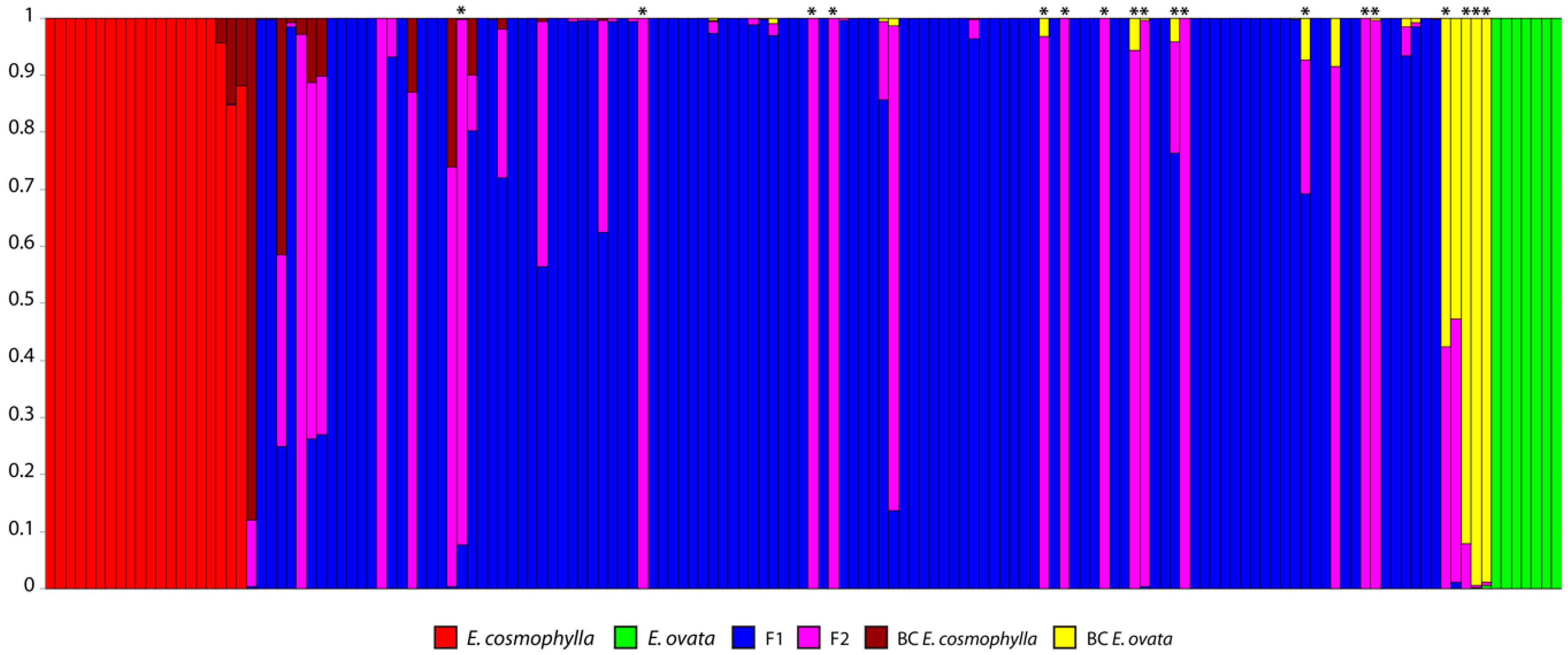
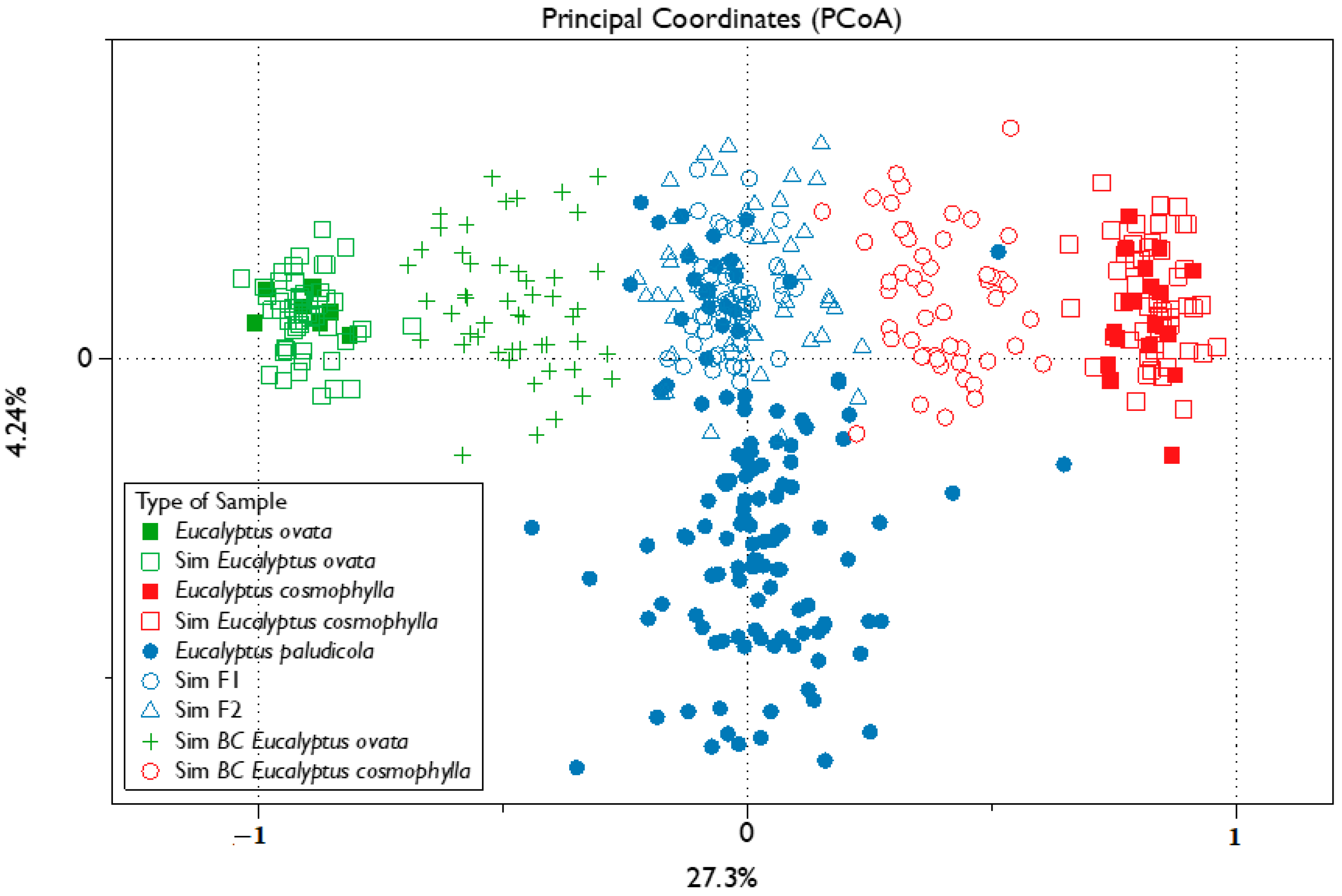
3. Results
3.1. Molecular Data
3.2. Admixture Analyses and Hybrid Class Assignment
4. Discussion
Implications for Conservation
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Abbott, R.; Albach, D.; Ansell, S.; Arntzen, J.W.; Baird, S.J.E.; Bierne, N.; Boughman, J.; Brelsford, A.; Buerkle, C.A.; Buggs, R.; et al. Hybridization and speciation. J. Evol. Biol. 2013, 26, 229–246. [Google Scholar] [CrossRef] [PubMed]
- Schumer, M.; Rosenthal, G.G.; Andolfatto, P. How common is homoploid hybrid speciation? Evolution 2014, 68, 1553–1560. [Google Scholar] [CrossRef] [PubMed]
- Nicolle, D. Classification of the Eucalypts (Angophora, Corymbia and Eucalyptus) Version 4. 2019. Available online: https://dn.com.au/Classification-Of-The-Eucalypts.pdf (accessed on 10 December 2020).
