Recent Trends in Research on the Genetic Diversity of Plants: Implications for Conservation
Abstract
1. Introduction
2. Methods
2.1. Scientometric Data
2.2. Genetic Diversity versus Phylogeny, and Ecological and Reproductive Traits
3. Results
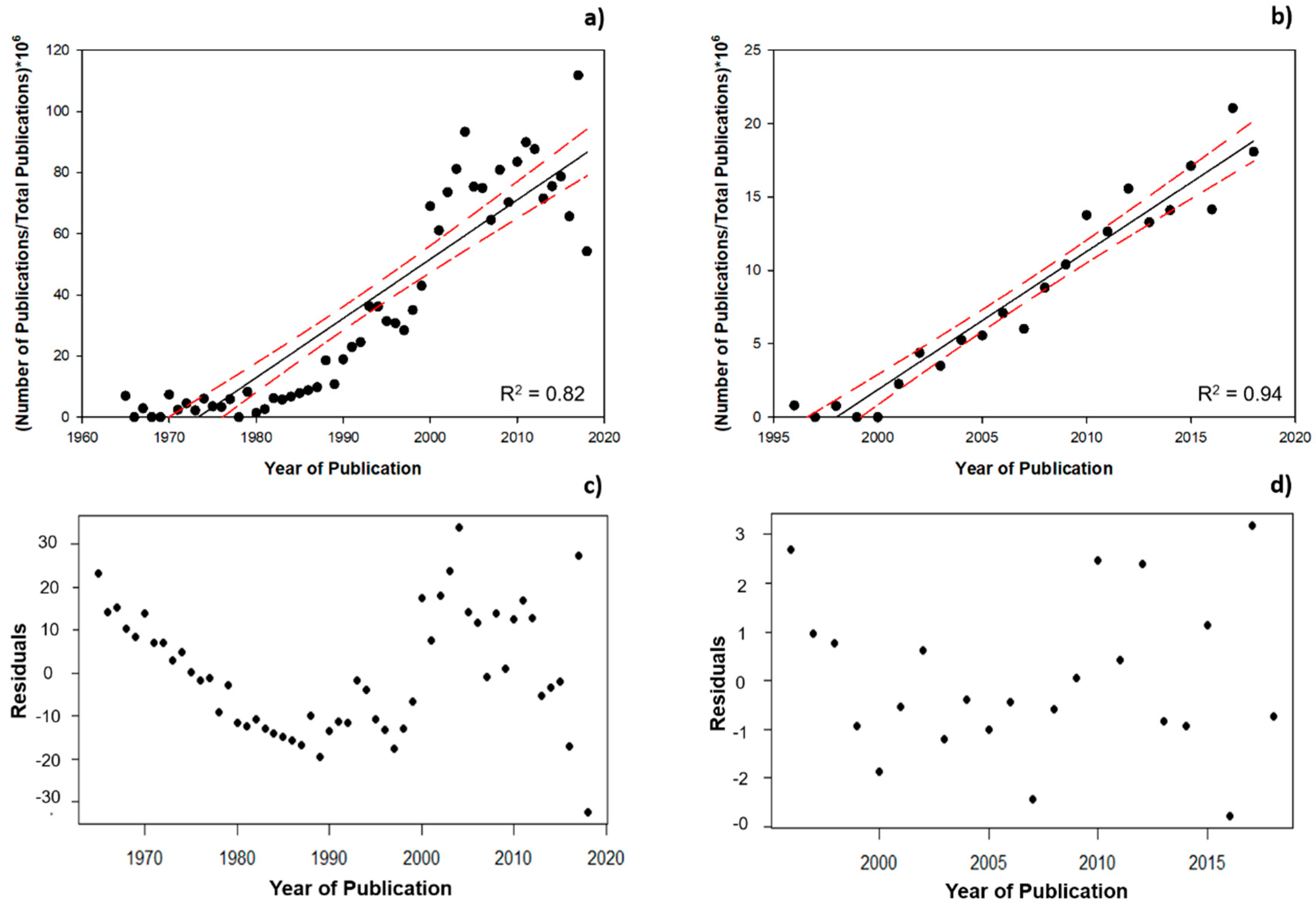
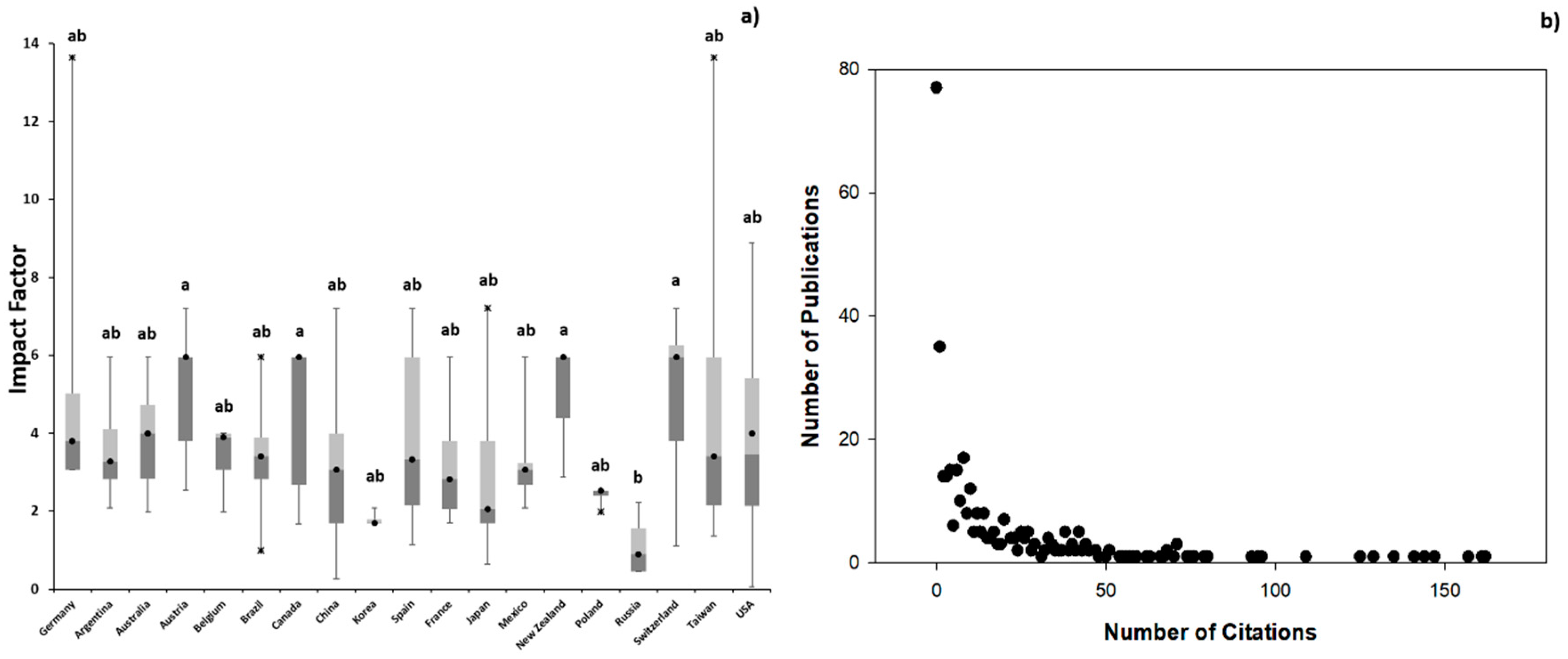
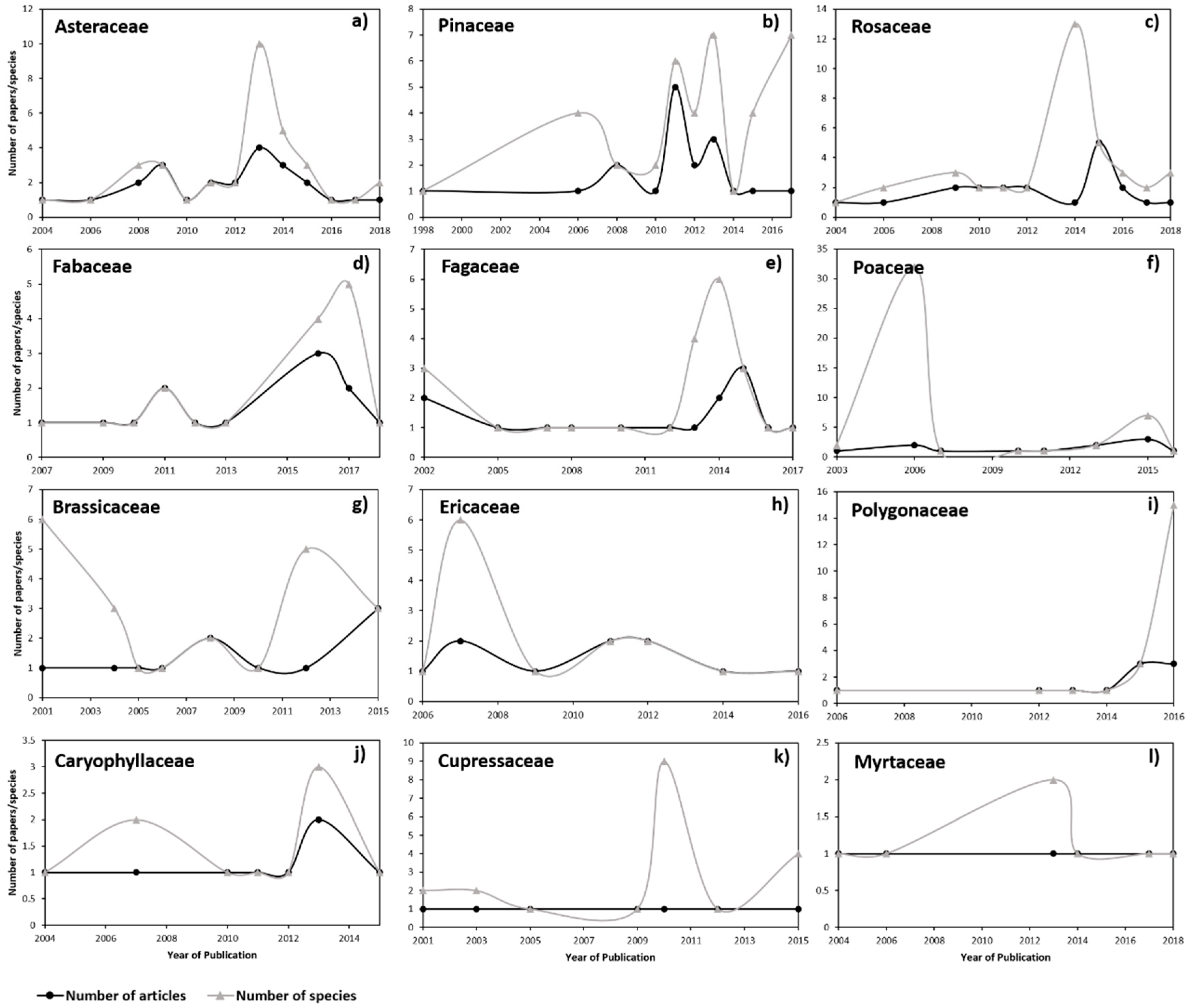
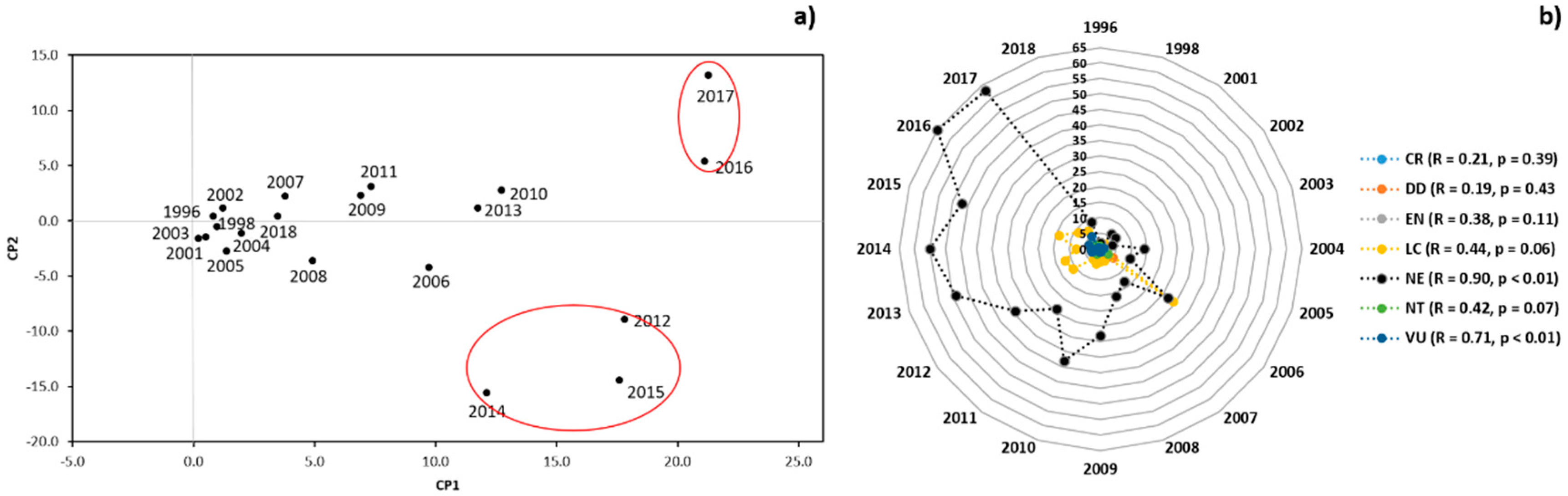
3.1. Scientometric Data
