Genetic Diversity and Population Structure of Bermudagrass [Cynodon dactylon (L.) Pers.] along Latitudinal Gradients and the Relationship with Polyploidy Level
Abstract
1. Introduction
2. Materials and Methods
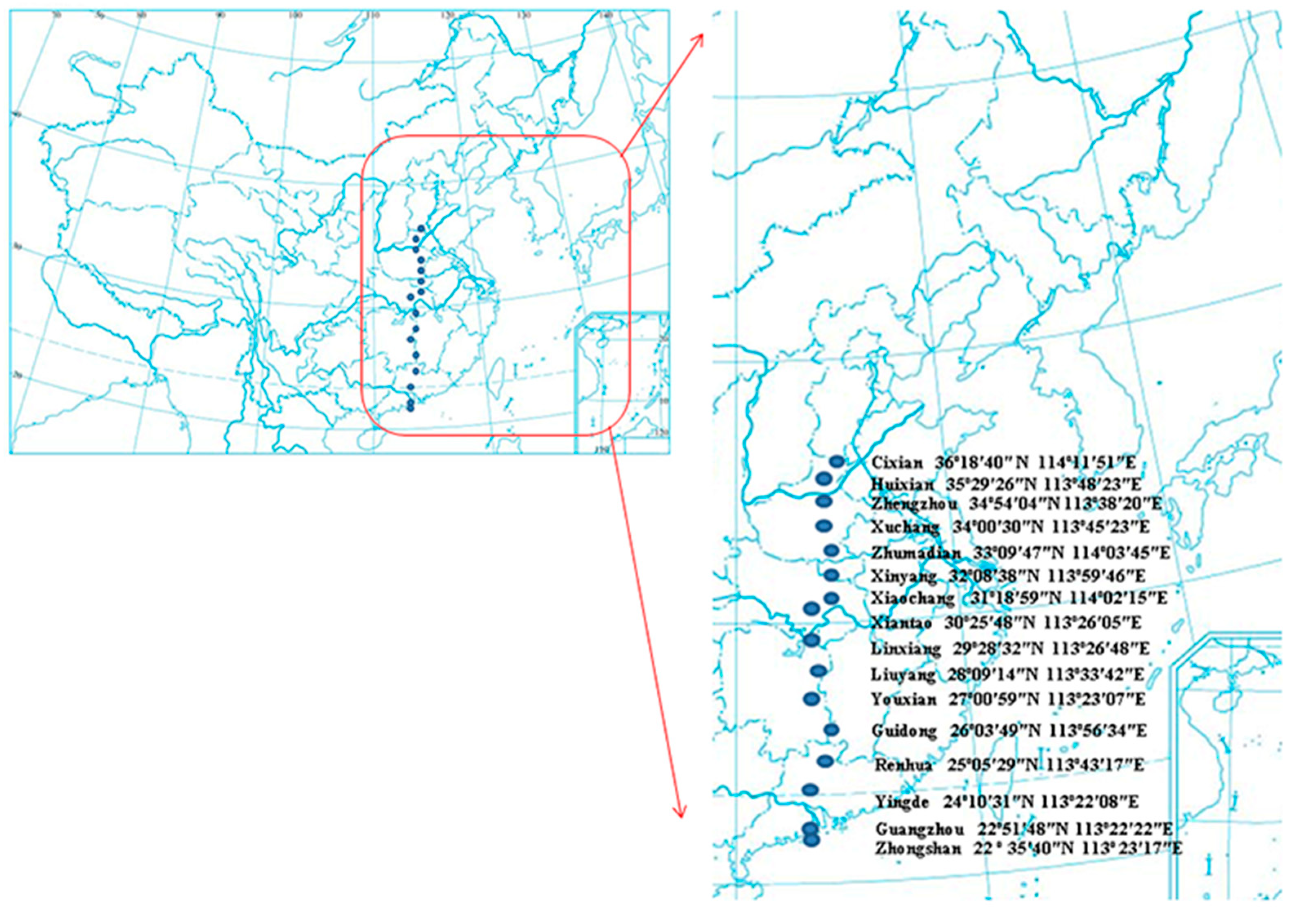
2.1. Plant Materials and EST-SSR Amplification
2.2. Genetic Diversity of C. dactylon along a Latitude Gradient
2.3. Correlation Analyses between Genetic Indexes and other Environmental Parameters
2.4. Separation and Cluster Analysis among all Populations of C. dactylon
2.5. Structure Model and Inferred Ancestry Population of Individuals
3. Results
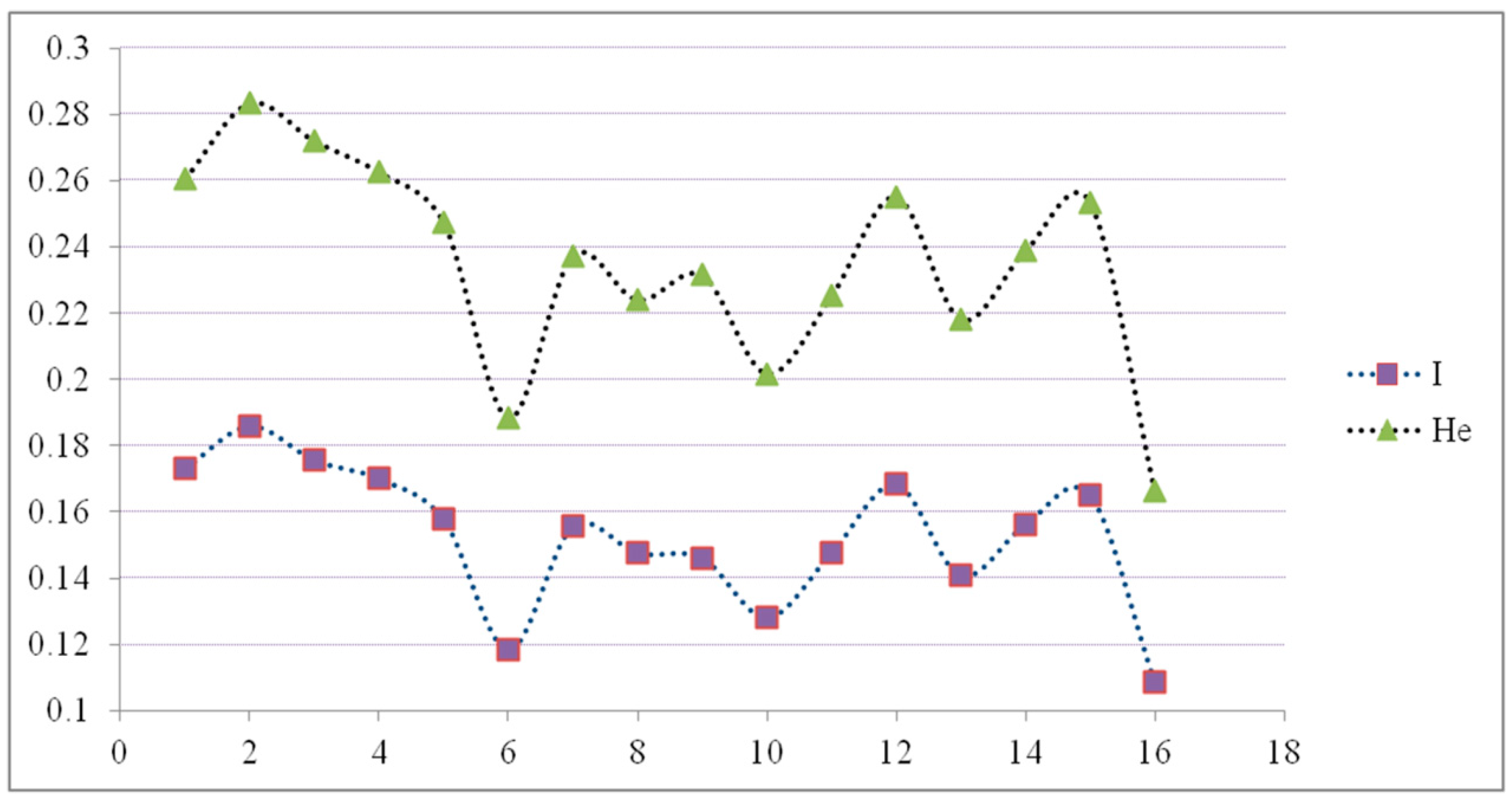
3.1. Intra-Population Genetic Diversity along Latitude
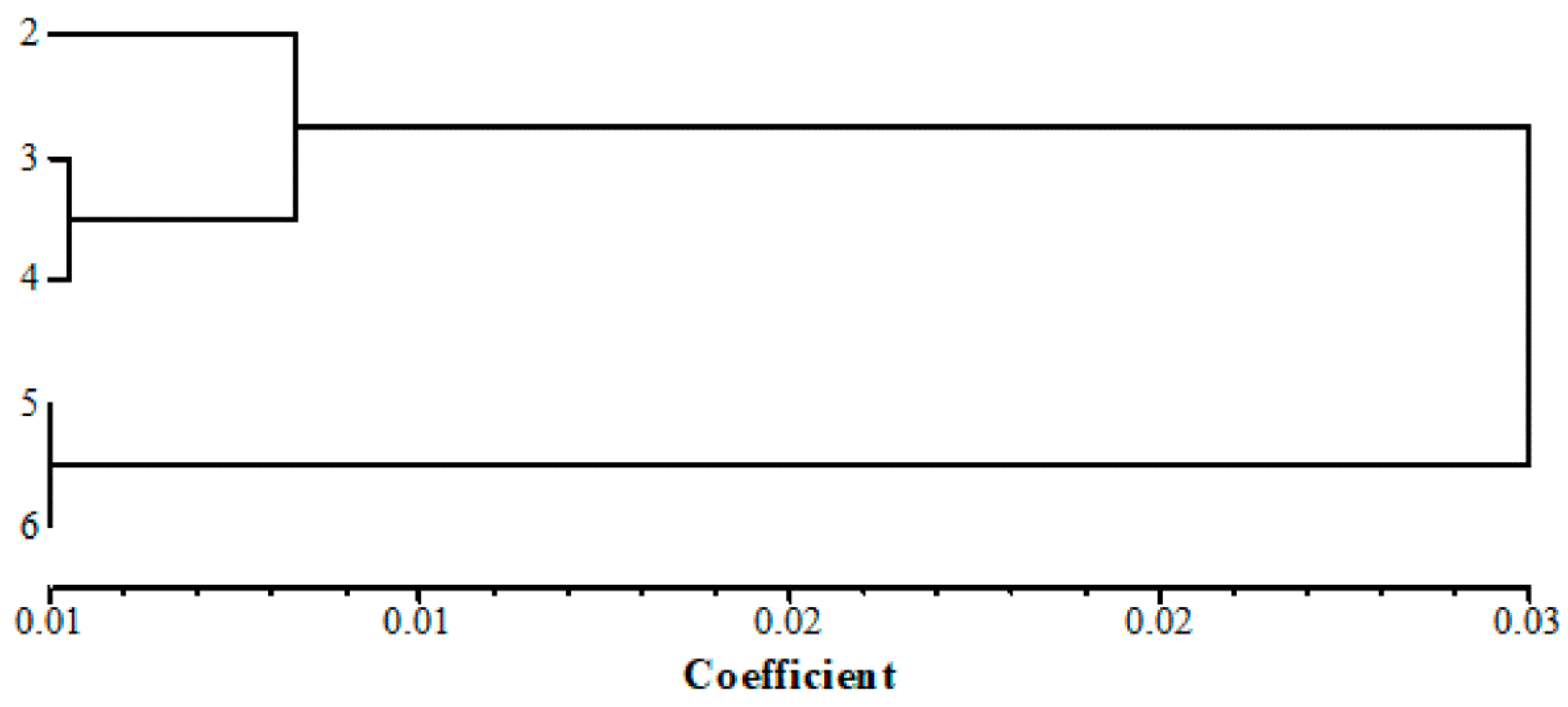
3.2. Genetic Differentiation and Clustering among Populations
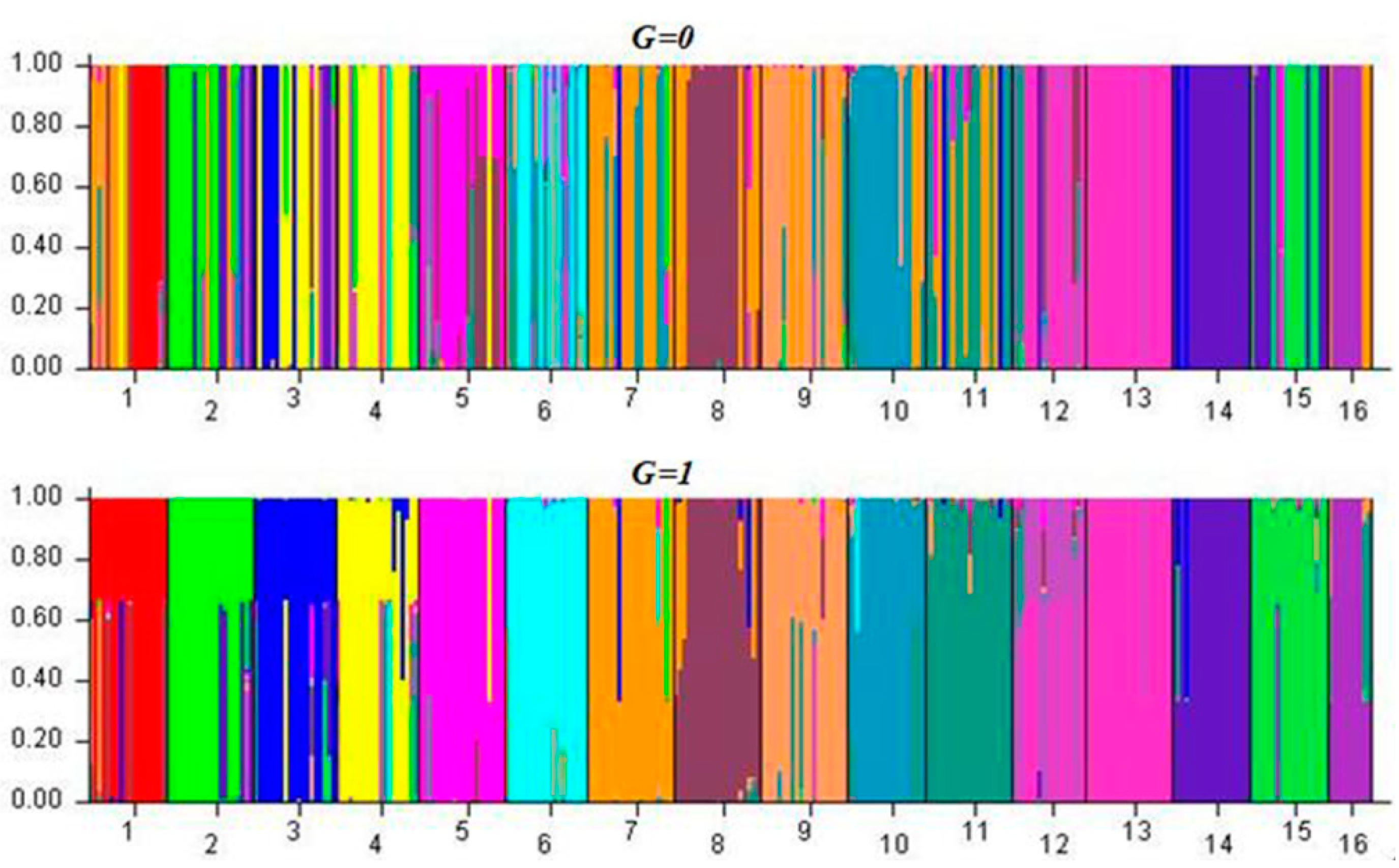
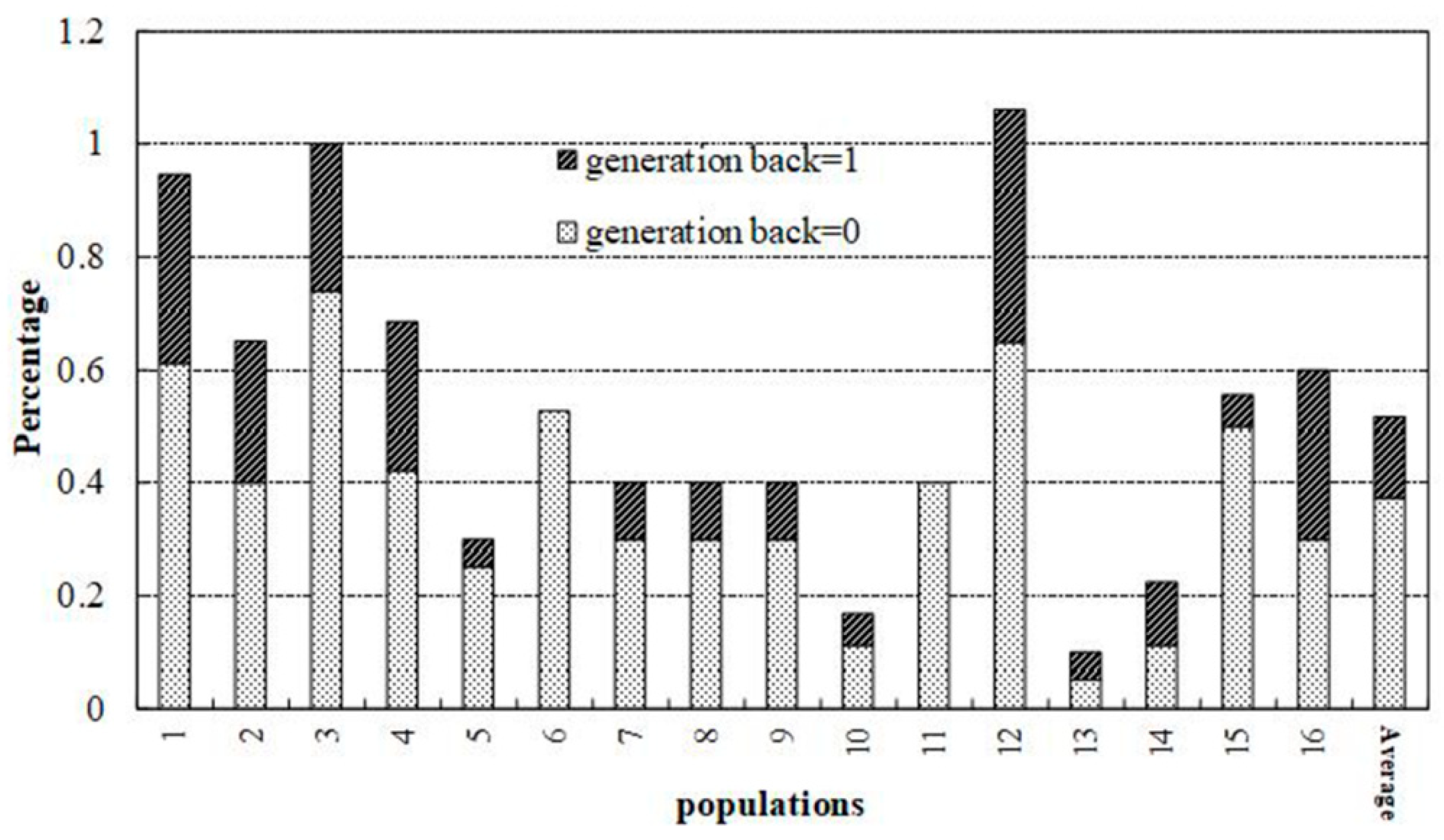
3.3. Genetic Structure and Gene Flow among Populations
4. Discusstion
4.1. Genetic Diversity along Latitudinal Gradients and Relationship with Polyploidy
4.2. Genetic Differentiation and Structure among Populations
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Frankham, R.; Ballou, J.D.; Briscoe, D.A.; Mcinnes, K.H. Introduction to conservation genetics: Glossary. Zool. Afr. 2002, 38, 192. [Google Scholar]
- Holderegger, R.; Kamm, U.; Gugerli, F. Adaptive versus neutral genetic diversity: Implications for landscape genetics. Landsc. Ecol. 2006, 21, 797–807. [Google Scholar] [CrossRef]
