Indications of Genetic Admixture in the Transition Zone between Fagus sylvatica L. and Fagus sylvatica ssp. orientalis Greut. & Burd
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. DNA Extraction and Genotyping
2.3. Data Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Denk, T. The taxonomy of Fagus in western Eurasia, 1: Fagus sylvatica subsporientalis (= F. orientalis). Feddes Repert. 1999, 110, 177–200. [Google Scholar] [CrossRef]
- Denk, T. The taxonomy of Fagus in western Eurasia. 2: Fagus sylvatica subsp sylvatica. Feddes Repert. 1999, 110, 381–412. [Google Scholar] [CrossRef]
- Renner, S.S.; Grimm, G.W.; Kapli, P.; Denk, T. Species relationships and divergence times in beeches: New insights from the inclusion of 53 young and old fossils in a birth-death clock model. Philos. Trans. R. Soc. B 2016, 371, 20150135. [Google Scholar] [CrossRef] [PubMed]
- Gömöry, D.; Paule, L.; Mačejovský, V. Phylogeny of beech in western Eurasia as inferred by approximate Bayesian computation. Acta Soc. Bot. Pol. 2018, 87, 3582. [Google Scholar] [CrossRef]
- Euro+Med PlantBase. Available online: http://www.emplantbase.org/home.html (accessed on 20 May 2019).
- Denk, T.; Grimm, G.; Stögerer, K.; Langer, M.; Hemleben, V. The evolutionary history of Fagus in western Eurasia: Evidence from genes, morphology and the fossil record. Plant Syst. Evol. 2002, 232, 213–236. [Google Scholar] [CrossRef]
- Hatziskakis, S.; Tsiripidis, I.; Papageorgiou, A.C. Leaf morphological variation in beech (Fagus sylvatica L.) populations in Greece and its relation to their post-glacial origin. Bot. J. Linn. Soc. 2011, 165, 422–436. [Google Scholar] [CrossRef]
- Hatziskakis, S.; Papageorgiou, A.C.; Gailing, O.; Finkeldey, R. High chloroplast haplotype diversity in Greek populations of beech (Fagus sylvatica L.). Plant Biol. 2009, 11, 425–433. [Google Scholar] [CrossRef]
- Papageorgiou, A.C.; Vidalis, A.; Gailing, O.; Tsiripidis, I.; Hatziskakis, S.; Boutsios, S.; Galatsidas, S.; Finkeldey, R. Genetic variation of beech (Fagus sylvatica L.) in Rodopi (N.E. Greece). Eur. J. For. Res. 2008, 127, 81–88. [Google Scholar] [CrossRef]
- Gailing, O.; von Wuehlisch, G. Nuclear markers (AFLPs) and chloroplast microsatellites differ between Fagus sylvatica and F. orientalis. Silv. Genet. 2004, 53, 105–110. [Google Scholar] [CrossRef]
- Gömöry, D.; Paule, L.; Brus, R.; Zhelev, P.; Tomović, Z.; Gračan, J. Genetic differentiation and phylogeny of beech on the Balkan peninsula. J. Evol. Biol. 1999, 12, 746–754. [Google Scholar] [CrossRef]
- Gömöry, D.; Paule, L.; Vyšný, J. Patterns of allozyme variation in western Eurasian Fagus. Bot. J. Linn. Soc. 2007, 154, 165–174. [Google Scholar] [CrossRef][Green Version]
- Magri, D.; Vendramin, G.G.; Comps, B.; Dupanloup, I.; Geburek, T.; Gömöry, D.; Latałowa, M.; Litt, T.; Paule, L.; Roure, J.M.; et al. A new scenario for the quaternary history of European beech populations: Palaeobotanical evidence and genetic consequences. New Phytol. 2006, 171, 199–221. [Google Scholar] [CrossRef] [PubMed]
- Von Wuehlisch, G.; Liesebach, M.; Muhs, H.-J.; Stephan, B.R. A Network of International Beech Provenance Trials. In First EUFORGEN Meeting on Social Broadleaves: 23–25 October 1997, Bordeaux, France; Turok, J., Kremer, A., De Vries, S.G.M., Eds.; IPGRI: Bordeaux, France, 1998; pp. 164–172. [Google Scholar]
- Seifert, S. Variation of Candidate Genes Related to Climate Change in European Beech (Fagus sylvatica L.). Ph.D. Thesis, University of Göttingen, Göttingen, Germany, 2012. [Google Scholar]
- Shorthouse, D. Simple Mappr, an Online Tool to Produce Publication-Quality Point Maps. Available online: http://www.simplemappr.net (accessed on 02 April 2019).
