Abstract
Collembola (springtails) are an early diverging class of apterygotes, and mark the first substantial radiation of hexapods on land. Despite extensive work, the relationships between major collembolan lineages are still debated and, apart from the Early Devonian fossil Rhyniella praecursor, which demonstrates their antiquity, the time frame of springtail evolution is unknown. In this study, we sequence two new mitochondrial genomes and reanalyze all known Collembola mt-genomes, including selected metagenomic data, to produce an improved phylogenetic hypothesis for the group, develop a tentative time frame for their differentiation, and provide a comprehensive overview of gene order diversity. Our analyses support most taxonomically recognized entities. We find support for an Entomobryomorpha + Symphypleona clade, while the position of Neelipleona could not be assessed with confidence. A Silurian time frame for their basal diversification is recovered, with an indication that divergence times may be fairly old overall. The distribution of mitochondrial gene order indicates the pancrustacean arrangement as plesiomorphic and dominant in the group, with the exception of the family Onychiuridae. We distinguished multiple instances of different arrangements in individual genomes or small clusters. We further discuss the opportunities and drawbacks associated with the inclusion of metagenomic data in a classic study on mitochondrial genome diversity.
Keywords:
springtails; basal hexapods; Neelus; Dicyrtomina; mitogenomics; metagenomics; divergence times 1. Introduction
Collembola, commonly referred to as springtails, is an ancient group of primitively wingless Hexapoda and represent the first substantial radiation of the group following the invasion of land. During their long evolutionary history, springtails have adapted to almost any ecosystem on Earth, from Antarctic ice-free areas to highlands in the Himalayas and the Australian deserts, dwelling in soil, in leaf litter, or on vegetation [1,2,3]. With 9000 described species, they are an important component of soil biodiversity, and recent data suggest that the current estimate of 50,000 undescribed species may, in fact, be a large underestimation [4]. The oldest fossils of Collembola, and hexapods in general, date back to the Early Devonian (~ 400 Millions of years ago, Mya), and were found in the Rhynie chert deposits of Scotland [5,6,7]. The taxonomic attribution of these fossils, generally identified as Rhyniella praecursor, has been the subject of some debate [8]. Currently, the identification of R. praecursor as a collembolan is widely accepted, although it remains to be clarified whether the Rhynie chert fossils can be identified as one species of the family Isotomidae [7], or may be assigned to as many as three different springtail groups [8] (D’Haese, personal communication).
Although the position of springtails within the arthropod tree has been challenged in the past [9,10], their monophyly has never been questioned and is supported by morphological and molecular data. Collembola are characterized by the presence of a springing organ (furca or furcula), derived from appendages of the fourth abdominal segment, and a ventral tube, derived from appendages of the first abdominal segment, mainly involved in fluid balance and sometimes used as a sticky organ [3].
Despite the many attempts to define the evolutionary relationships between major lineages, the application of both morphological and molecular data has not been successful, to date, in resolving inter-order relationships conclusively, as their reciprocal position in springtail phylogenetic reconstructions differs based on the morphological and/or molecular character set applied. Traditionally, two main groups were identified within Collembola: Arthropleona (Entomobryomorpha + Poduromorpha), characterized by an elongated body and the abdominal segments (I–IV) generally separated by an intersegmental membrane; and Symphypleona sensu lato (Symphypleona sensu stricto + Neelipleona), which instead display a globular-shaped body with fused abdominal segments [11]. Since the work of Cassagnau [12] and Massoud [13], it is nevertheless now widely accepted that four orders of Collembola should be considered: Entomobryomorpha, Neelipleona, Poduromorpha, and Symphypleona. D’Haese [14], studying the distribution of 131 different morphological characters, recovered Poduromorpha as basal and Entomobryomorpha as a sister group to a Symphypleona + Neelipleona clade, thus hypothesizing a paraphyletic status of Arthropleona. More recently, based on the study of tibiotarsal chaetotaxy, Zhang and Deharveng [15] suggested that Poduromorpha may occupy a basal position in the class and that Entomobryomorpha may be paraphyletic, as the family Entomobryidae clusters, in their reconstructions, with the Symphypleona. This possibility was also supported by Carapelli [16], based on the analysis of concatenated 13 mitochondrial protein-coding genes. Noteworthy, no representative of Neelipleona was included in the two latter analyses. Globular-shaped Collembola (i.e., Neelipleona and Symphypleona) have been generally considered the most derived springtail groups based on morphology, while molecular analyses repeatedly suggested a basal position for both groups [17,18,19]. When species from all the four orders of Collembola are included in phylogenetic analyses using nuclear ribosomal genes as markers, Neelipleona are recovered as the most primitive taxon, sister group to all others: (Neelipleona, (Symphypleona, (Poduromorpha, Entomobryomorpha))) [19,20,21].
Although the development and application of molecular techniques have provided new insights into the order-level phylogeny of Collembola, evolutionary relationships between major lineages are still far from being resolved with confidence, and the amount of phylogenetic information is highly unbalanced—dedicated to solving the relationships among recently derived, rather than early-diverging, taxonomic groups. One possible hurdle may be unbalanced taxon sampling, as most studies included a large proportion of sequences from Poduromorpha and Entomobryomorpha, with a more limited sampling of Symphypleona and only scant data from Neelipleona. In fact, of the 15 complete or almost complete mitochondrial genomes (mtDNAs) determined to date, only two are from Symphypleona (i.e., Bourletiella arvalis and Sminthurus viridis) and none from Neelipleona. Furthermore, even though ribosomal sequences are available from both large and small nuclear subunits, the use of different domains by different authors prevents the assembly of a taxon-balanced and taxon-rich dataset.
The usefulness of mitochondrial DNA for phylogenetic purposes is well-established and associated to its genetic and biological properties. Mitogenomes are, with minor exceptions, maternally inherited, haploid, homologous, characterized by different rates of nucleotide substitution in different genes that make them suitable for resolving relationships at different taxonomic levels. The mitochondrial genome typically includes 37 genes: 13 protein-coding genes (PCGs) involved in the oxidative phosphorylation process (atp6 and atp8, cox1-3, cob, nad1-6, and nad4L), 22 genes encoding for transfer RNAs (trnX, where X stands for the amino acid one-letter code), and two genes encoding for ribosomal subunits (rrnS and rrnL), alongside a non-coding region, referred to as A + T-rich or control region, in which the sequences for initiation of replication and transcription are present [22]. Although the number/type of mitochondrial genes is almost invariable within major metazoan groups, the order in which they are arranged along the organelle chromosome can vary. Modifications in the gene order (GO) are nevertheless rare events, and the chance of one and the same arrangement arising by convergence in two unrelated lineages is extremely low. As such, a shared re-arrangement can be interpreted as a solid indication for common ancestry [22,23,24]. So far, only four different gene arrangements have been identified in Collembola [10,16,25]: GO1, which is the most frequent gene order observed among springtails and corresponds to the ancestral gene order for Pancrustracea; GO2, which is shared among species of the family Onychiuridae and characterized by the translocation of two tRNAs (i.e., trnSuga and trnQ); GO3, described in S. viridis and distinguished from the ancestral one by three translocations (trnF, trnT and trnP), and one translocation plus inversion (trnD); GO4, observed in Podura aquatica, characterized by two tRNA translocations (trnW and trnY).
In the present study, an enlarged mitogenome data set of 31 springtail species, including two new mitochondrial genomes from the species Dicyrtomina saundersi Lubbock, 1862 (Symphypleona) and Neelus murinus Folsom, 1896 (Neelipleona), as well as metagenomic data from [26], is used to investigate the evolutionary relationships between lineages at the order/family level and to identify the time frame of Collembola initial diversification and evolution. A larger dataset of complete/semicomplete genomes is also used to identify the possible occurrence of new gene orders.
This study was designed to create the blueprints necessary to shed light on the polarity of changes in the collembolan body plan that occurred in early-diverging lineages (e.g., whether globular or elongated shape of the specimens’ body is plesiomorphic for the class), as well as to contextualize major shifts in collembolan evolution in the correct geological/ecological background (e.g., transition to terrestrial environment and collembolan origin/diversification). Finally, the utility and possible drawbacks of the inclusion of metagenomic data into a classic mitochondrial genomic study were evaluated. The results will contribute to enriching our understanding of hexapod evolution, in its earliest stages, a frequently neglected issue in modern phylogenetic studies.