- Bolliger, J.; Lander, T.; Balkenhol, N. Landscape genetics since 2003: Status, challenges and future directions. Landsc. Ecol. 2014, 29, 361–366. [Google Scholar] [CrossRef]
- Landguth, E.L.; Cushman, S.A.; Schwartz, M.K.; Mckelvey, K.S.; Murphy, M.; Luikart, G. Quantifying the lag time to detect barriers in landscape genetics. Mol. Ecol. 2010, 19, 4179–4191. [Google Scholar] [CrossRef] [PubMed]
- Blair, C.; Weigel, D.E.; Balazil, M.; Keeley, A.T.H.; Walker, F.M.; Landguth, E.; Cushman, S.; Murphy, M.; Waits, L.; Balkenhol, N. A simulation-based evaluation of methods for inferring linear barriers to gene flow. Mol. Ecol. Res. 2012, 12, 822–833. [Google Scholar] [CrossRef] [PubMed]
- Angeloni, F.; Ouborg, N.J.; Leimu, R. Meta-analysis on the association of population size and life history with inbreeding depression in plants. Biol. Conserv. 2011, 144, 35–43. [Google Scholar] [CrossRef]
- Harlan, J.R.; De Wet, J.M.J. Sources of variation in Cynodon dactylon (L.). Pers. 1. Crop Sci. 1969, 9, 774–778. [Google Scholar] [CrossRef]
- Taliaferro, C.M. Diversity and Vulnerability of Bermuda Turfgrass Species. Crop Sci. 1995, 35, 327–332. [Google Scholar] [CrossRef]
- Taliaferro, C.M. Bermudagrass (Cynodon (L.) Rich). In Turfgrass Biology, Genetics and Breeding; Casler, M.D., Duncan, R.R., Eds.; John Wiley and Sons Inc.: Hoboken, NJ, USA, 2003; pp. 235–257. [Google Scholar]