- Tsiripidis, I.; Athanasiadis, N. Contribution to the knowledge of the vascular flora of NE Greece: Floristic composition of the beech (Fagus sylvatica L.) forests in the Greek Rodopi. Willdenowia 2003, 33, 273–297. [Google Scholar] [CrossRef]
- Burger, K.; Müller, M.; Gailing, O. Characterization of EST-SSRs for European beech (Fagus sylvatica L.) and their transferability to Fagus orientalis Lipsky, Castanea dentata Bork., and Quercus rubra L. Silv. Genet. 2018, 67, 127–132. [Google Scholar] [CrossRef]
- Kubisiak, T.; Carey, D.; Burdine, C.; Koch, J. Characterization of ten EST-based polymorphic SSR loci isolated from American beech, Fagus grandifolia Ehrh. Permanent genetic resources note added to Molecular Ecology Resources. Mol. Ecol. Resour. 2009, 9, 1460–1466. [Google Scholar] [CrossRef]
- Kubisiak, T.L.; Nelson, C.D.; Staton, M.E.; Zhebentyayeva, T.; Smith, C.; Olukolu, B.A.; Fang, G.C.; Hebard, F.V.; Anagnostakis, S.; Wheeler, N.; et al. A transcriptome-based genetic map of Chinese chestnut (Castanea mollissima) and identification of regions of segmental homology with peach (Prunus persica). Tree Genet. Genomes 2013, 9, 557–571. [Google Scholar] [CrossRef]
- Schuelke, M. An economic method for the fluorescent labeling of PCR fragments. Nat. Biotechnol. 2000, 18, 233–234. [Google Scholar] [CrossRef] [PubMed]
- Brownstein, M.J.; Carpten, J.D.; Smith, J.R. Modulation of non-templated nucleotide addition by Taq DNA polymerase: Primer modifications that facilitate genotyping. BioTechniques 1996, 20, 1004–1006. [Google Scholar] [CrossRef] [PubMed]
- Peakall, R.; Smouse, P.E. Genalex 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Chesser, R.K. Estimation of fixation indices and gene diversities. Ann. Hum. Genet. 1983, 47, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Meirmans, P.G.; Hedrick, P.W. Assessing population structure: F(ST) and related measures. Mol. Ecol. Resour. 2011, 11, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Goudet, J. FSTAT (Version 1.2): A computer program to calculate F-statistics. J. Hered. 1995, 86, 485–486. [Google Scholar] [CrossRef]
- Rice, W.R. Analyzing tables of statistical tests. Evolution 1989, 43, 223–225. [Google Scholar] [CrossRef] [PubMed]
- Van Oosterhout, C.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes 2004, 4, 535–538. [Google Scholar] [CrossRef]
- Antao, T.; Lopes, A.; Lopes, R.J.; Beja-Pereira, A.; Luikart, G. LOSITAN: A workbench to detect molecular adaptation based on a Fst-outlier method. BMC Bioinform. 2008, 9, 323. [Google Scholar] [CrossRef] [PubMed]
- Foll, M.; Gaggiotti, O. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: A Bayesian perspective. Genetics 2008, 180, 977–993. [Google Scholar] [CrossRef]
- Langella, O. Populations Version 1.2.32. Available online: http://www.bioinformatics.org/project/?group_id=84 (accessed on 13 March 2019).