- Potts, B.M.; Dungey, H.S. Interspecific hybridization of Eucalyptus: Key issues for breeders and geneticists. New For. 2004, 27, 115–138. [Google Scholar] [CrossRef]
- Griffin, A.R.; Burgess, I.P.; Wolf, L. Patterns of natural and manipulated hybridisation in the genus Eucalyptus L’Herit—A review. Aust. J. Bot. 1988, 36, 41–66. [Google Scholar] [CrossRef]
- Rossetto, M.; Lucarotti, F.; Hopper, S.D.; Dixon, K.W. DNA fingerprinting of Eucalyptus graniticola: A critically endangered relict species or a rare hybrid? Heredity 1997, 79, 310–318. [Google Scholar] [CrossRef][Green Version]
- Stokoe, R.L.; Shepherd, M.; Lee, D.J.; Nikles, D.G.; Henry, R.J. Natural Inter-subgeneric Hybridization Between Eucalyptus acmenoides Schauer and Eucalyptus cloeziana F. Muell (Myrtaceae) in Southeast Queensland. Ann. Bot. 2001, 88, 563–570. [Google Scholar] [CrossRef]
- Field, D.L.; Ayre, D.J.; Whelan, R.J.; Young, A.G. Molecular and morphological evidence of natural interspecific hybridization between the uncommon Eucalyptus aggregata and the widespread E. rubida and E. viminalis. Conserv. Genet. 2009, 10, 881–896. [Google Scholar] [CrossRef]
- Walker, E.; Byrne, M.; Macdonald, B.; Nicolle, D. Clonality and hybrid origin of the rare Eucalyptus bennettiae (Myrtaceae) in Western Australia. Aust. J. Bot. 2009, 57, 180–188. [Google Scholar] [CrossRef]
- Delaporte, K.L.; Conran, J.G.; Sedgley, M. Interspecific Hybridization within Eucalyptus (Myrtaceae): Subgenus Symphyomyrtus, Sections Bisectae and Adnataria. Int. J. Plant. Sci. 2001, 162, 1317–1326. [Google Scholar] [CrossRef]
- Potts, B.; Barbour, R.; Hingston, A.B.; Vaillancourt, R. Genetic pollution of native eucalypt gene pools—Identifying the risks. Aust. J. Bot. 2003, 51, 1–25. [Google Scholar] [CrossRef]
- Dickinson, G.R.; Wallace, H.M.; Lee, D.J. Reciprocal and advanced generation hybrids between Corymbia citriodora and C. torelliana: Forestry breeding and the risk of gene flow. Ann. For. Sci. 2013, 70, 1–10. [Google Scholar] [CrossRef]
- Schuster, T.M.; Setaro, S.D.; Tibbits, J.F.G.; Batty, E.L.; Fowler, R.M.; McLay, T.G.B.; Wilcox, S.; Ades, P.K.; Bayly, M.J. Chloroplast variation is incongruent with classification of the Australian bloodwood eucalypts (genus Corymbia, family Myrtaceae). PLoS ONE 2018, 13, e0195034. [Google Scholar] [CrossRef] [PubMed]
- Ellis, M.F.; Sedgley, M.; Gardner, J.A. Interspecific Pollen-Pistil Interaction in Eucalyptus L’Hér. (Myrtaceae): The Effect of Taxonomic Distance. Ann. Bot. 1991, 68, 185–194. [Google Scholar] [CrossRef]
- Gore, P.L.; Potts, B.M.; Volker, P.W.; Megalos, J. Unilateral cross-incompatibility in Eucalyptus: The case of hybridisation between E. globulus and E. nitens. Aust. J. Bot. 1990, 38, 383–394. [Google Scholar] [CrossRef]
- Costa e Silva, J.; Potts, B.M.; Tilyard, P. Epistasis causes outbreeding depression in eucalypt hybrids. Tree Genet. Genomes 2012, 8, 249–265. [Google Scholar] [CrossRef]
- Larcombe, M.J.; Holland, B.; Steane, D.A.; Jones, R.C.; Nicolle, D.; Vaillancourt, R.E.; Potts, B.M. Patterns of reproductive isolation in Eucalyptus—A phylogenetic perspective. Mol. Biol. Evol. 2015, 32, 1833–1846. [Google Scholar] [CrossRef]
- Nicolle, D. A new series, Incognitae, of Eucalyptus L’Hér., including a new species endemic to Fleurieu Peninsula and Kangaroo Island, South Australia. J. Adel. Bot. Gard. 1995, 16, 73–78. [Google Scholar]
- T.S.S.C. Commonwealth Listing Advice on Eucalyptus paludicola. 2006. Available online: http://www.environment.gov.au/biodiversity/threatened/species/pubs/eucalyptus-paludicola-advice.pdf (accessed on 10 December 2020).