- Wu, Y.Q.; Taliaferro, C.M.; Bai, G.H.; Anderson, M.P. AFLP analysis of Cynodon dactylon (L.) Pers. var. dactylon genetic variation. Genome 2004, 47, 689–696. [Google Scholar] [CrossRef]
- de Silva, P.H.A.U.; Snaydon, R.W. Chromosome number in Cynodon dactylon in relation to ecological conditions. Ann. Bot. 1995, 76, 535–537. [Google Scholar] [CrossRef]
- Wu, Y.Q.; Taliaferro, C.M.; Martin, D.L.; Goad, C.L.; Anderson, J.A. Genetic variability and relationships for seed yield and its components in Chinese Cynodon accessions. Field Crop Res. 2006, 98, 245–252. [Google Scholar] [CrossRef]
- Vlachou, D.; Konsolaki, M.; Tolias, P.P.; Kafatos, F.C.; Komitopoulou, K. The austosomal chorion locus of the medfly Ceratitis capitata. 1. Conserved synteny, amplification and tissue specificity but sequence divergence and altered temporal regulation. Genetics 1997, 147, 1829–1842. [Google Scholar]
- Jewell, M.C.; Frere, C.H.; Prentis, P.J.; Lambrides, C.J.; Godwin, I.D. Characterisation and multiplexing of EST-SSR primers in Cynodon (Poaceae) species. Am. J. Bot. 2010, 97, e99–e101. [Google Scholar] [CrossRef]
- Ho, C.Y.; Mcmaugh, S.J.; Wilton, A.N.; Mcfarlane, I.J.; Mackinlay, A.G. DNA amplification variation within cultivars of turf-type Couch grasses (Cynodon spp.). Plant Cell Rep. 1997, 16, 797–801. [Google Scholar] [CrossRef]