- Nei, M. Genetic distance between populations. Am. Nat. 1972, 106, 283–292. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Earl, D.A.; vonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. CLUMPAK: A program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef] [PubMed]
- Rajendra, K.C.; Seifert, S.; Prinz, K.; Gailing, O.; Finkeldey, R. Subtle human impacts on neutral genetic diversity and spatial patterns of genetic variation in European beech (Fagus sylvatica). For. Ecol. Manag. 2014, 319, 138–149. [Google Scholar] [CrossRef]
- Dounavi, A.; Netzer, F.; Celepirovic, N.; Ivanković, M.; Burger, J.; Figueroa, A.G.; Schön, S.; Simon, J.; Cremer, E.; Fussi, B.; et al. Genetic and physiological differences of European beech provenances (F. sylvatica L.) exposed to drought stress. For. Ecol. Manag. 2016, 361, 226–236. [Google Scholar] [CrossRef]
- Lind, J.F.; Gailing, O. Genetic structure of Quercus rubra L. and Quercus ellipsoidalis E. J. Hill populations at gene-based EST-SSR and nuclear SSR markers. Tree Genet. Genomes 2013, 9, 707–722. [Google Scholar] [CrossRef]
- Beaumont, M.A.; Nichols, R.A. Evaluating loci for use in the genetic analysis of population structure. Proc. R. Soc. B 1996, 263, 1619–1626. [Google Scholar] [CrossRef]
- Flanagan, S.P.; Jones, A.G. Constraints on the FST–heterozygosity outlier approach. J. Hered. 2017, 108, 561–573. [Google Scholar] [CrossRef]