- Quarmby, J. Action Plan for Mount Compass Swamp Gum (Eucalyptus paludicola) 2011–2016; Government of South Australia, Department of Environment and Natural Resources: Adelaide, Australia, 2016; p. 17.
- Ottewell, K.; Bickerton, D.; Quarmby, J.; Lowe, A.J. Genetic Status of the Endangered Mount Compass Swamp Gum (Eucalyptus paludicola); University of Adelaide, Department of Environmental and Natural Resources: Adelaide, Australia, 2011. [Google Scholar]
- Jones, R.C.; Nicolle, D.; Steane, D.A.; Vaillancourt, R.E.; Potts, B.M. High density, genome-wide markers and intra-specific replication yield an unprecedented phylogenetic reconstruction of a globally significant, speciose lineage of Eucalyptus. Mol. Phylogenet. Evol. 2016, 105, 63–85. [Google Scholar] [CrossRef]
- Nicolle, D. Myrtaceae (partly) (version 1). In Flora South Australia, 5th ed.; State Herbarium of South Australia: Adelaide, Australia, 2014; p. 102. [Google Scholar]
- Myburg, A.A.; Grattapaglia, D.; Tuskan, G.A.; Hellsten, U.; Hayes, R.D.; Grimwood, J.; Jenkins, J.; Lindquist, E.; Tice, H.; Bauer, D.; et al. The genome of Eucalyptus grandis. Nature 2014, 510, 356–362. [Google Scholar] [CrossRef]
- Cross, H.; Biffin, E.; van Dijk, K.-J.; Lowe, A.; Waycott, M. Effective application of next-generation sequencing (NGS) approaches in systematics and population genetics: Case studies in Eucalyptus and Acacia. Aust. Syst. Bot. 2016, 29, 235–246. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, E.M.; Schwartz, R.; Bullock, C.T.; Williams, J.S.; Shaffer, H.B.; Aguilar-Miguel, X.; Parra-Olea, G.; Weisrock, D.W. Parallel tagged amplicon sequencing reveals major lineages and phylogenetic structure in the North American tiger salamander (Ambystoma tigrinum) species complex. Mol. Ecol. 2013, 22, 111–129. [Google Scholar] [CrossRef] [PubMed]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in EXCEL. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Earl, D.; von Holdt, B. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Anderson, E.C.; Thompson, E.A. A Model-Based Method for Identifying Species Hybrids Using Multilocus Genetic Data. Genetics 2002, 160, 1217–1229. [Google Scholar]
- Nielsen, E.E.; Bach, L.A.; Kotlicki, P. HYBRIDLAB (version 1.0): A program for generating simulated hybrids from population samples. Mol. Ecol. Notes 2006, 6, 971–973. [Google Scholar] [CrossRef]
- Hudson, C.J.; Freeman, J.S.; Myburg, A.A.; Potts, B.M.; Vaillancourt, R.E. Genomic patterns of species diversity and divergence in Eucalyptus. New Phytol. 2015, 206, 1378–1390. [Google Scholar] [CrossRef]
- Crisp, M.D.; Burrows, G.E.; Cook, L.G.; Thornhill, A.H.; Bowman, D.M.J.S. Flammable biomes dominated by eucalypts originated at the Cretaceous–Palaeogene boundary. Nat. Commun. 2011, 2, 193. [Google Scholar] [CrossRef] [PubMed]
- Thornhill, A.H.; Crisp, M.D.; Külheim, C.; Lam, K.E.; Nelson, L.A.; Yeates, D.K.; Miller, J.T. A dated molecular perspective of eucalypt taxonomy, evolution and diversification. Aust. Syst. Bot. 2019, 32, 29–48, 20. [Google Scholar] [CrossRef]