2. Materials and Methods
2.1. Sequencing of the Mt-genomes of Dicyrtomina saundersi and Neelus murinus
Soil samples were collected near Castello di Belcaro (Siena, Italy: 43°18′25.31″ N; 11°17′26.56″ E) and the microfauna extracted in a Berlese-Tullgreen funnel. Several specimens of D. saundersi and N. murinus were morphologically identified and stored at -80 °C until molecular analyses. DNA extraction, amplification, sequencing, and assembly/annotation of the genome closely followed [27]. PAUP* (v 4.0, Sinauer Associates, Sunderland, Massachusetts) [28] was used to calculate base frequencies along each entire genome, for concatenated PCGs oriented on the same strand and for each codon position separately. Strand asymmetry was computed following the formulae proposed by Hassanin et al. [29]: AT-skew: [A%] − [T%]/[A%] + [T%]; CG-skew: [C%] − [G%]/[C%] + [G%].
2.2. Phylogenetic Analysis
All available complete or semi-complete mitochondrial genomes of Collembola were downloaded from GenBank in April 2019 alongside those of Daphnia pulex (Crustacea, Branchiopoda), Japyx solifugus (Diplura, Japygidae), and Trigoniophthalmus alternatus (Microcoryphia, Machilidae) as outgroups (see GenBank records for sequence reference). The two new sequences determined in this study, D. saundersi and N. murinus, were added to the collection. Individual PCGs were extracted based on the original annotations using an in-house perl script. Scaffolds from the metagenomic study in Cicconardi et al. [26] were revised to identify sequences that, based on the original annotations, (a) correspond to complete or semi-complete mitochondrial genomes, with a minimum of 32 annotated genes; and (b) include taxonomic information at least to the family level (Table 1).

Table 1.
Data set used for the phylogenetic and dating analyses. See Table S1 for complete information.
Original automatic annotations were revised by: (a) resubmitting raw sequences to the MITOS webserver [30] to obtain complete automatic annotations and related statistics; (b) comparing automatically annotated sequences of PCGs with preliminary alignments of manually annotated genomes, and (c) overlaying all genes (including those encoding for rRNAs and tRNAs) throughout the genome to visualize overlaps/spacers. The taxonomic attribution of scaffold sequences was updated comparing cox1 sequences with the species level- and all-barcode collections in the Barcode of Life Data System (BOLD) [31], and considering sequences with a similarity > 99%. DNA sequences of individual PCGs were retro-aligned using RevTrans (2.0 b) [32] based on their amino acid alignment produced using MAFFT (v. 7.309) [33]. A total of 15 single base uncertainties were fixed to the most common state observed in related sequences to guarantee compatibility with downstream software/scripts. Each gene alignment was filtered with Gblocks (v. 0.91 b) [34] to eliminate regions of unreliable alignment under ‘strict’ settings (i.e., all characters with a gap/missing base in at least one sequence were removed). The absence of three sequence segments (one gene in the nad1 data set, two partial genes in cob and nad5), due to incomplete sequences and not uncertainties in the alignment process, was allowed. Single gene alignments were concatenated to produce the final data set.
The most appropriate partitioning scheme and evolutionary model associated to each charset (see below) were determined using PartitionFinder (v. 2.1.1) [35]. Initial blocks were defined dividing the data by strand, by codon position, and by gene family (ATPases, Cytochrome oxidases, cob and NADH dehydrogenases), for a total of 15 initial blocks, and then re-associated using the greedy search algorithm based on corrected Akaike Information Criterion values into an optimal set of 12 partitions and evolutionary models: GTR + I + Γ in all partitions, except for third codon positions in ATPases (HKY + I + Γ) and third positions in NADH dehydrogenases (GTR + Γ). Bayesian phylogenetic analyses were conducted on the complete data set and on first and second codon positions only using MrBayes (v. 3.2.7a) [36]. Two parallel runs of four chains each were conducted for 50 million generations, and the resulting trees and posterior probabilities were summarized, following analysis of run traces in tracer (v. 1.7.1) [37], excluding the initial 20% generations as burn-in. The resulting trees were visualized using Figtree (v. 1.4.4) [38].
2.3. Dating Analysis
The two data sets described above were used for a series of dating analyses using a relaxed clock in BEAST (v. 1.10.4) [38]. Different calibration points were applied to test the sensibility of the analysis to different priors (Table 2). In all analyses, the split between D. pulex and Hexapoda (i.e., the root of the tree) was set at 510 Mya (monophyletic, normal prior: 510 ± 7 Mya) and the split between J. solifugus + T. alternatus and Collembola was set at 485 Mya (monophyletic, normal prior: 485 ± 6 Mya). These correspond to dates, with confidence intervals, obtained from the analysis of a large phylogenomic data set in Rehm et al. [39], corroborated by almost identical estimates, based on a similarly extensive data set, in Rota-Stabelli et al. [40]. Based on the observation that in our analyses J. solifugus clusters with T. alternatus (at variance with the aforementioned studies that would suggest a closer relationship of Collembola and Diplura [17]), and given the open discussion on the relationships/monophyly of Entognatha and the hypothesis that Diplura may in fact be an early offshoot on the branch leading to Insecta [41,42], the second calibration point was assigned to the split between J. solifugus + T. alternatus and Collembola. Different combinations of calibration points were used in ingroup taxa.

Table 2.
Calibration points and estimated age, with 95% Highest Posterior Density (HPD), in My of key groups. Dates refer to the crown group, not including the stem.
The occurrence of the fossil R. praecursor [5,6,7] which, with some uncertainty [3,8], is generally considered a representative of family Isotomidae or Entomobryomorpha at large from the Pragian/Givetian period of the early/middle Devonian [14,43,44], was used to calibrate the basal split of Entomobryomorpha (i.e., with R. praecursor interpreted as an isotomid) or the split between Entomobryomorpha and Symphypleona (i.e., with R. praecursor interpreted as a basal Entomobryomorpha) as older than 391 Mya (gamma prior: shape 1.1, scale 20, offset 391). The occurrence of the fossil Permobrya mirabilis [45], identified as belonging to the family Entomobryidae, with some resemblance to modern Lepidocyrtinae, and dated to the upper Permian middle Ecca age of South Africa [46] was used to calibrate the split of Isotomidae vs. Entomobryidae (i.e., with P. mirabilis interpreted as an entomobryid) or the split between Orchesellinae and Lepidocyrtinae (i.e., with P. mirabilis interpreted as related to Lepidocyrtinae with the exclusion of Orchesellinae) as older than 280 Mya (gamma prior: shape 1.1, scale 35, offset 280; see Table 2 for complete information). Each analysis consisted of 50 million generations in BEAST (v. 1.10.4) [38] with partitioning and evolutionary model estimated in PartitionFinder (see above), an uncorrelated log-normal relaxed clock and a Yule tree prior to starting dates set at peak estimates (normal calibration points) or at 5 My before the limit (gamma calibration points). Runs were evaluated for convergence in Tracer (v. 1.7.1) [37] and the resulting trees and posterior probabilities summarized using Logcombiner after discarding 10% of generations as burn-in. The resulting tree was visualized using Figtree (v. 1.4.4).
2.4. Gene Order
The gene order of publicly available mitochondrial genomes of Collembola was obtained from GenBank and/or from the original publications. Automatic annotations of scaffolds sequenced in [26] were transformed in gene order data using an in-house perl script. Following the observation that largely incomplete scaffolds tend to have annotation errors (unpublished observation, [30]), only scaffolds with more than half annotated genes (i.e., 19) were retained for analysis, and all scaffolds displaying a gene order different from known ones were manually revised. Some shorter scaffolds were analyzed for comparison (see Results). The occurrence and distribution of different gene orders were plotted over the phylogenetic tree in [26]. Given the peculiarities of the data set, a less-stringent approach was applied in the interpretation of gene order data: (a) scaffold with 37 annotated genes in known order were regarded as ‘complete’ although many were missing part of the A + T rich region; (b) incomplete genomes were deemed ‘compatible’ with a known gene order, unless they display gene order differences; (c) two genomes were deemed as ‘sharing a translocation’ even if only one among the source or endpoint of one translocation was available in one of the genomes (see Discussion for a justification of this approach).