- Modesto, I.S.; Miguel, C.; Pina-Martins, F.; Glushkova, M.; Veloso, M.; Paulo, O.S.; Batista, D. Identifying signatures of natural selection in cork oak (Quercus suber L.) genes through SNP analysis. Tree Genet. Genomes 2014, 10, 1645–1660. [Google Scholar] [CrossRef]
- Homolka, A.; Schueler, S.; Burg, K.; Fluch, S.; Kremer, A. Insights into drought adaptation of two European oak species revealed by nucleotide diversity of candidate genes. Tree Genet. Genomes 2013, 9, 1179–1192. [Google Scholar] [CrossRef]
- Derory, J.; Leger, P.; Garcia, V.; Schaeffer, J.; Hauser, M.T.; Salin, F.; Luschnig, C.; Plomion, C.; Glossl, J.; Kremer, A. Transcriptome analysis of bud burst in sessile oak (Quercus petraea). New Phytol. 2006, 170, 723–738. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.; Gailing, O. Abiotic genetic adaptation in the Fagaceae. Plant Biol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Varsamis, G.; Papageorgiou, A.C.; Merou, T.; Takos, I.; Malesios, C.; Manolis, A.; Tsiripidis, I.; Gailing, O. Adaptive diversity of beech seedlings under climate change scenarios. Front. Plant Sci. 2019, 9, 1918. [Google Scholar] [CrossRef] [PubMed]
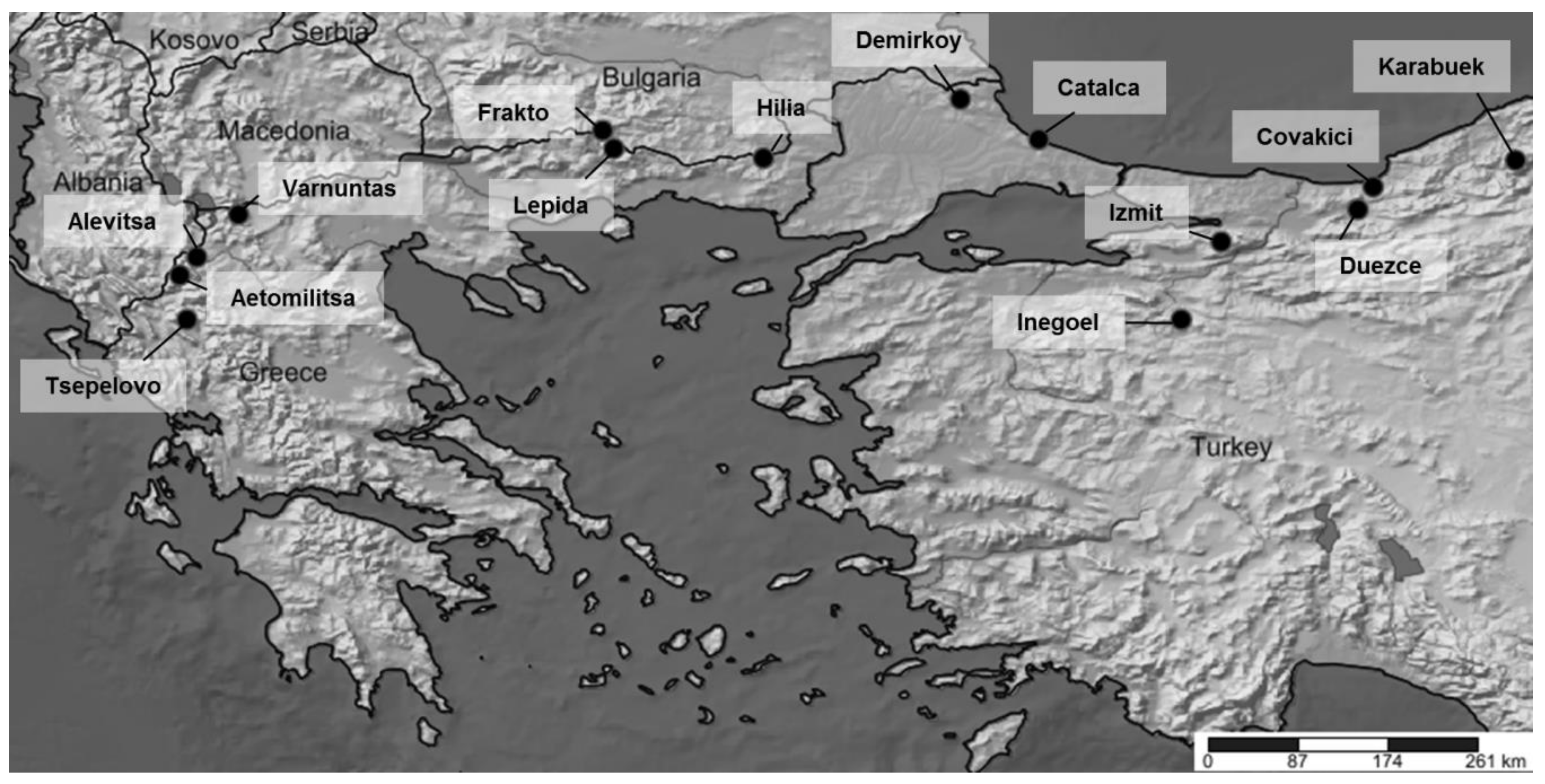
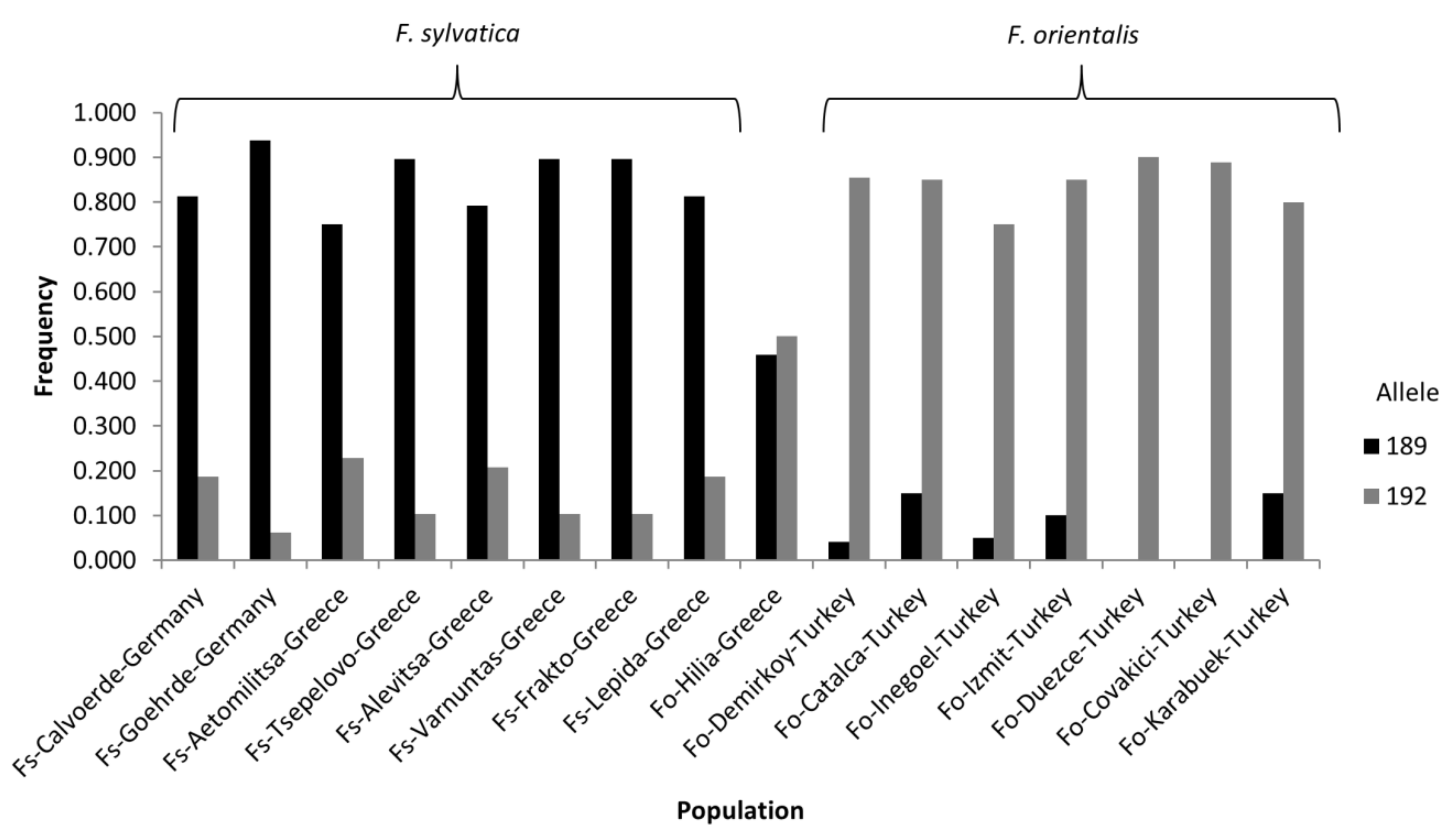
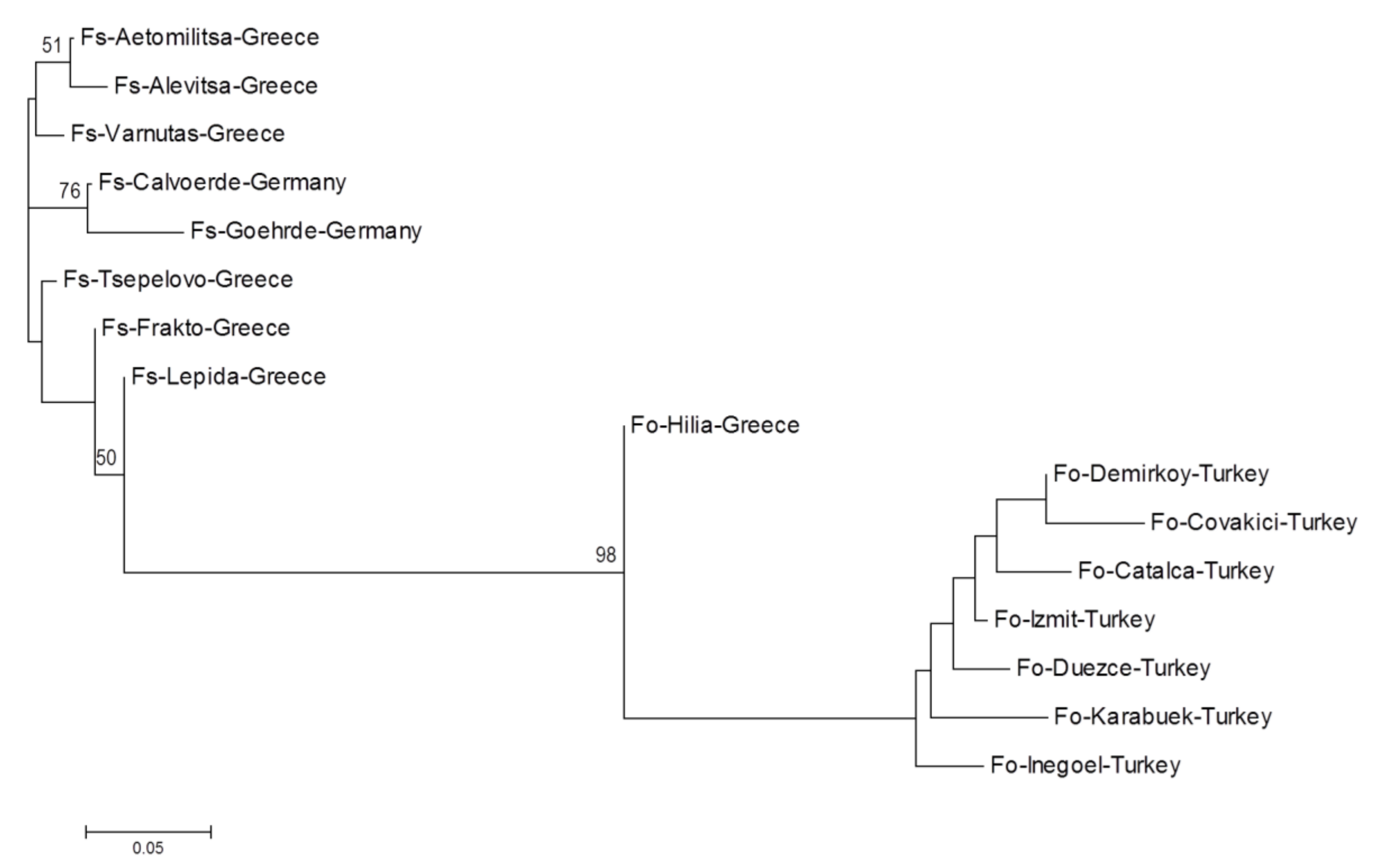
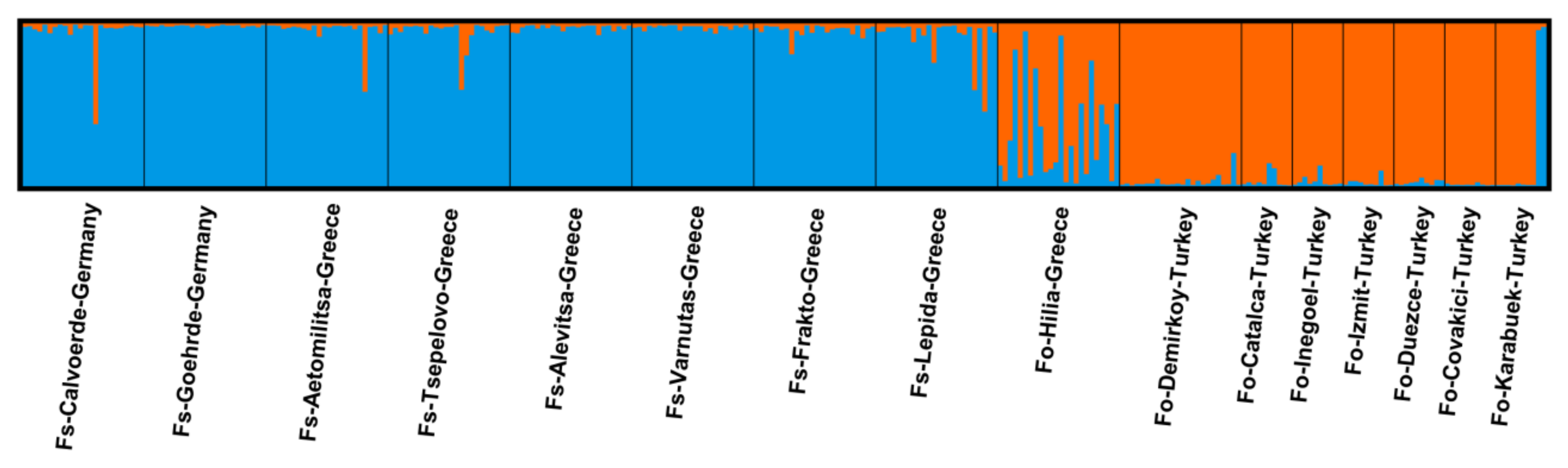
| Population Name | Origin | No. of Samples | Subspecies | Latitude | Longitude |
|---|---|---|---|---|---|
| Calvoerde | Germany | 24 | F. sylvatica | 52.403967 | 11.261017 |
| Goehrde | Germany | 24 | F. sylvatica | 53.122983 | 10.820400 |