- Caetano-Anollés, G. DNA Analysis of Turfgrass Genetic Diversity. Crop Sci. 1998, 38, 1415–1424. [Google Scholar] [CrossRef]
- Zhang, L.; Ozias-Akins, P.; Kochert, G.; Kresovich, S.; Dean, R.; Hanna, W. Differentiation of bermudagrass (Cynodon spp.) genotypes by AFLP analyses. Theor. Appl. Genet. 1999, 98, 895–902. [Google Scholar] [CrossRef]
- Anderson, M.P.; Taliaferro, C.M.; Martin, D.L.; Anderson, C.S. Comparative DNA Profiling of U-3 Turf Bermudagrass Strains. Crop Sci. 2001, 41, 1184–1189. [Google Scholar] [CrossRef]
- Karaca, M. Genetic diversity among forage bermudagrass (Cynodon spp.): Evidence from chloroplast and nuclear DNA fingerprinting. Crop Sci. 2002, 42, 2118–2127. [Google Scholar] [CrossRef]
- Roodt, R.; Spies, J.J.; Burger, T.H. Preliminary DNA fingerprinting of the turf grass Cynodon dactylon (Poaceae: Chloridoideae). Bothalia 2002, 32, 117–122. [Google Scholar] [CrossRef]
- Etemadi, N.; Sayed-Tabatabaei, B.E.; Zamanni, Z.; Razmjoo, K.; Khalighi, A.; Lessani, H. Evaluation of diversity among Cynodon dactylon (L.) Pers. using RAPD markers. Int. J. Agric. Biol. 2005, 8, 198–202. [Google Scholar]
- Tan, J.Q. Chinese Ground Covers of Lawn; Science and Technology Literature Press: Beijing, China, 1993. [Google Scholar]
- Harrisshultz, K.R.; Schwartz, B.M.; Hanna, W.W.; Brady, J.A. Development, linkage mapping, and use of microsatellites in bermudagrass. J. Am. Soc. Hortic. Sci. 2010, 135, 511–520. [Google Scholar] [CrossRef]