3. Results
3.1. Description of Two New Genomes
The mitochondrial genome of D. saundersi (Table S2) is a circular molecule of 15,045 bp. The sequence of the mitochondrial genome of N. murinus (Table S3) could not be completely determined. The sequenced portion—13,992 bp in length—is missing three tRNAs (trnI, trnQ and trnM), as well as a portion of the rrnS and the A+T-rich region. Notably, although the genome of N. murinus was not sequenced completely, amplifications were successfully performed which cross the undetermined portion, thus confirming the circularity of the genome. The annotated genome sequences were deposited in GenBank under accession numbers: MG701393 (D. saundersi) and MH155200 (N. murinus). In both genomes, most of the mitochondrial genes are oriented on the same strand (i.e., the J-strand). The canonical start codon (ATA/ATG, encoding for Methyonine) is used in most of the PCGs in both mtDNAs (7/13 in D. saundersi and 8/13 in N. murinus), whereas in all other instances, a codon for Isoleucine is observed (Tables S2 and S3). Incomplete stop codons (TA-/T--), most likely post-transcriptionally restored into complete stop codons [47], are observed in most PCGs of the D. saundersi genome and six genes of N. murinus. The occurrence of intergenic spacers is limited in both genomes, with a maximum of 7 and 3 intergenic nucleotides, respectively, in D. saundersi and N. murinus. Gene overlaps are observed in both mitogenomes, with the longest being a 46-nucleotides overlap between atp6 and cox3 in N. murinus mtDNA (Tables S2 and S3). In both mitochondrial genomes, genes are oriented along the organelle chromosome, following the ancestral Pancrustacea arrangement. The overall nucleotide composition of both genomes is biased toward a higher content of As (38.5% and 35.4% in D. saundersi and N. murinus, respectively) and Ts (33.1% and 30.6%, respectively; Tables S2 and S3).
The strand bias commonly observed between the J- and N- filaments of animal mtDNAs (that are usually A+C- and T+G-rich, respectively, and therefore display AT- and CG-skew values that are positive for the J-strand and negative for the N-strand [29]) is generally observed in both genomes. Exceptions are: a) J strand AT-skew in both genomes, that is strongly negative for second codon positions and reflects on a mildly negative value overall; and b) J strand CG-skew of D. saundersi, limited to first codon positions, that is negative (Figures S1 and S2).
3.2. Data Set
Revision of the taxonomic attribution of sequence scaffolds led to the improvement of three annotations and the correction of one. Scaffold s6653 was identified as Folsomia candida, scaffold s7289 as Desoria trispinata, scaffold s6543 as Thalassophorura sp. Scaffold s6802, originally automatically annotated as Entomobryidae gen. sp. based on two dubious records in the all-barcode collection, was reannotated as Dicyrtomidae gen. sp. (Table 1). Starting with 12,127 aligned sites and after the exclusion of 27% on the total data set (5–90% gene-wise), the final data set used for the phylogenetic and dating analyses was composed of 34 sequences by 8862 characters, with a completeness of 99.5%.
3.3. Phylogenetic Analysis
The phylogenetic analysis produced two trees, one based on the complete data set and one on the first and second codon positions only. In this latter tree (Figure 1), Collembola are recovered as monophyletic, as well as the four orders (Entomobryomorpha, Neelipleona, Poduromorpha, and Symphypleona). Three species (Folsomia candida, Friesea antarctica, and Parisotoma notabilis), 5 genera and 6 out of 7 families for which more than one representative was included were similarly recovered as monophyletic. The only exception was the family Hypogastruridae, which appears to be paraphyletic, as scaffold s6241 (Hypogastruridae) is basal to the Gomphiocephalus hodgsoni + Neanuridae + Poduridae clade. Intermediate and recent nodes show full support (posterior probability = 1), while the basalmost nodes are less supported (1 > p.p. > 0.86). Orders Symphypleona and Entomobryomorpha form a monophyletic clade that is, in turn, associated with Poduromorpha. Neelipleona is recovered in a basal position in Collembola. Podura aquatica, whose position has been debated at length [14], is recovered as the sister group of Neanuridae.
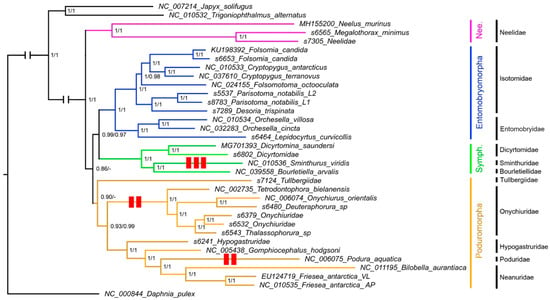
Figure 1.
Phylogenetic reconstruction based on first/second codon positions. Numbers at nodes indicate support (posterior probability) for the node based on the first/second positions data set and the complete data set. - node not present in the second tree. Red rectangles indicate gene order variations. See Table 1 for complete information.
The tree obtained based on the full data set (i.e., including third codon positions) is identical to the former, with similar support values, with two exceptions: (a) scaffold s7124 (Tullbergiidae) is recovered as the sister group of Neelipleona, making Poduromorpha paraphyletic; and (b) Neelipleona (inclusive of scaffold s7124) is recovered as the sister group of Poduromorpha.
3.4. Dating Analysis
The dating analysis produced 10 dated trees, each arising from the combination of one data set and one set of calibration points (Table 2). Run statistics were adequate in all instances, with Effective Sample Size (ESS) values always over 100 and generally well over 1000.
The tree shown in Figure 2 was obtained applying two outgroup calibration points and two ingroup calibrations, with R. praecursor interpreted as an isotomid and P. mirabilis as associated with Lepidocyrtinae (following the separation of Orchesellinae) to the first and second codon position data set, that is in turn the most credible a priori data set and set of assumptions. All orders, genera, species, and 6 out of 7 families for which more than one representative is present are recovered as monophyletic. The only exception was the family Hypogastruridae, that appears paraphyletic as scaffold s6241 (Hypogastruridae) is basal to the G. hodgsoni + Neanuridae + Poduridae clade, as in the aforementioned analysis. Order relationships suggest a basal dichotomy between Poduromorpha+Neelipleona and Entomobryomorpha+Symphypleona. In terms of timing, the basal node encompassing all Collembola is placed at 437 Mya (426–452 95% Highest Posterior Density (HPD)), and the time frame for the diversification of the four collembolan orders is 421–437 Mya (411–452, 95% HPD). Family level diversification begins at 414 Mya (395–433, 95% HPD) and all 7 families for which more than one representative is included were in essence by 184 Mya (150–218, 95% HPD). The three pairs of individuals that, based on current taxonomy, belong to single species, differentiated at 88 Mya (70–108, 95% HPD: F. antarctica), 92 Mya (73–114, 95% HPD: P. notabilis), and 47 Mya (36–59, 95% HPD: F. candida).
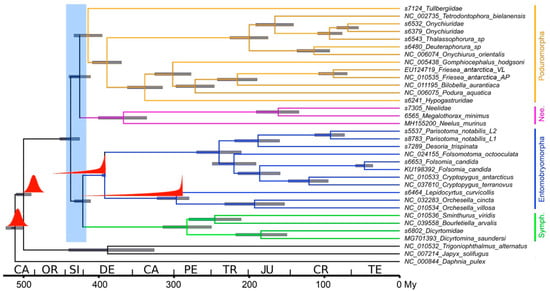
Figure 2.
Dated tree based on first/second positions, see Table 2 (4th row) for complete prior information. Red insets indicate prior probability for node age, left to right: D. pulex vs. Hexapoda, Collembola vs. higher insects, R. praecursor, P. mirabilis. Blue inset indicates the time range corresponding to the diversification of collembolan orders.