| Aetomilitsa | Greece | 24 | F. sylvatica ** | 40.272958 | 20.813221 |
| Tsepelovo | Greece | 24 | F. sylvatica ** | 39.882826 | 20.876105 |
| Alevitsa | Greece | 24 | F. sylvatica ** | 40.432116 | 20.963033 |
| Varnuntas | Greece | 24 | F. sylvatica ** | 40.806592 | 21.328064 |
| Frakto | Greece | 24 | F. sylvatica ** | 41.545455 | 24.523565 |
| Lepida | Greece | 24 | F. sylvatica ** | 41.386269 | 24.621609 |
| Hilia | Greece | 24 | F. orientalis ** | 41.302195 | 25.934421 |
| Demirkoy | Turkey | 24 | F. orientalis ** | 41.819996 | 27.662091 |
| Catalca | Turkey * | 10 | F. orientalis | 41.466670 | 28.350000 |
| Inegoel | Turkey * | 10 | F. orientalis | 39.883330 | 29.600000 |
| Izmit | Turkey * | 10 | F. orientalis | 40.566670 | 29.950000 |
| Duezce | Turkey * | 10 | F. orientalis | 40.850000 | 31.150000 |
| Covakici | Turkey * | 10 | F. orientalis | 41.050000 | 31.283330 |
| Karabuek | Turkey * | 10 | F. orientalis | 41.283330 | 32.533330 |
| Marker | N | Na | Ho | He | FIS | GST |
|---|---|---|---|---|---|---|
| FgSI0006 | 18.6 | 3.0 | 0.345 | 0.349 | 0.029 | 0.244 * |
| FgSI0024 | 18.6 | 4.6 | 0.589 | 0.600 | 0.057 | 0.014 |
| FS_C1968 | 18.4 | 9.6 | 0.769 | 0.761 | 0.019 | 0.032 * |
| FS_C2361 | 18.4 | 3.8 | 0.564 | 0.534 | −0.032 | 0.118 * |
| FgSI0009 | 17.4 | 3.0 | 0.447 | 0.473 | 0.085 | 0.099 * |
| FS_C6785 | 18.7 | 2.6 | 0.284 | 0.278 | 0.012 | 0.469 * |
| FS_C7377 | 18.6 | 3.2 | 0.521 | 0.509 | −0.006 | 0.158 * |
| FS_C7797 | 18.5 | 3.6 | 0.415 | 0.346 | −0.145 | 0.016 * |