- Jewell, M.C.; Yi, Z.; Loch, D.S.; Godwin, I.D.; Lambrides, C.J. Maximizing Genetic, Morphological, and Geographic Diversity in a Core Collection of Australian Bermudagrass. Crop Sci. 2012, 52, 879–889. [Google Scholar] [CrossRef]
- Bennett, M.D.; Smith, J.B. Nuclear DNA amounts in angiosperms. Philos. Trans. R. Soc. B 1976, 274, 227–274. [Google Scholar] [CrossRef]
- Yeh, F.C.; Yang, R.C.; Boyle, T.; Ye, Z.H.; Mao, J.X. POPGENE, the User Friendly Shareware for Population Genetic Analysis; Molecular Biology and Biotechnology Center, University of Alberta: Edmonton, AB, Canada, 1997. [Google Scholar]
- Duncan, D.B. Multiple Range and Multiple F Tests. Biometrics 1955, 11, 1–42. [Google Scholar] [CrossRef]
- Khan, A.; Nawaz, M.; Khan, S.; Steele, R. Detection and characterization of erythromycin-resistant methylase genes in Gram-positive bacteria isolated from poultry litter. Appl. Microbiol. Biot. 2002, 59, 377–381. [Google Scholar]
- Peakall, R.; Smouse, P. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Rohlf, F.J. NTSYS-pc numerical taxonomy and multivariate analysis system version 2.1 manual. N. Y. Appl. Biostat. 2000. Available online: https://www.scienceopen.com/document?vid=8071739f-efd0-4990-9970-47e831f11f8b (accessed on 14 August 2019).
- STRUCTURE V.2.3. Available online: http://web.stanford.edu/group/pritchardlab/structure.html (accessed on 4 July 2019).