Comparing distinct analyses, differing in the inclusion of third codon positions and/or in the use of alternative calibration points in the ingroup, it is possible to observe that phylogenetic relationships between orders, in line with the phylogenetic analysis, are recovered according to two models: (a) (((Symphypleona+Entomobryomorpha), Poduromorpha), Neelipleona) in 6/10 cases and (b) ((Symphypleona+Entomobryomorpha), (Poduromorpha+Neelipleona)) in 4/10. In the first case, scaffold s7124 (Tullbergiidae gen. sp.) generally clusters at the base of Poduromorpha; and in the second at the base of Neelipleona, making Poduromorpha paraphyletic. A pattern can be observed where runs with no or weak ingroup calibrations tend to produce the first model, with s7124 within Poduromorpha, while runs where one or two ingroup constraints are imposed that force the associated nodes backward in time tend to produce the second model, with s7124 associated with Neelipleona. Phylogenetic relationships at the family/genus level tend to be stable and in line with the phylogenetic analysis.
In terms of dates, the time range for the basal diversification of Collembola orders is placed, in the analyses based on different calibration points, in the Silurian or Devonian. The analysis with only outgroup calibration points produces the most recent dates (late Devonian to early Carboniferous). The addition of ingroup calibration points (minimum ages) tend to gradually shift nodes backwards. The addition of P. mirabilis associated to Lepidocyrtinae places the diversification of collembolan orders in the middle Devonian; the further addition of R. praecursor associated to basal Entomobryomorpha further moves the node to the early Devonian. The interpretation of R. praecursor as a representative of family Isotomidae and of P. mirabilis as associated to Lepidocyrtinae after the separation of Orchesellinae shifts the node to the Silurian, as described above. The use of the complete data set vs. first/second positions had a limited impact on estimated dates, with differences further diminishing as more/stronger ingroup calibration points were added.
3.5. Gene Order
Seventy-four mitochondrial genomes were analyzed in terms of genome organization (Table 1 and Table S4; Figure 3 and Figure 4). Fifty-seven are scaffolds that include 19 or more annotated genes, 15 are complete or semi-complete mitochondrial genomes from the literature, and two were determined in this study. Their gene order was compared with the four already known from Collembola: the pancrustacean ancestral gene order (AGO), exemplified by Drosophila; and arrangements observed in Podura, Sminthurus, and Tetrodontophora (Figure 3 and Figure 4).

Figure 3.
Comparison between the ancestral gene order (AGO) and variant gene orders described. * indicates missing sequence information.
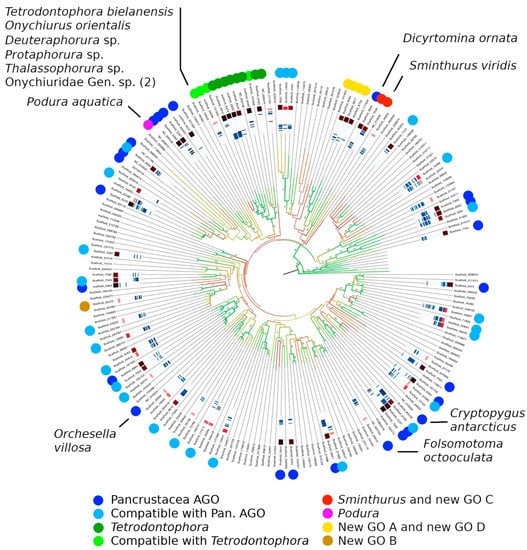
Figure 4.
Gene orders observed in Collembola (color coded), overlaid over the phylogenetic tree of [26].
Thirty sequences (18 scaffolds, 11 sequences from the literature, one new sequence) represent complete genomes and display the pancrustacean AGO. These include 12 sequences with a clear taxonomic identification and 8 that were identified in [26] (with modifications), with representatives from all four orders (see Table S4 for a complete list). Sixteen sequences (15 scaffolds, one new sequence) represent incomplete genomes whose gene order is compatible, limited to the four arrangements known from Collembola, only with the pancrustacean AGO. These include N. murinus, as well as two scaffolds identified as Tullbergiidae and one as Parisotoma notabilis L1. Thirteen scaffolds represent incomplete genomes that are compatible with the pancrustacean AGO, as well as one or more additional gene orders. Only one was identified as Paronellinae. The pancrustacean AGO is dominant in Collembola and is observed in all domains of the springtail diversity tree (Figure 4).
Eight sequences represent genomes displaying the Tetrodontophora gene order: T. bielanensis, six scaffolds corresponde to complete genomes and O. orientalis, incomplete but compatible with Tetrodontophora only. Besides T. bielanensis and O. orientalis, some of these scaffolds were identified: two as Onychiuridae, one as Deuteraphorura sp., and one as Thalassophorura sp. This gene order, the second more common in Collembola, is restricted to a single monophyletic cluster that accounts for ~6% of the collembolan diversity sampled in [26]. Besides the aforementioned complete or semi-complete genomes, the cluster includes four fragments of 4–14 annotated genes, all compatible with the Tetrodontophora gene order. Based on the taxonomic identification of the source material (at varying level of uncertainty), this cluster can be tentatively identified with the family Onychiuridae. The cluster emerges from a large assemblage of sequences characterized by, or compatible with, the AGO gene order, and its closest relatives display the AGO gene order. As such, the data suggest a single origin of this variant gene order, possibly at the base of family Onychiuridae, and its maintenance in all sampled sequences of the group.
Podura aquatica, nearly complete, displays a unique gene order. Considering that no other genome that shares any of its two translocations was identified, and that it emerges from a cluster characterized by the AGO, it is possible to hypothesize this gene order as an autapomorphy of P. aquatica or a small group therein.
Scaffold s6702 (complete genome), together with scaffolds s6817 (35 genes), s8054 (30 genes), and s191403 (2 genes), display two novel and related gene orders that are here referred to as novel GO A and novel GO D. The gene order GO A, exemplified by scaffold s6702, differs from the pancrustacean AGO in the position of three segments, namely for the translocation of trnSuga and trnQ and the translocation plus inversion of the cluster trnSgcu-trnE-trnF. GO D derives from GO A following the additional exchange between trnI and trnM. This translocation is observed in the scaffold s8054, whereas it is not possible to determine its status in scaffolds s6817 and s191403. The four sequences emerge as a monophyletic clade in [26]. None of the four sequences could be identified, but the lineage emerges from a larger assemblage that contains at least one Dicyrtomidae, suggesting that it represents a subgroup of Symphypleona alternative to both Dicyrtomidae (characterized by the typical pancrustacean AGO) and S. viridis + allies, (identified by a unique and unrelated gene order).
Scaffold s35470 displays a novel gene order here referred to as novel GO B. The sequence includes the genomic portion between trnI and trnE, with a total of 20 annotated genes. The gene order differs from the pancrustacean AGO in the position of trnW and trnC that are not observed in the area between trnY and nad2, nor in other parts of the sequenced portion. This suggests that they have relocated elsewhere in the portion of the genome not analyzed. The position of this sequence in the phylogenetic tree suggests that the new gene order evolved directly from the pancrustacean AGO.
Scaffold s7540, with a total of 34 annotated genes in the region between trnQ and rrnL, displays a novel gene order that is here referred to as novel GO C. Based on [26], this sequence is related to S. viridis, which is, in turn, characterized by an unique order, as already identified in [16], and the two sequences emerge together from an assemblage characterized by the typical pancrustacean AGO. The novel gene order differs from the pancrustacean AGO for a translocation with inversion of trnD, that is here found between trnSuga and nad1, and the contiguous exchange between trnT and trnP. The gene order of S. viridis, in turn, differs from novel GO C for the position of trnF, which is observed in the former between the A+T-rich region and trnI. The position in the phylogenetic tree and the comparison of gene order strongly suggest that scaffold s7540 may mark an intermediate step between the pancrustacean AGO and the arrangement observed in Sminthurus, with two translocations having taken place along the branch leading to Sminthurus plus scaffold s7540 and one on the branch leading to Sminthurus after the divergence of scaffold s7540.