| FS_C4971 | 18.7 | 5.1 | 0.706 | 0.636 | −0.078 | 0.095 * |
| Mean | 18.4 | 4.3 | 0.515 | 0.498 | −0.007 | 0.132 * |
| Population | N | Na | AR | Ho | He | FIS |
|---|---|---|---|---|---|---|
| Fs-Calvoerde-Germany | 24 | 3.8 | 3.1 | 0.500 | 0.464 | −0.056 |
| Fs-Goehrde-Germany | 24 | 3.4 | 2.9 | 0.421 | 0.402 | −0.026 |
| Fs-Aetomilitsa-Greece | 24 | 5.0 | 3.5 | 0.551 | 0.528 | −0.022 |
| Fs-Tsepelovo-Greece | 24 | 4.2 | 3.4 | 0.516 | 0.519 | 0.028 |
| Fs-Alevitsa-Greece | 24 | 4.4 | 3.3 | 0.506 | 0.499 | 0.009 |
| Fs-Varnuntas-Greece | 24 | 4.8 | 3.4 | 0.527 | 0.512 | −0.007 |
| Fs-Frakto-Greece | 24 | 4.7 | 3.3 | 0.532 | 0.503 | −0.034 |
| Fs-Lepida-Greece | 24 | 4.1 | 3.4 | 0.485 | 0.546 | 0.133 |
| Fo-Hilia-Greece | 24 | 4.8 | 3.8 | 0.563 | 0.582 | 0.053 |
| Fo-Demirkoy-Turkey | 24 | 5.2 | 3.7 | 0.569 | 0.500 | −0.117 |
| Fo-Catalca-Turkey | 10 | 3.9 | 3.7 | 0.460 | 0.472 | 0.079 |
| Fo-Inegoel-Turkey | 10 | 4.4 | 4.2 | 0.468 | 0.512 | 0.143 |
| Fo-Izmit-Turkey | 10 | 4.2 | 3.9 | 0.543 | 0.490 | −0.054 |
| Fo-Duezce-Turkey | 10 | 4.0 | 3.8 | 0.558 | 0.500 | −0.062 |
| Fo-Covakici-Turkey | 10 | 3.2 | 3.1 | 0.447 | 0.421 | −0.002 |
| Fo-Karabuek-Turkey | 10 | 4.0 | 3.8 | 0.599 | 0.522 | −0.094 |
| Mean | 18.8 | 4.3 | 3.5 | 0.515 | 0.498 | −0.002 |
| Populations | Fs-Calvoerde-Germany | Fs-Goehrde-Germany | Fs-Aetomilitsa-Greece | Fs-Tsepelovo-Greece | Fs-Alevitsa-Greece | Fs-Varnuntas-Greece | Fs-Frakto-Greece | Fs-Lepida-Greece | Fo-Hilia-Greece | Fo-Demirkoy-Turkey | Fo-Catalca-Turkey | Fo-Inegoel-Turkey | Fo-Izmit-Turkey | Fo-Duezce-Turkey | Fo-Covakici-Turkey | Fo-Karabuek-Turkey |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fs-Calvoerde-Germany | 0.000 | |||||||||||||||