- Steane, D.A.; Nicolle, D.; Sansaloni, C.P.; Petroli, C.D.; Carling, J.; Kilian, A.; Myburg, A.A.; Grattapaglia, D.; Vaillancourt, R.E. Population genetic analysis and phylogeny reconstruction in Eucalyptus (Myrtaceae) using high-throughput, genome-wide genotyping. Mol. Phylogenetics Evol. 2011, 59, 206–224. [Google Scholar] [CrossRef] [PubMed]
- Barbour, R.C.; Potts, B.M.; Vaillancourt, R.E.; Tibbits, W.N. Gene flow between introduced and native Eucalyptus species: Flowering asynchrony as a barrier to F1 hybridisation between exotic E. nitens and native Tasmanian Symphyomyrtus species. For. Ecol. Manag. 2006, 226, 9–21. [Google Scholar] [CrossRef]
- Shepherd, M.; Lee, D.J. Gene flow from Corymbia hybrids in northern New South Wales. For. Ecol. Manag. 2016, 362, 205–217. [Google Scholar] [CrossRef]
- Lopez, G.A.; Potts, B.M.; Tilyard, P.A. F1 hybrid inviability in Eucalyptus: The case of E. ovata × E. globulus. Heredity 2000, 85 Pt 3, 242–250. [Google Scholar] [CrossRef]
- Volker, P.W.; Potts, B.M.; Borralho, N.M.G. Genetic parameters of intra- and inter-specific hybrids of Eucalyptus globulus and E. nitens. Tree Genet. Genomes 2008, 4, 445–460. [Google Scholar] [CrossRef]
- Lindtke, D.; Gompert, Z.; Lexer, C.; Buerkle, C.A. Unexpected ancestry of Populus seedlings from a hybrid zone implies a large role for postzygotic selection in the maintenance of species. Mol. Ecol. 2014, 23, 4316–4330. [Google Scholar] [CrossRef]
- Grattapaglia, D.; Bradshaw, H.D., Jr. Nuclear DNA content of commercially important Eucalyptus species and hybrids. Can. J. For. Res. 1994, 24, 1074–1078. [Google Scholar] [CrossRef]
- Oudjehih, B.; Abdellah, B. Chromosome numbers of the 59 species of Eucalyptus L’Herit. (Myrtaceae). Caryologia 2006, 59, 207–212. [Google Scholar] [CrossRef]
- Grattapaglia, D.; Vaillancourt, R.E.; Shepherd, M.; Thumma, B.R.; Foley, W.; Külheim, C.; Potts, B.M.; Myburg, A.A. Progress in Myrtaceae genetics and genomics: Eucalyptus as the pivotal genus. Tree Genet. Genomes 2012, 8, 463–508. [Google Scholar] [CrossRef]
- Yakimowski, S.B.; Rieseberg, L.H. The role of homoploid hybridization in evolution: A century of studies synthesizing genetics and ecology. Am. J. Bot. 2014, 101, 1247–1258. [Google Scholar] [CrossRef] [PubMed]
- Moritz, C. Conservation units and translocations: Strategies for conserving evolutionary processes. Hereditas 1999, 130, 217–228. [Google Scholar] [CrossRef]
- Allendorf, F.W.; Leary, R.F.; Spruell, P.; Wenburg, J.K. The problems with hybrids: Setting conservation guidelines. Trends Ecol. Evol. 2001, 16, 613–622. [Google Scholar] [CrossRef]
- Jackiw, R.N.; Mandil, G.; Hager, H.A. A framework to guide the conservation of species hybrids based on ethical and ecological considerations. Conserv. Biol. 2015, 29, 1040–1051. [Google Scholar] [CrossRef] [PubMed]
- Dungey, H.S.; Potts, B.M.; Whitham, T.G.; Li, H.-F. Plant genetics affects arthropod community richness and composition: Evidence from a synthetic eucalypt hybrid population. Evolution 2000, 54, 1938–1946. [Google Scholar] [CrossRef]