- Hubisz, M.J.; Falush, D.; Stephens, M.; Pritchard, J.K. Inferring weak population structure with the assistance of sample group information. Mol. Ecol. Res. 2009, 9, 1322–1332. [Google Scholar] [CrossRef]
- Rosenberg, N.A.; Burke, T.; Elo, K.; Feldman, M.W.; Freidlin, P.J.; Groenen, M.A.; Hillel, J.; Mäki-Tanila, A.; Tixier-Boichard, M.; Vignal, A.; et al. Empirical evaluation of genetic clustering methods using multilocus genotypes from 20 chicken breeds. Genetics 2001, 159, 699. [Google Scholar]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Yan, X.B.; Guo, Y.X.; Liu, F.Y.; Zhao, C.; Liu, Q.L.; Lu, B.R. Population structure affected by excess gene flow in self-pollinating Elymus nutansand E. Popul. Ecol. 2010, 52, 233–241. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Rosenberg, N.A.; Donnelly, P. Association mapping in structured populations. Am. J. Hum. Genet. 2000, 67, 170. [Google Scholar] [CrossRef]
- Ellegren, H.; Galtier, N. Determinants of genetic diversity. Nat. Rev. Genet. 2016, 17, 422. [Google Scholar] [CrossRef]
- Welch, D.B.M.; Meselson, M. Evidence for the evolution of bdelloid rotifers without sexual reproduction or genetic exchange. Science 2000, 288, 1211–1215. [Google Scholar] [CrossRef]
- François, B.; Laurent, L.; de Thierry, M. The population genetics of clonal and partially clonal diploids. Genetics 2003, 164, 1635–1644. [Google Scholar]
- Hollister, J.D.; Greiner, S.; Wang, W.; Wang, J.; Zhang, Y.; Wong, G.K.S.; Wright, S.I.; Johnson, M.T.J. Recurrent loss of sex is associated with accumulation of deleterious mutations in oenothera. Mol. Biol. Evol. 2014, 32, 896–905. [Google Scholar] [CrossRef]
- Hamrick, J.L.; Godt, M.J.W. Allozyme diversity in plant species. In Plant Population Genetics, Breeding and Genetic Resources; Brown, A.H.D., Clegg, M.T., Kahler, A.L., Weir, B.S., Eds.; Sinauer Associates Inc.: Sunderland, MA, USA, 1989. [Google Scholar]
- Borba, E.L.; Semir, J.; Shepherd, G.J. Self-incompatibility, Inbreeding Depression and Crossing Potential in Five Brazilian Pleurothallis (Orchidaceae) Species. Ann. Bot. 2001, 88, 89–99. [Google Scholar] [CrossRef]
- Nybom, H. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol. Ecol. 2010, 13, 1143–1155. [Google Scholar] [CrossRef]
- Hamrick, J.L.; Godt, M.J.W. Effects of Life History Traits on Genetic Diversity in Plant Species. Philos. Trans. Biol. Sci. 1996, 351, 1291–1298. [Google Scholar]
- Nevo, E. Genetic diversity in wild cereals: Regional and local studies and their bearing on conservation ex situ and in situ. Genet. Res. Crop Evol. 1998, 45, 355–370. [Google Scholar] [CrossRef]
- Gulsen, O.; Sever-Mutlu, S.; Mutlu, N.; Tuna, M.; Karaguzel, O.; Shearman, R.C.; Riordan, T.P.; Heng-Moss, T.M. Polyploidy creates higher diversity among Cynodon accessions as assessed by molecular markers. Theor. Appl. Genet. 2009, 118, 1309–1319. [Google Scholar] [CrossRef]
- Gulsen, O.; Shearman, R.C.; Vogel, K.P.; Lee, D.J.; Baenziger, P.S.; Heng-Moss, T.M.; Budak, H. Nuclear genome diversity and relationships among naturally occurring buffalograss genotypes determined by sequence-related amplified polymorphism. HortScience 2005, 40, 537–541. [Google Scholar] [CrossRef]
- Budak, H.; Shearman, R.C.; Gulsen, O.; Dweikat, I. Understanding ploidy complex and geographic origin of the Buchloe dactyloides genome using cytoplasmic and nuclear marker systems. Theor. Appl. Genet. 2005, 111, 1545–1552. [Google Scholar] [CrossRef]
- Johnson, P.G.; Kenworthy, K.E.; Auld, D.L.; Riordan, T.P. Distribution of buffalograss polyploid variation in the southern great plains. Cropence 2001, 41, 909–913. [Google Scholar] [CrossRef]
- Ferrero, V.; Barrett, S.C.; Castro, S.; Caldeirinha, P.; Navarro, L.; Loureiro, J.; Rodríguez-Echeverría, S. Invasion genetics of the Bermuda buttercup (Oxalis pes-caprae): Complex intercontinental patterns of genetic diversity, polyploidy and heterostyly characterize both native and introduced populations. Mol. Ecol. 2015, 24, 2143–2155. [Google Scholar] [CrossRef]
- Zhang, J.X.; Wang, M.L.; Guo, Z.P.; Guan, Y.Z.; Guo, Y.X.; Yan, X.B. Variation in ploidy level and genome size of Cynodon dactylon (L.) Pers. along a latitudinal gradient. Folia Geobot. under review.