4. Discussion
4.1. Structure and Compositional Biases in the Two New Genomes
The two newly sequenced genomes (D. saundersi and N. murinus) conform with the standard features of hexapod mitochondrial genome, with a compact array of gene sequences and limited intergenic regions. In terms of gene order, both mtDNAs comply with the typical plesiomorphic state of Pancrustacea, which is also the most frequently observed in Collembola. Hexapod mitochondrial genomes are usually characterized by three different biases. Generally, they show an overall nucleotide content strongly skewed toward a higher frequency of As and Ts, and the two mtDNAs herein described are not an exception (Tables S2 and S3). The second difference is observed in nucleotide composition between the J- and N-strand that are AC- and GT-rich, respectively. This has been explained as a consequence of the peculiar asynchronous and asymmetric replication process, during which it is mainly the N-strand that persists as single strand, thus being much more affected by deaminations than the J-strand [29]. While CG-skew values of both strands and genomes conform, with minor exceptions, with the expected trend, the AT-skew appears to be markedly negative for second codon positions on the J strand, that in D. saundersi and N. murinus extends to the overall strand (Figures S1 and S2). These negative values can be explained by taking into account the third bias generally observed in mitochondrial PCGs, the codon bias. Proteins encoded by the mitochondrial chromosome are generally to be positioned within the organelle inner membrane, thus requiring mostly hydrophobic amino acids. During the translational process, the hydrophobic nature of amino acid is associated with the presence of pyrimidines, and especially of Ts, at second codon positions [48]; accordingly, the AT- and CG-skew values can be respectively negative or positive, irrespective of the strand in which the PCGs are oriented (Figures S1 and S2).
4.2. Phylogenetic Analysis
Overall, the topologies obtained from the phylogenetic and dating analyses are grossly congruent, as well as generally in line with the current taxonomy of the group. Specifically, all lower-level groups (families and below) are well supported and in line with current taxonomic hypotheses, with the possible exception of the monophyly of Hypogastruridae and the erratic position of s7124_Tullbergiidae. Higher-level groups are somehow more problematic, as basal nodes are less supported and some inter-order relationships can vary in different reconstructions. Focusing on the monophyly and relationships between the four orders, which is possibly one of the most interesting issues, our reconstructions support the monophyly of all four groups (not considering the erratic placement of s7124_Tullbergiidae), and the sister group relationship between Entomobryomorpha and Symphypleona. This is an interesting result per se, as it negates the validity of Arthropleona, i.e., a unique origin of all elongated Collembola (Poduromorpha and Entomobryomorpha), a super-order group that, although long dismissed [13,49], has been at the core of our interpretation of springtail evolution for decades. Noteworthy, a relationship between Entomobryomorpha and Symphypleona has already found some support in the analysis of retro-cerebral endocrine structures and chaetotaxy, as well as the shared reduction of thorax I [12,15,49]. The relative position of Neelipleona and Poduromorpha with respect to Entomobryomorpha+Symphypleona is more problematic, as support values are not conclusive for the relevant nodes and different analyses identify two different scenarios, namely: (a) (((Symphypleona+Entomobryomorpha), Poduromorpha), Neelipleona) and (b) ((Symphypleona+Entomobryomorpha), (Poduromorpha+Neelipleona)) (see Results section). This is a limitation of the current analysis, as it makes it difficult to compare body plan of four orders. Notably, both scenarios have been proposed in previous studies [20,50].
As a further outlook on phylogeny, gene order data is liable to provide strong evidence for the monophyly of a group when more than one sequence share a derived gene order. Nevertheless, the paucity of gene order rearrangements observed in Collembola, their distribution, and the exclusion of scaffold with an uncertain taxonomic identification from the phylogenetic analysis, limited the utility of this marker to only one node, namely Onychiuridae, that receives support from sequence-based analyses as well (Figure 1).
At shallower taxonomic levels, our analysis supports a derived position of P. aquatica (Poduridae), associated with Neanuridae within Poduromorpha. Given the debate over an aquatic origin of Hexapoda and, among these, Collembola, associated with the early colonization of land and based on its semi-aquatic status, P. aquatica has been at length considered a primitive springtail and an indication of the aquatic origin of these latter. The position of P. aquatica was revised in [14], which suggested its placement within a paraphyletic Hypogastruridae that was, in turn, a sister group to the Neanuridae, therefore arguing that P. aquatica could not be interpreted as a primarily semi-aquatic species, nor a proof of the semi-aquatic origin of the class as a whole. Our results support this latter view.
4.3. Dating Analysis
The dating analysis produced a hypothetical time frame for the collembolan evolution. Limited to key events, the diversification of springtail orders may have taken place in the Silurian period, followed by family level diversification that occupied the rest of the Paleozoic and extended through the Triassic. Species-level diversification is recovered as Cenozoic, with some older instances. Following the finding of fossil R. praecursor from the Devonian, the notion that Collembola are an old taxon is well acknowledged. Nevertheless, it is important to note that R. praecursor is not to be identified as a basal springtail, associated with the first appearance of the taxon, but rather represents a fairly derived form, being identified as belonging to modern family Isotomidae. Accordingly, the appearance and basal diversification of Collembola predates the fossil and is here hypothesized as having taken place in the Silurian. This observation is in line with the presence, in upper Silurian strata, of coprolites of likely collembolan origin [51], which suggest that springtails may have been common since that period. These dates are further in line with the hypothesis, presented in [3], that the diversification of Collembola is to be related with the diffusion of vascular plants in the Silurian period and the associated formation of significant layers of soil, the typical habitat of modern Collembola, in the Devonian.
The inclusion of three pairs of taxa that belong to the same species (F. antarctica, F. candida, and P. notabilis) opens to the possibility to date the emergence of species, an uncommon possibility in genome-wide dating analyses [52,53]. The dates obtained for species diversification are fairly old, namely 47–92 Mya. While the identification of very old lineages of Collembola that persisted through extensive evolutionary time, well into the Miocene, is not new [54,55], the dates obtained in this study tend to be significantly older, in the early Tertiary. A possible technical justification for this difference is that the aforementioned studies are based on a rate of 2.3%-5%/My, while the current study relies on an internal calibration point. This interpretation of very old species may nevertheless be also biased by the fact that alpha-biodiversity in Collembola is notoriously heavily underestimated [4], and it is not inconceivable that these pairs of conspecific taxa may in the future be described as separate entities. Specifically, the two individuals of F. antarctica come from different bioregions of Antarctica, an unlikely occurrence that is leading to the revision of the entire group [56,57] and the interpretation of F. candida, as well as P. notabilis, as single species is not unproblematic [55,58].
Some of the difficulties encountered in the present analysis suggest a possible prospect for dating analyses in Collembola. Dating is heavily based on the availability of calibration points, that in turn require, if a fossil is used as a calibration point: (a) a fossil; (b) a date for the fossil; and (c) the possibility to relate the fossil to a specific node on the tree. While recent fossils for Collembola are available, Paleozoic fossils, of primary importance here, are exceedingly rare, in fact limited to R. praecursor, P. mirabilis, and a series of coprolites tentatively interpreted as of collembolan origin. Dates for the aforementioned fossils are available, although the antiquity of R. praecursor has been hotly debated in the past. The attribution of the two aforementioned fossils to specific taxonomic groups is, on the other hand, somehow more problematic. Although further morphological analyses are still ongoing, R. praecursor is currently recognized as an isotomid [7], but has been variously associated to different modern taxa, including Neanuridae/Poduromorpha [59,60], or basal Entomobryomorpha [61,62]. P. mirabilis is similarly described as an entomobryid, but displays a number of features that would suggest its placement in a derived entomobryid group, namely Lepidocyrtinae. These uncertainties become further exacerbated by two aspects: a) sequence data sets are taxonomically incomplete, because of sample limitations (i.e., sequence data is not available for all taxa) as well as the obvious impossibility to sample extinct taxa; and b) the uncertainty in the attribution of the fossil to the crown group (i.e., a node enclosed by modern available taxa) or to the stem group (i.e., including the stem up to the closest available neighbor that does not belong to the group). This is of key importance in the current (and foreseeable) dating analyses of Collembola, as fossils are used to define the minimal age of a group in a context where, in the absence of the calibrator, nodes tend to be more recent than the fossil would suggest. As such, the importance of paleontological studies cannot be underestimated in this context, as they address these aspects explicitly, besides providing a description of the fossil and a tentative taxonomic attribution.