| Taxon | Location | n | Latitude | Longitude |
|---|---|---|---|---|
| E. cosmophylla | Three Chain Rd (KI) | 6 | −35.829 | 137.704 |
| E. cosmophylla | Crafers West (FL) | 2 | −34.990 | 138.681 |
| E. cosmophylla | Kyeema (FL) | 4 | −35.270 | 138.674 |
| E. cosmophylla | Mt Billy Cp (FL) | 2 | −35.448 | 138.600 |
| E. cosmophylla | Burnfoot (FL) | 4 | −35.400 | 138.557 |
| E. ovata | Burnfoot (FL) | 3 | −35.400 | 138.557 |
| E. ovata | Cleland Gully Rd (FL) | 2 | −35.368 | 138.638 |
| E. ovata | Stipiturus CP (FL) | 1 | −35.371 | 138.551 |
| Undetermined | Stipiturus CP (FL) | 1 | −35.371 | 138.551 |
| E. paludicola | Stipiturus CP (FL) | 1 | −35.371 | 138.551 |
| E. paludicola | Kelly Hill Caves CP (KI) | 15 | −35.997 | 136.873 |
| E. paludicola | Short’s property, original tree (KI) | 1 | −35.739 | 137.029 |
| E. paludicola | Short’s property, revegetation (KI) | 9 | −35.739 | 137.028 |
| E. paludicola | Edwards Lagoon (KI) | 4 | −35.808 | 137.038 |
| E. paludicola | O’Donnell property (KI) | 5 | −35.963 | 136.999 |
| E. paludicola | Rocky River, natural (KI) | 4 | −35.946 | 136.753 |
| E. paludicola | Rocky River revegetation (KI) | 9 | −35.947 | 136.741 |
| E. paludicola | Nangkita (FL) | 11 | −35.345 | 138.711 |
| E. paludicola | Burnfoot (FL) | 8 | −35.400 | 138.557 |
| E. paludicola | Gold Diggings Swamp (FL) | 2 | −35.591 | 138.372 |
| E. paludicola | Hindmarsh Valley (FL) | 5 | −35.402 | 138.518 |
| E. paludicola | Range Rd (FL) | 5 | −35.567 | 138.469 |
| E. paludicola | Parawa (FL) | 2 | −35.591 | 138.372 |
| E. paludicola | Mosquito Hill Rd (FL) | 19 | −35.446 | 138.646 |
| E. paludicola | Kokoda Rd (FL) | 10 | −35.407 | 138.688 |
| E. paludicola | Cox Scrub CP (FL) | 9 | −35.331 | 138.747 |
| E. paludicola | Proctor Road (FL) | 7 | −35.326 | 138.621 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
van Dijk, K.-j.; Waycott, M.; Quarmby, J.; Bickerton, D.; Thornhill, A.H.; Cross, H.; Biffin, E. Genomic Screening Reveals That the Endangered Eucalyptus paludicola (Myrtaceae) Is a Hybrid. Diversity 2020, 12, 468. https://doi.org/10.3390/d12120468
van Dijk K-j, Waycott M, Quarmby J, Bickerton D, Thornhill AH, Cross H, Biffin E. Genomic Screening Reveals That the Endangered Eucalyptus paludicola (Myrtaceae) Is a Hybrid. Diversity. 2020; 12(12):468. https://doi.org/10.3390/d12120468
Chicago/Turabian Stylevan Dijk, Kor-jent, Michelle Waycott, Joe Quarmby, Doug Bickerton, Andrew H. Thornhill, Hugh Cross, and Edward Biffin. 2020. "Genomic Screening Reveals That the Endangered Eucalyptus paludicola (Myrtaceae) Is a Hybrid" Diversity 12, no. 12: 468. https://doi.org/10.3390/d12120468
APA Stylevan Dijk, K.-j., Waycott, M., Quarmby, J., Bickerton, D., Thornhill, A. H., Cross, H., & Biffin, E. (2020). Genomic Screening Reveals That the Endangered Eucalyptus paludicola (Myrtaceae) Is a Hybrid. Diversity, 12(12), 468. https://doi.org/10.3390/d12120468