- Francisco-Ortega, J.; Santos-Guerra, A.; Kim, S.C.; Crawford, D.J. Plant genetic diversity in the Canary Islands: A conservation perspective. Am. J. Bot. 2000, 87, 909–919. [Google Scholar] [CrossRef]
- Bolibok-Brągoszewska, H.; Targońska, M.; Bolibok, L.; Kilian, A.; Rakoczy-Trojanowska, M. Genome-wide characterization of genetic diversity and population structure in Secale. Bmc Plant Biol. 2014, 14, 1–15. [Google Scholar] [CrossRef]
- Zhang, C.; Huang, J.; Xiao, Y.; Lian, C.; Li, X. Genetic diversity and population structure of sour jujube, Ziziphus acidojujuba. Tree Genet. Genomes 2015, 11, 809. [Google Scholar] [CrossRef]
- Zheng, Y.; Xu, S.; Liu, J.; Zhao, Y.; Liu, J. Genetic diversity and population structure of Chinese natural bermudagrass [Cynodon dactylon (L.) Pers.] germplasm based on SRAP markers. PLoS ONE 2017, 12, e0177508. [Google Scholar] [CrossRef]
- Ling, Y.; Zhang, X.Q.; Ma, X.; Chen, S.Y.; Chen, T.T.; Liu, W. Analysis of genetic diversity among wild bermudagrass germplasm from southwest China using SSR markers. Genet. Mol. Res. 2012, 11, 4598. [Google Scholar] [CrossRef]
- Zong, J.W.; Zhao, T.T.; Ma, Q.H.; Liang, L.S.; Wang, G.X. Assessment of Genetic Diversity and Population Genetic Structure of Corylus mandshurica in China Using SSR Markers. PLoS ONE 2015, 10, e0137528. [Google Scholar] [CrossRef]
- Ladizinsky, G. Founder effect in crop-plant evolution. Econ. Bot. 1985, 39, 191–199. [Google Scholar] [CrossRef]
- Rosenblum, E.B. Convergent evolution and divergent selection: Lizards at the White Sands ecotone. Am. Nat. 2006, 167, 1–15. [Google Scholar] [CrossRef]
- Nelson, E.C. Phytogeography of southern Australia. In Ecological Biogeography of Australia; Keast, A., Ed.; Junk: The Hague, The Netherlands, 1981; pp. 735–759. [Google Scholar]
- Rius, M.; Darling, J.A. How important is intraspecific genetic admixture to the success of colonising populations? Trends Ecol. Evol. 2014, 29, 233–242. [Google Scholar] [CrossRef]
- Tate, J.A.; Soltis, D.E.; Soltis, P.S. CHAPTER 7–Polyploidy in Plants. Evol. Genome 2005, 2005, 371–426. [Google Scholar]
- Durka, W.; Bossdorf, O.; Prati, D.; Auge, H. Molecular evidence for multiple introductions of garlic mustard (Alliaria petiolata, Brassicaceae) to North America. Mol. Ecol. 2005, 14, 1697–1706. [Google Scholar] [CrossRef]


| Accession No. | Primer Sequence (5′ – 3′) | Repeat | Size (bp) | Ta (°C) |
|---|---|---|---|---|
| ES293996 | F: GAGCGAACCTCAGAGTAGCC | (CT)11 | 99 | 50 |
| R: CAGAAGCAATCGCGGAAAC | ||||
| ES297444 | F: GCCTACTCCATGGAAGAGTCAG | (CAG)7 | 93 | 50 |
| R: CGACCACTGCTTCATCACC | ||||
| ES295368 | F: TCGGTTACCTGCTTCACACC | (CT)14 | 107 | 60 |
| R: GGGAAGACGAAGACCTCCAG | ||||
| ES295577 | F: TGATTGTCCGAGGTCAGCA | (GGC)7 | 118 | 50 |
| R: GGTGTGAACCATCAGCAGTT | ||||
| ES297517 | F: GCGCAGCTGATAGAGGTTGT | (AGA)4 | 159 | 50 |
| R: ACAGGCGGCTTTCATCTTCT | ||||
| ES302010 | F: CTACAACAGCTTCCCCAGCA | (AAC)7 | 128 | 50 |
| R: TGACGTCGTTGGTGTAGAGC | ||||
| ES297043 | F: TTGGCCAGTCATGTATTTTCC | (TTC)4 | 163 | 50 |
| R: AACATGGAGTCTGGTGCTCA | ||||
| ES305182 | F: AAGGTTGTGGACAAGGATGG | (TGC)4 | 144 | 50 |
| R: GGGGTCCAACAACTCAAAAG | ||||
| ES303634 | F: GCGTTCCAGAAGTTGCAGTT | (CCT)6, (GCG)6 | 166 | 50 |
| R: AGATCTTCTCTCCGGCCTTG | ||||
| ES296960 | F: ACATGTCTCCGTCCCATCA | CCG | 167 | 62 |
| R: ATGAGTCGGTCCTTCTTGG | ||||
| ES300915 | F: GGCAGCTCCTCTCTCCTTAAA | (CGG)8 | 166 | 62 |
| R: ACCATGACCTTGTCCTCGTC | ||||
| ES295668 | F: ATCCAATGAGTGGGACTCCA | (CAA)10 | 167 | 58 |
| R: CCAGCTTGCTTGGGATTAAA | ||||
| ES294037 | F: TTGTGCCAGCTATCTTGGTG | 159 | 58 | |
| R: TGCGAAGCAAGAAGAGTACG | ||||
| ES294662 | F: CAAGGACCACATCACCATCA | (CGAT)6 | 204 | 58 |
| R: CGGCCATTGATTATCTGTGA | ||||
| ES300599 | F: AGGGGAAGAAGGGTAAGCAG | CAG | 189 | 62 |
| R: CACCAAATCCACCAAAGGAG | ||||
| ES296415 | F: GGACTCAAAAATGCTCAGAAA | (CAAA)6 | 202 | 58 |
| R: TTTGCAGAGCCCGTAACTCT | ||||
| ES300198 | F: TGACGTCGTTGGTGTAGAGC | (TTG)10 | 239 | 62 |
| R: CGACTCCATCTGGTCCAACT | ||||
| ES306918 | F: AGATCGGGGTGGGGAAGA | (CGA)6 | 199 | 58 |
| R: GTACATCTCCAGCAGCGACA | ||||
| ES303889 | F: TCCGTTGCCTATACGGTTG | CGG | 225 | 62 |
| R: GGAACCGATAATCACTCCA | ||||
| ES295349 | F: CCAAGAGCTAGGCCATCAAC | (CT)10 | 150 | 50 |
| R: CACAAATCTTGGAACCATGTG |
| Population ID | Origin | Sample Number | P | I | He |
|---|---|---|---|---|---|
| 1 | Zhongshan | 18 | 51.63 | 0.173 | 0.2605 |