4.4. Gene Order
The present analysis of gene order in Collembola, based on new data (a revision of previously described genomes and the integration of a large metagenomic data [26]), marks a significant increase in the amount of available information. The number of analyzed genomes increased from 17 (15 complete, i.e., all 37 genes mapped) to 74 (39 complete), with a 4.4 × increase, while the number of different gene orders recorded increased from 4 (3 complete) to 8 (4 complete), with a 2 × increase. The inclusion of metagenomic data, nevertheless, came at a cost, with only 42% of the new genomes being complete and only 31% having and associated taxonomic identification, with different degrees of uncertainty. Although (a) the sampling of Collembola is by no means complete (as more than 9000 species have been described for the group [63] and the vast majority remains unexplored in terms of mitochondrial gene order), and (b) samples are not evenly distributed across known collembolan diversity (as tropical island species dominate metagenomic data and Isotomidae/Onychiuridae/Antarctic species are over-represented in classical studies due to a specific interest of some of the research groups involved), the sizable—for a group of this dimension—overall coverage (0.8%) allows for some consideration of larger breadth and the identification of a general pattern in the data. The Pancrustacea AGO appears to be the plesiomorphic state for Collembola—it is numerically dominant in the group and distributed all across the diversity tree. Variant gene orders, on the other hand, emerge in scattered positions on the tree, and in every major instance, it is possible to hypothesize that the novel gene order arose independently from others by direct modification of the ancestral Pancrustacea GO. Of the seven variant gene orders, only one is found in a substantial number of species/scaffolds, namely that which is already described for T. bielanensis [25]. Based on available taxonomic information, this order may be associated with the family Onychiuridae [64]. The remaining six variant gene orders, which include those previously described for S. viridis [10] and P. aquatica [64], are on the other hand restricted to 1–4 species/scaffolds each. Hence, it seems that only one rearrangement took place during the early diversification of Collembola, namely at the onset of the family Onychiuridae, while the rest may have originated more recently and may be restricted to groups at a shallow level of diversity. The distribution of gene order changes along the tree leads to some additional considerations. Based on (a) the notion that such changes are rare events and (b) a simple model where changes are randomly distributed along the tree, the occurrence of gene rearrangements in two contiguous nodes would be expected to be an extremely unlikely event. Nevertheless, it is possible to observe two instances of this pattern in our reconstruction: the mutation of AGO to new GO C to the Sminthurus arrangement and the mutation of AGO to new GO A and then to new GO D. Albeit with limited numbers (which in turn does not suggest the possibility of a formal statistical analysis), the reconstruction presented here suggests that the occurrence of a first gene order modification may increase the chance of a second modification occurring in the short term along the same line. Although it is not possible at present to provide a satisfactory interpretation of this observation, a possible line of reasoning may be suggested. The occurrence, throughout Metazoa, of a number of different gene orders, with apparently no preference over a specific order as long as all genes are present and complete, suggests that different gene orders are equally functional/viable. Nevertheless, the mitochondrial genome is a strongly integrated system, with mechanisms such as genome duplication, transcription, and regulation, which act over sets of multiple genes. As such, the first event may create a viable gene order, confirmed by the fact that this is propagated to successive generations, but somehow disturb the internal equilibrium of the genome, creating the condition for a second event on the short term. If this proves to be the case, it would be possible that, as sampling becomes more and more dense, gene order changes that are now interpreted as one mutation composed of more than one translocation/inversion (e.g., two in Tetrodontophora and two in Podura) may in fact be reinterpreted as two mutation events occurring along a same lineage. This is, in turn, visible in our reconstruction with new GO C, which marks an intermediate step between AGO and the Sminthurus arrangement and new GO A that marks an intermediate step between AGO and new GO D. In terms of phylogenetic utility (i.e., the use of shared rearrangements to support a monophyletic origin of different lineages), it is possible to hypothesize that no information is liable to be produced for inter-order relationships, as it is now well established that the basal gene order of the four orders is the same as Collembola. Possible information at deep-to-intermediate (e.g., family) level is likely to be produced for Onychiuridae per se or for groups associated with this latter, while it is unlikely that an equally large assemblage, to date undetected, may exist at deep taxonomic levels that share an additional variant gene order. Information at shallow (e.g., genus) level is equally likely to be produced, as variant gene orders have been, and may be expected to be, described in roughly 9% of species. These will, nevertheless, most likely be shared by groups of closely related species at shallow taxonomic levels, and their finding in different areas of collembolan diversity is totally unpredictable. The occurrence of multiple changes along a line, discussed above, may further increase the phylogenetic utility of gene order changes.
Given the peculiarities of the data set—which includes a large number of genomes, which although partly incomplete, possibly provide a substantial coverage of mitogenome diversity in the class at variance with more typical approaches (where a limited number of genomes is included that, although complete, are by no means representative)—we evaluated the opportunity of a less-stringent approach to gene order comparison as the best option to valorize the specificities of the current data set (see Methods section). Most importantly, partially incomplete genomes were analyzed in terms of ‘compatibility’ to other known complete genomes, indicating as ‘compatible’ any partial genome that does not display differences from a known gene order. While incomplete genomes compatible with more than one known arrangement (i.e., those that do not cover an area where two known gene order differ) were observed and identified as such, these were tentatively interpreted as sharing the gene order of their closest relatives. This approach is, in our view, more appropriate, in order to take full advantage of the data in case of large and taxonomically representative gene order data sets, and justified under the assumption that gene order rearrangements are rare events, and therefore absence of evidence of gene order differences, in a sufficiently representative data set between two phylogenetically related genomes can be taken as a preliminary indication, with due caution, of a shared gene order.
5. Conclusions
One of the explicit aims of this study was to evaluate the possibilities and possible drawbacks of the inclusion of mitochondrial metagenomic data in more traditional studies on mitochondrial genome diversity. With costs dropping for Next Generation Sequencing, and the increasing difficulty in developing morphology-based studies due to the specificity of expertise required, it is foreseeable that the production and availability of mitochondrial metagenomic data for biodiversity studies will increase significantly in the near future. On the other hand, the classic strategy of determining the sequence of individual genomes from taxonomically selected and identified material is becoming less competitive in terms of time and costs.
Metagenomic data sets differ from classic data sets in a number of aspects, both in the methodology for data production and in the nature of the data produced [65]. They tend to be extremely rich in terms of the amount of sequences produced but, at the same time (a) are generally incomplete, i.e., only a subset of genomes is closed and includes all 37 genes; (b) have some degree of uncertainty associated with sequence assembly and gene annotation; and (c) have no a priori taxonomic attribution, although this information can be inferred a posteriori. These issues can be effectively approached by filtering data for the presence/quality of a taxonomic identification and for completeness, followed by a quality check on the assembled data set (i.e., length distribution in single gene alignments, low score alignments, presence of stop codons) and/or on the results (i.e., manually revising all scaffold that present a unique and novel gene order). While these filtering steps are liable to reduce the number of usable sequences substantially, the generally very large amount of starting data may counterbalance for data filtering. In our case, the starting data set included 183 scaffolds with an annotated gene content of 14 ± 12 genes (mean, standard deviation), and only 19 (~10%) had an associated taxonomic identification, at least to the family level. Filtering led to the inclusion of 13 genomes (~7%) in the phylogenetic analysis and 57 (~31%) genomes for gene order analysis. Diminutive as this may look, this produced a significant increase (1.7 × in the phylogenetic analysis, 4.4 × in gene order analysis) compared to classic data produced in 18 years since the sequencing of the first complete mitochondrial genome of a springtail. As such, we envision the deployment of metagenomic data sets, possibly developed for other purposes, as a feasible strategy to complement classic mitochondrial genome data for the study of phylogeny and gene order evolution, and the development of improved bioinformatic approaches to help standardize the procedure and limit the need of manual intervention.