| Fs-Goehrde-Germany | 0.016 | 0.000 | ||||||||||||||
| Fs-Aetomilitsa-Greece | 0.014 | 0.039 | 0.000 | |||||||||||||
| Fs-Tsepelovo-Greece | 0.014 | 0.026 | 0.001 + | 0.000 | ||||||||||||
| Fs-Alevitsa-Greece | 0.015 | 0.046 | 0.000 + | 0.010 | 0.000 | |||||||||||
| Fs-Varnuntas-Greece | 0.009 | 0.032 | 0.003 + | 0.001 + | 0.006 | 0.000 | ||||||||||
| Fs-Frakto-Greece | 0.009 | 0.032 | 0.004 + | 0.002 + | 0.012 | 0.005 + | 0.000 | |||||||||
| Fs-Lepida-Greece | 0.020 | 0.043 | 0.005 + | 0.009 | 0.013 | 0.010 | 0.000 + | 0.000 | ||||||||
| Fo-Hilia-Greece | 0.073 | 0.104 | 0.054 | 0.047 | 0.069 | 0.062 | 0.039 | 0.035 | 0.000 | |||||||
| Fo-Demirkoy-Turkey | 0.150 | 0.190 | 0.125 | 0.121 | 0.143 | 0.140 | 0.116 | 0.106 | 0.019 | 0.000 | ||||||
| Fo-Catalca-Turkey | 0.159 | 0.205 | 0.119 | 0.120 | 0.139 | 0.141 | 0.114 | 0.099 | 0.024 | 0.013 | 0.000 | |||||
| Fo-Inegoel-Turkey | 0.124 | 0.158 | 0.111 | 0.103 | 0.127 | 0.121 | 0.101 | 0.097 | 0.018 | 0.006 + | 0.019 | 0.000 | ||||
| Fo-Izmit-Turkey | 0.139 | 0.183 | 0.115 | 0.112 | 0.136 | 0.131 | 0.102 | 0.092 | 0.012 | 0.004 + | 0.000 + | 0.000 + | 0.000 | |||
| Fo-Duezce-Turkey | 0.137 | 0.172 | 0.117 | 0.115 | 0.136 | 0.136 | 0.106 | 0.092 | 0.015 | 0.002 + | 0.017 + | 0.000 + | 0.000 + | 0.000 | ||