| 2 | Guangzhou | 20 | 58.82 | 0.1858 | 0.2833 |
| 3 | Yingde | 19 | 59.48 | 0.1755 | 0.2719 |
| 4 | Renhua | 19 | 58.17 | 0.1699 | 0.2625 |
| 5 | Guidong | 20 | 56.86 | 0.1576 | 0.2473 |
| 6 | Youxian | 19 | 45.75 | 0.1183 | 0.1883 |
| 7 | Liuyang | 20 | 50.98 | 0.1555 | 0.2373 |
| 8 | Linxiang | 20 | 45.75 | 0.1475 | 0.2239 |
| 9 | Xiantao | 20 | 55.56 | 0.146 | 0.2314 |
| 10 | Xiaochang | 18 | 45.75 | 0.128 | 0.2016 |
| 11 | Xinyang | 20 | 46.41 | 0.1475 | 0.2251 |
| 12 | Zhumadian | 17 | 52.29 | 0.1682 | 0.2551 |
| 13 | Xuchang | 20 | 49.67 | 0.1407 | 0.218 |
| 14 | Zhengzhou | 18 | 49.67 | 0.1561 | 0.2387 |
| 15 | Huixian | 18 | 54.25 | 0.1649 | 0.2534 |
| 16 | Cixian | 10 | 33.33 | 0.1086 | 0.1663 |
| Total | 296 | 100 | 0.2003 | 0.3197 |
| Group | P | He | I |
|---|---|---|---|
| 1 | 57.025 A | 0.17605 A | 0.26955 A |
| 2 | 49.835 B | 0.144725 B | 0.2242 B |
| 3 | 50.0025 B | 0.147425 B | 0.2283 B |
| 4 | 46.73 C | 0.142575 B | 0.2191 B |
| Ploidy Level | N | P | He | I |
|---|---|---|---|---|
| 2 | 16 | 58.17 C | 0.1702 C | 0.2647 C |
| 3 | 37 | 67.32 B | 0.1846 B | 0.2876 B |
| 4 | 100 | 85.62 A | 0.1804 B | 0.2909 B |
| 5 | 23 | 69.93 B | 0.2053 A | 0.3157 A |
| 6 | 35 | 71.24 B | 0.2086 A | 0.3204 A |
| Overall | 211 | 100 | 0.1995 | 0.3166 |
| Source | df | SS | MS | Estimate of Variation | % |
|---|---|---|---|---|---|
| Among Pops | 15 | 1054.470 | 70.298 | 3.125 | 20% |
| Within Pops | 280 | 3511.601 | 12.541 | 12.541 | 80% |
| Total | 295 | 4566.071 | 15.667 | 100% |
| K | L(K) | StDev | L’(K) | L″(K) | [″(K)] | Delta K |
|---|---|---|---|---|---|---|
| 1 | −14,576.2 | 73.6 | ||||
| 2 | −13,211 | 235.9 | 1365.2 | \ | \ | \ |
| 3 | −12,602.8 | 459.4 | 608.2 | −757 | 757 | 1.6478 |
| 4 | −12,079 | 486.8 | 523.8 | −84.4 | 84.4 | 0.1734 |
| 5 | −11,794.9 | 648.4 | 284.1 | −239.7 | 239.7 | 0.3697 |
| 6 | −11,575.2 | 669.8 | 219.7 | −64.4 | 64.4 | 0.0961 |
| 7 | −11,394.3 | 765.7 | 180.9 | −38.8 | 38.8 | 0.0507 |
| 8 | −11,187.9 | 790.1 | 206.4 | 25.5 | 25.5 | 0.0323 |
| 9 | −10,980.5 | 938.9 | 207.4 | 1 | 1 | 0.0011 |
| 10 | −10,802.8 | 945.6 | 177.7 | −29.7 | 29.7 | 0.0314 |
| 11 | −10,840.4 | 863.9 | −37.6 | −215.3 | 215.3 | 0.2492 |
| 12 | −10,770.6 | 1027.4 | 69.8 | 107.4 | 107.4 | 0.1045 |
| 13 | −10,609.7 | 986.1 | 160.9 | 91.1 | 91.1 | 0.0924 |
| 14 | −10,460.1 | 1236.1 | 149.6 | −11.3 | 11.3 | 0.0091 |
| 15 | −10,353.5 | 1187.2 | 106.6 | −43 | 43 | 0.0362 |
| 16 | −10,228.7 | 1370.8 | 124.8 | 18.2 | 18.2 | 0.0133 |
| 17 | −10,285.8 | 1611.3 | −57.1 | −181.9 | 181.9 | 0.1129 |
| 18 | −10,215 | 1466.3 | 70.8 | 127.9 | 127.9 | 0.0872 |
| 19 | −10,057 | 1572.1 | 158 | 87.2 | 87.2 | 0.0555 |
| 20 | −9928.5 | 1504.1 | 128.5 | −29.5 | 29.5 | 0.0196 |
| 21 | −9542.8 | 1492.3 | 385.7 | 257.2 | 257.2 | 0.1724 |
| 22 | −9632.7 | 1606.2 | −89.9 | −475.6 | 475.6 | 0.2961 |
| 23 | −9285 | 1608.9 | 347.7 | 437.6 | 437.6 | 0.2720 |
| 24 | −9754.2 | 2023.1 | −469.2 | −816.9 | 816.9 | 0.4038 |
| 25 | −10,053.5 | 1880.9 | −299.3 | 169.9 | 169.9 | 0.0903 |
| 26 | −9408.1 | 1704.9 | 645.4 | 944.7 | 944.7 | 0.5541 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Wang, M.; Guo, Z.; Guan, Y.; Liu, J.; Yan, X.; Guo, Y. Genetic Diversity and Population Structure of Bermudagrass [Cynodon dactylon (L.) Pers.] along Latitudinal Gradients and the Relationship with Polyploidy Level. Diversity 2019, 11, 135. https://doi.org/10.3390/d11080135
Zhang J, Wang M, Guo Z, Guan Y, Liu J, Yan X, Guo Y. Genetic Diversity and Population Structure of Bermudagrass [Cynodon dactylon (L.) Pers.] along Latitudinal Gradients and the Relationship with Polyploidy Level. Diversity. 2019; 11(8):135. https://doi.org/10.3390/d11080135
Chicago/Turabian StyleZhang, Jingxue, Miaoli Wang, Zhipeng Guo, Yongzhuo Guan, Jianyu Liu, Xuebing Yan, and Yuxia Guo. 2019. "Genetic Diversity and Population Structure of Bermudagrass [Cynodon dactylon (L.) Pers.] along Latitudinal Gradients and the Relationship with Polyploidy Level" Diversity 11, no. 8: 135. https://doi.org/10.3390/d11080135
APA StyleZhang, J., Wang, M., Guo, Z., Guan, Y., Liu, J., Yan, X., & Guo, Y. (2019). Genetic Diversity and Population Structure of Bermudagrass [Cynodon dactylon (L.) Pers.] along Latitudinal Gradients and the Relationship with Polyploidy Level. Diversity, 11(8), 135. https://doi.org/10.3390/d11080135