Supplementary Materials
The following are available online at https://www.mdpi.com/1424-2818/11/9/169/s1, Figure S1: Compositional biases in Dicyrtomina saundersi, Figure S2: Compositional bias in Neelus murinus, Table S1: Taxa under study, detail of authority and sampling location, Table S2: Genome annotation of Dicyrtomina saundersi, Table S3: Genome annotation of Neelus murinus, Table S4: Gene order in genomes and scaffolds.
Author Contributions
Conceptualization, C.L., A.C., F.N.; Formal analysis, C.L., A.C., F.C., F.N.; Resources, A.C., F.F.; Data curation, C.L., F.C., F.N.; Writing—original draft preparation, C.L., F.N.; Writing—review and editing, C.L., A.C., F.C., F.F., F.N.; Visualization, F.C., F.N.; Supervision, A.C., F.F. Funding acquisition, A.C., F.F. All authors approved the final manuscript.
Funding
This study was funded by the Italian Program of Research in Antarctica (PNRA16_00234). Partial support was also provided by the University of Siena.
Acknowledgments
The authors wish to thank Alessandro Donati for providing the computational power to conduct dating analyses and two anonymous reviewers for their insightful comments.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Yoshii, R. Collembola of Himalaya. J. Coll. Arts Sci. Chiba Univ. 1966, 4, 461–531. [Google Scholar]
- Wise, K.A.J. Collembola (Springtails). In Entomology of Antarctica; Antarctic Research Series; American Geophysical Union (AGU): Washington, DC, USA, 1967; Volume 10, pp. 123–148. ISBN 978-1-118-66869-6. [Google Scholar]
- Hopkin, S.P. Biology of the Springtails: (Insecta: Collembola); OUP Oxford: Oxford, UK, 1997; pp. 1–330. ISBN 978-0-19-158925-6. [Google Scholar]
- Cicconardi, F.; Fanciulli, P.P.; Emerson, B.C. Collembola, the biological species concept and the underestimation of global species richness. Mol. Ecol. 2013, 22, 5382–5396. [Google Scholar] [CrossRef] [PubMed]
- Hirst, S.; Maulik, S. On some Arthropod Remains from the Rhynie Chert (Old Red Sandstone). Geol. Mag. 1926, 63, 69–71. [Google Scholar] [CrossRef]
- Whalley, P.; Jarzembowski, E.A. A new assessment of Rhyniella, the earliest known insect, from the Devonian of Rhynie, Scotland. Nature 1981, 291, 317. [Google Scholar] [CrossRef]
- Greenslade, P.; Whalley, P. The systematic position of Rhyniella praecursor hirst and maulik (Collembola), the earliest known hexapod. In Proceedings of the Second International Seminar on Apterygota, Siena, Italy, 4–6 September 1986; pp. 319–323. [Google Scholar]
- Greenslade, P.J.M. Reply to R. A. Crowson’s Comments on Insecta of the Rhynie Chert (1985 Entomol. Gener. 11 (1/2): 097–098). Entomol. Gen. 1988, 13, 115–117. [Google Scholar] [CrossRef]
- Nardi, F.; Spinsanti, G.; Boore, J.L.; Carapelli, A.; Dallai, R.; Frati, F. Hexapod origins: Monophyletic or Paraphyletic? Science 2003, 299, 1887–1889. [Google Scholar] [CrossRef] [PubMed]
- Carapelli, A.; Liò, P.; Nardi, F.; van der Wath, E.; Frati, F. Phylogenetic analysis of mitochondrial protein coding genes confirms the reciprocal paraphyly of Hexapoda and Crustacea. BMC Evol. Biol. 2007, 7, S8. [Google Scholar] [CrossRef]
- Borner, C. Das System der Collembolen, nebst beschreibungen neuer Collembolen des Hamburger Naturhistorichen Museums. Mitteilungen aus dem Naturhistorischen Museum in Hamburg 1906, 23, 147–188. [Google Scholar]
- Cassagnau, P. La Phylogenie des Collemboles à la lumiere des structures endocrines retrocerebrales. In Proceedings of the I Symposio International de Zoofilogenia. Facultad de Ciencias; Universidad de Salamanca: Salamanca, Spain, 1971; pp. 333–349. [Google Scholar]
- Massoud, Z. Essai de synthese sur la phylogenie des Collemboles. Rev. Ecol. Biol. Sol. 1976, 13, 241–252. [Google Scholar]
- D’Haese, C.A. Morphological appraisal of Collembola phylogeny with special emphasis on Poduromorpha and a test of the aquatic origin hypothesis. Zool. Scr. 2003, 32, 563–586. [Google Scholar] [CrossRef]
- Zhang, F.; Deharveng, L. First instar tibiotarsal chaetotaxy supports the Entomobryidae and Symphypleona (Collembola) forming a cluster in a phylogenetic tree. Zootaxa 2015, 3955, 487–504. [Google Scholar] [CrossRef] [PubMed]
- Carapelli, A.; Convey, P.; Nardi, F.; Frati, F. The mitochondrial genome of the antarctic springtail Folsomotoma octooculata (Hexapoda; Collembola), and an update on the phylogeny of collembolan lineages based on mitogenomic data. Entomologia 2014, 2, 46–55. [Google Scholar] [CrossRef]
- Luan, Y.-X.; Mallatt, J.M.; Xie, R.-D.; Yang, Y.-M.; Yin, W.-Y. The phylogenetic positions of three basal-hexapod groups (Protura, Diplura, and Collembola) based on ribosomal RNA gene sequences. Mol. Biol. Evol. 2005, 22, 1579–1592. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.; Cruaud, C.; D’Haese, C.A. Unexpected diversity in Neelipleona revealed by molecular phylogeny approach (Hexapoda, Collembola). Soil Org. 2011, 83, 383–398. [Google Scholar]
- Von Reumont, B.M.; Meusemann, K.; Szucsich, N.U.; Dell’Ampio, E.; Gowri-Shankar, V.; Bartel, D.; Simon, S.; Letsch, H.O.; Stocsits, R.R.; Luan, Y.; et al. Can comprehensive background knowledge be incorporated into substitution models to improve phylogenetic analyses? A case study on major arthropod relationships. BMC Evol. Biol. 2009, 9, 119. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Bu, Y.; Luan, Y.-X. Phylogenetic Relationships of Basal Hexapods Reconstructed from Nearly Complete 18S and 28S rRNA Gene Sequences. Zoolog. Sci. 2008, 25, 1139–1145. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Gao, Y.; Yin, W.; Luan, Y. Molecular phylogeny of Collembola inferred from ribosomal RNA genes. Mol. Phylogenet. Evol. 2008, 49, 728–735. [Google Scholar] [CrossRef] [PubMed]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef]
- Boore, J.L.; Lavrov, D.V.; Brown, W.M. Gene translocation links insects and crustaceans. Nature 1998, 392, 667. [Google Scholar] [CrossRef]
- Boore, J.L.; Brown, W.M. Big trees from little genomes: Mitochondrial gene order as a phylogenetic tool. Curr. Opin. Genet. Dev. 1998, 8, 668–674. [Google Scholar] [CrossRef]
- Nardi, F.; Carapelli, A.; Fanciulli, P.P.; Dallai, R.; Frati, F. The complete mitochondrial DNA sequence of the basal hexapod Tetrodontophora bielanensis: Evidence for heteroplasmy and tRNA translocations. Mol. Biol. Evol. 2001, 18, 1293–1304. [Google Scholar] [CrossRef] [PubMed]
- Cicconardi, F.; Borges, P.A.V.; Strasberg, D.; Oromí, P.; López, H.; Pérez Delgado, A.J.; Casquet, J.; Caujapé Castells, J.; Fernández Palacios, J.M.; Thébaud, C.; et al. MtDNA metagenomics reveals large-scale invasion of belowground arthropod communities by introduced species. Mol. Ecol. 2017, 26, 3104–3115. [Google Scholar] [CrossRef] [PubMed]
- Carapelli, A.; Fanciulli, P.P.; Frati, F.; Leo, C. Mitogenomic data to study the taxonomy of Antarctic springtail species (Hexapoda: Collembola) and their adaptation to extreme environments. Polar Biol. 2019, 42, 715–732. [Google Scholar] [CrossRef]