| Fo-Covakici-Turkey | 0.185 | 0.231 | 0.152 | 0.145 | 0.177 | 0.168 | 0.144 | 0.136 | 0.034 | 0.001 + | 0.010 + | 0.011 + | 0.005 + | 0.014 + | 0.000 | |
| Fo-Karabuek-Turkey | 0.136 | 0.173 | 0.114 | 0.103 | 0.129 | 0.126 | 0.109 | 0.106 | 0.024 | 0.015 | 0.017 | 0.009 + | 0.013 + | 0.014 + | 0.004 + | 0.000 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Müller, M.; Lopez, P.A.; Papageorgiou, A.C.; Tsiripidis, I.; Gailing, O. Indications of Genetic Admixture in the Transition Zone between Fagus sylvatica L. and Fagus sylvatica ssp. orientalis Greut. & Burd. Diversity 2019, 11, 90. https://doi.org/10.3390/d11060090
Müller M, Lopez PA, Papageorgiou AC, Tsiripidis I, Gailing O. Indications of Genetic Admixture in the Transition Zone between Fagus sylvatica L. and Fagus sylvatica ssp. orientalis Greut. & Burd. Diversity. 2019; 11(6):90. https://doi.org/10.3390/d11060090
Chicago/Turabian StyleMüller, Markus, Precious Annie Lopez, Aristotelis C. Papageorgiou, Ioannis Tsiripidis, and Oliver Gailing. 2019. "Indications of Genetic Admixture in the Transition Zone between Fagus sylvatica L. and Fagus sylvatica ssp. orientalis Greut. & Burd" Diversity 11, no. 6: 90. https://doi.org/10.3390/d11060090
APA StyleMüller, M., Lopez, P. A., Papageorgiou, A. C., Tsiripidis, I., & Gailing, O. (2019). Indications of Genetic Admixture in the Transition Zone between Fagus sylvatica L. and Fagus sylvatica ssp. orientalis Greut. & Burd. Diversity, 11(6), 90. https://doi.org/10.3390/d11060090