- Swofford, D.L. Phylogenetic Analysis Using Parsimony (*and Other Methods); Sinauer Associates: Sunderland, MA, USA, 2003. [Google Scholar]
- Hassanin, A.; Léger, N.; Deutsch, J. Evidence for Multiple Reversals of Asymmetric Mutational Constraints during the Evolution of the Mitochondrial Genome of Metazoa, and Consequences for Phylogenetic Inferences. Syst. Biol. 2005, 54, 277–298. [Google Scholar] [CrossRef] [PubMed]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Ratnasingham, S.; Hebert, P.D.N. BOLD: The Barcode of Life Data System. Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Wernersson, R.; Pedersen, A.G. RevTrans: Multiple alignment of coding DNA from aligned amino acid sequences. Nucleic Acids Res. 2003, 31, 3537–3539. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New Methods for Selecting Partitioned Models of Evolution for Molecular and Morphological Phylogenetic Analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [PubMed]
- Suchard, M.A.; Lemey, P.; Baele, G.; Ayres, D.L.; Drummond, A.J.; Rambaut, A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018, 4, vey016. [Google Scholar] [CrossRef] [PubMed]
- Rehm, P.; Borner, J.; Meusemann, K.; von Reumont, B.M.; Simon, S.; Hadrys, H.; Misof, B.; Burmester, T. Dating the arthropod tree based on large-scale transcriptome data. Mol. Phylogenet. Evol. 2011, 61, 880–887. [Google Scholar] [CrossRef] [PubMed]
- Rota-Stabelli, O.; Daley, A.C.; Pisani, D. Molecular timetrees reveal a Cambrian colonization of land and a new scenario for ecdysozoan evolution. Curr. Biol. 2013, 23, 392–398. [Google Scholar] [CrossRef]
- Koch, M. Monophyly and phylogenetic position of the Diplura (Hexapoda). Pedobiologia 1997, 41, 9–12. [Google Scholar]
- Leo, C.; Nardi, F.; Frati, F.; Fanciulli, P.P.; Cucini, C.; Vitale, M.; Brunetti, C.; Carapelli, A. The mitogenome of the jumping bristletail Trigoniophthalmus alternatus (Insecta, Microcoryphia) and the phylogeny of insect early-divergent lineages. Mitochondrial DNA B. 2019, 4, 2855–2856. [Google Scholar] [CrossRef]
- Westoll, T.S. Northern Britain. In A correlation of the Devonian Rocks of the British Isles; Geological Society of London Special Report; Geological Society: London, UK, 1977; Volume 8, pp. 66–93. [Google Scholar]
- Garrouste, R.; Clément, G.; Nel, P.; Engel, M.S.; Grandcolas, P.; D’Haese, C.; Lagebro, L.; Denayer, J.; Gueriau, P.; Lafaite, P.; et al. A complete insect from the Late Devonian period. Nature 2012, 488, 82–85. [Google Scholar] [CrossRef]
- Rick, E.F. An entomobryid collembolan (Hexapoda: Collembola) from the Lower Permian of Southern Africa. Paleontol. Afr. 1976, 19, 141–143. [Google Scholar]
- Belica, M.E.; Tohver, E.; Poyatos-Moré, M.; Flint, S.; Parra-Avila, L.A.; Lanci, L.; Denyszyn, S.; Pisarevsky, S.A. Refining the chronostratigraphy of the Karoo Basin, South Africa: Magnetostratigraphic constraints support an early Permian age for the Ecca Group. Geophys. J. Int. 2017, 211, 1354–1374. [Google Scholar] [CrossRef]
- Lavrov, D.V. Key transitions in animal evolution: A mitochondrial DNA perspective. Integr. Comp. Biol. 2007, 47, 734–743. [Google Scholar] [CrossRef]
- Bradshaw, P.C.; Rathi, A.; Samuels, D.C. Mitochondrial-encoded membrane protein transcripts are pyrimidine-rich while soluble protein transcripts and ribosomal RNA are purine-rich. BMC Genomics 2005, 6, 136. [Google Scholar] [CrossRef] [PubMed]
- Henning, W. Insect Phylogeny; Wiley: New York, NY, USA, 1981; pp. 1–514. [Google Scholar]
- Yu, D.; Zhang, F.; Stevens, M.I.; Yan, Q.; Liu, M.; Hu, F. New insight into the systematics of Tomoceridae (Hexapoda, Collembola) by integrating molecular and morphological evidence. Zool. Scr. 2016, 45, 286–299. [Google Scholar] [CrossRef]
- Edwards, D.; Selden, P.A.; Richardson, J.B.; Axe, L. Coprolites as evidence for plant–animal interaction in Siluro–Devonian terrestrial ecosystems. Nature 1995, 377, 329–331. [Google Scholar] [CrossRef]
- Nardi, F.; Carapelli, A.; Boore, J.L.; Roderick, G.K.; Dallai, R.; Frati, F. Domestication of olive fly through a multi-regional host shift to cultivated olives: Comparative dating using complete mitochondrial genomes. Mol. Phylogenet. Evol. 2010, 57, 678–686. [Google Scholar] [CrossRef] [PubMed]
- Torricelli, G.; Carapelli, A.; Convey, P.; Nardi, F.; Boore, J.L.; Frati, F. High divergence across the whole mitochondrial genome in the pan-Antarctic springtail Friesea grisea: Evidence for cryptic species? Gene 2010, 449, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Cicconardi, F.; Nardi, F.; Emerson, B.C.; Frati, F.; Fanciulli, P.P. Deep phylogeographic divisions and long-term persistence of forest invertebrates (Hexapoda: Collembola) in the North-Western Mediterranean basin. Mol. Ecol. 2010, 19, 386–400. [Google Scholar] [CrossRef]
- Von Saltzwedel, H.; Scheu, S.; Schaefer, I. Genetic structure and distribution of Parisotoma notabilis (Collembola) in Europe: Cryptic diversity, split of lineages and colonization patterns. PLoS ONE 2017, 12, e0170909. [Google Scholar] [CrossRef]
- Greenslade, P. An antarctic biogeographical anomaly resolved: The true identity of a widespread species of Collembola. Polar Biol. 2018, 41, 969–981. [Google Scholar] [CrossRef]
- Greenslade, P. A new species of Friesea (Collembola: Neanuridae) from the Antarctic Continent. J. Nat. Hist. 2018, 52, 2197–2207. [Google Scholar] [CrossRef]
- Tully, T.; D’Haese, C.A.; Richard, M.; Ferrière, R. Two major evolutionary lineages revealed by molecular phylogeny in the parthenogenetic collembola species Folsomia candida. Pedobiologia 2006, 50, 95–104. [Google Scholar] [CrossRef]
- Tillyard, R.J. Some remarks on the Devonian Fossil insects from the Rhynie Chert Beds, Old Red Sandstone. Trans. R. Entomol. Soc. Lond. 1928, 76, 65–71. [Google Scholar] [CrossRef]
- Massoud, Z. Contribution à l’étude de Rhyniella praecursor Hirst et Maulik. Rev. Ecol. Biol. Sol 1967, 4, 497–505. [Google Scholar]
- Scourfield, D.J. The oldest known fossil insect (Rhyniella praecursor Hirst & Maulik)—Further details from additional specimens. Proc. Linn. Soc. Lond. 1940, 152, 113–131. [Google Scholar]
- Scourfield, D.J. The oldest known fossil insect. Nature 1940, 145, 799–801. [Google Scholar] [CrossRef]
- Janssens, F. Checklist of the Collembola of the World. Available online: https://www.collembola.org/ (accessed on 17 July 2019).
- Cook, C.E.; Yue, Q.; Akam, M. Mitochondrial genomes suggest that hexapods and crustaceans are mutually paraphyletic. Proc. R. Soc. B Biol. Sci. 2005, 272, 1295–1304. [Google Scholar] [CrossRef] [PubMed]
- Crampton-Platt, A.; Yu, D.W.; Zhou, X.; Vogler, A.P. Mitochondrial metagenomics: Letting the genes out the bottle. GigaScience 2016, 5, 15. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).