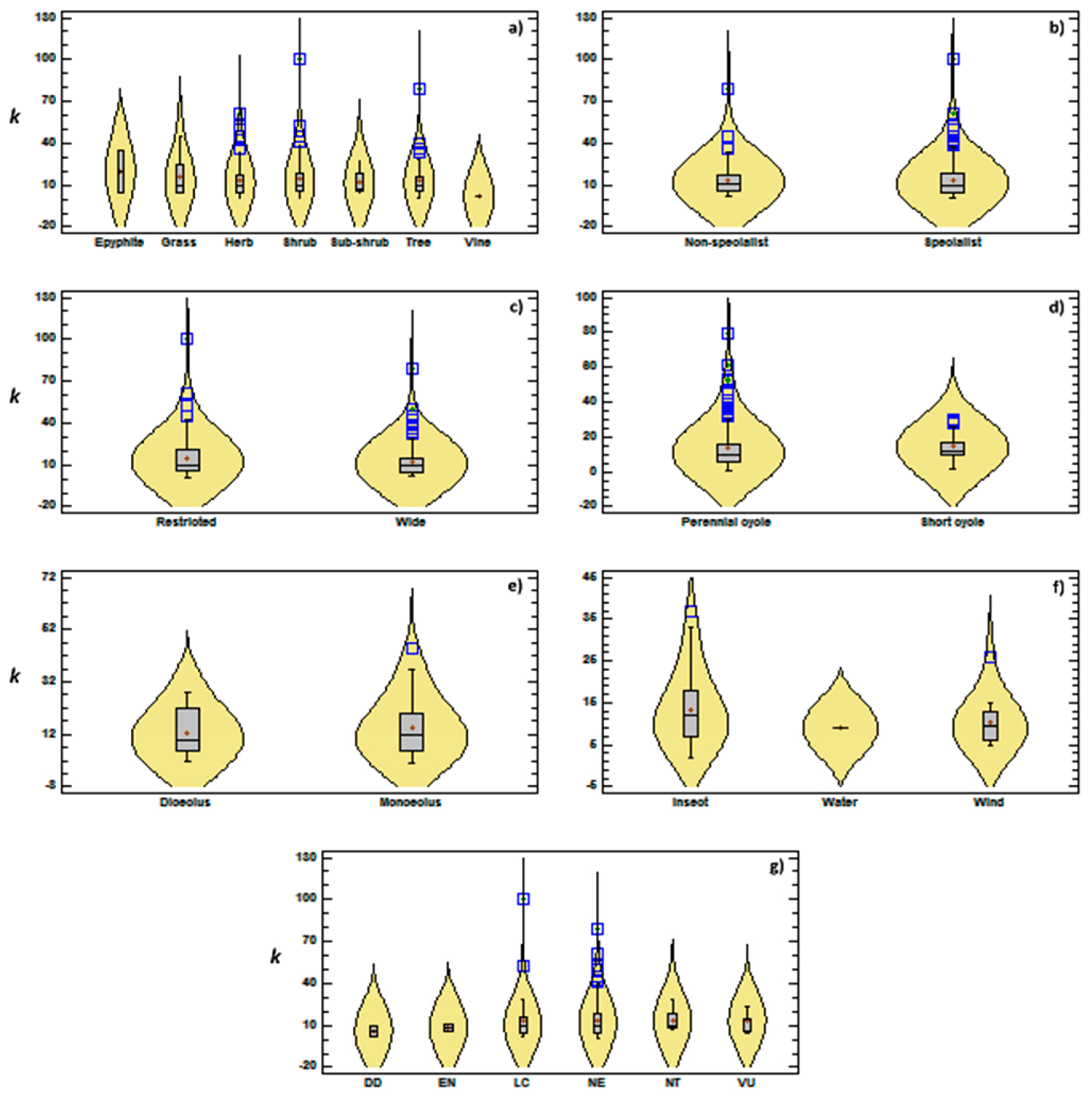
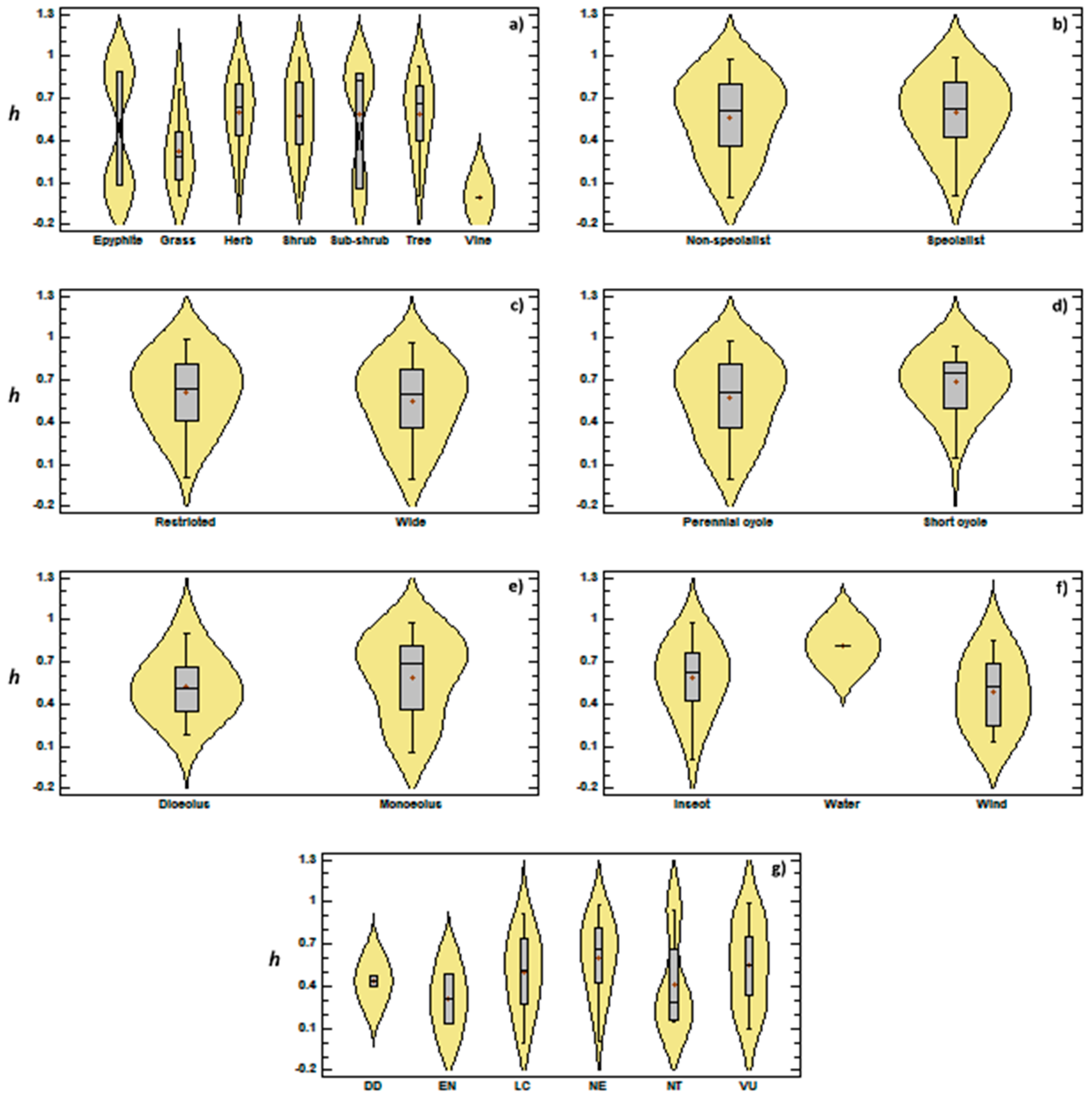
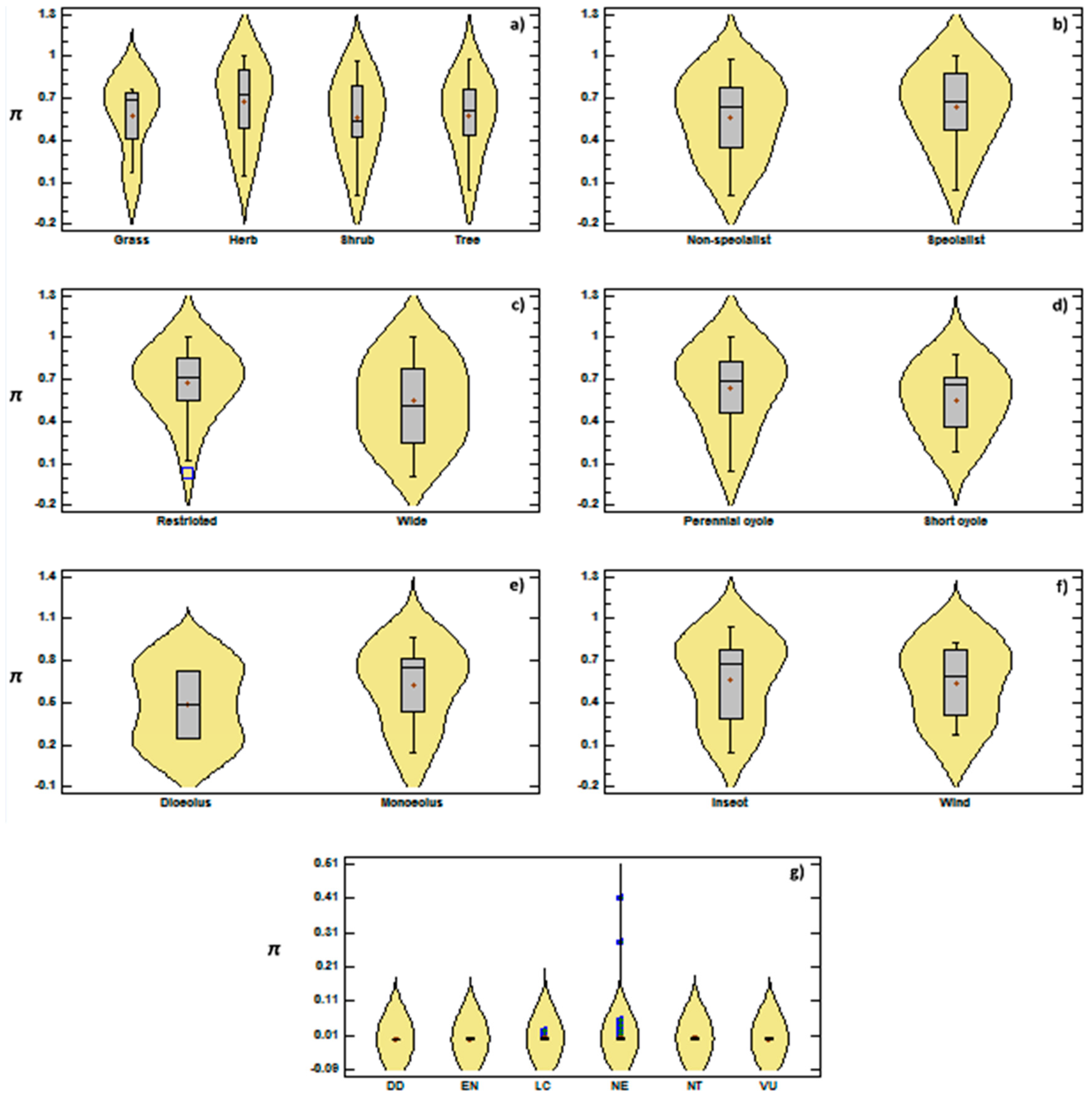
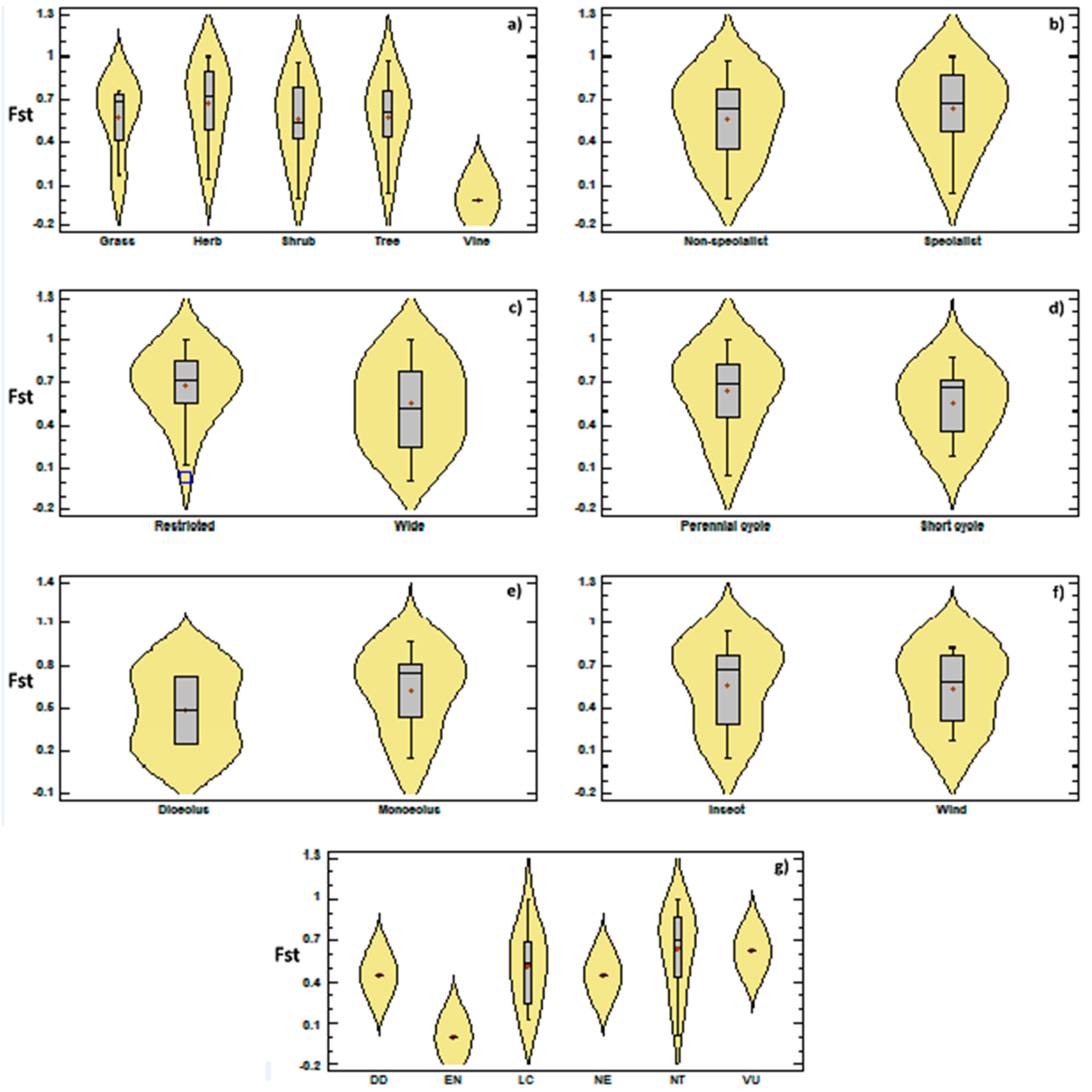
3.2. Genetic Diversity vs. Phylogeny, and Ecological and Reproductive Traits
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hughes, A.; Inouye, B.D.; Johnson, M.T.; Underwood, N.; Vellend, M. Ecological consequences of genetic diversity. Ecol. Lett. 2008, 11, 609–623. [Google Scholar] [CrossRef] [PubMed]
- Leffler, E.M.; Bullaughey, K.; Matute, D.R.; Meyer, W.K.; Ségurel, L.; Venkat, A.; Andolfatto, P.; Przeworski, M. Revisiting an old riddle: What determines genetic diversity levels within species? PLoS Biol. 2012, 10, e1001388. [Google Scholar] [CrossRef] [PubMed]
- Tanksley, S.D. Molecular markers in plant breeding. Plant Mol. Biol. Rep. 1983, 1, 3–8. [Google Scholar] [CrossRef]
- Mohan, M.; Nair, S.; Bhagwat, A.; Krishna, T.G.; Yano, M.; Bhatia, C.R.; Sasaki, T. Genome mapping, molecular markers and marker-assisted selection in crop plants. Mol. Breed. 1997, 3, 87–103. [Google Scholar] [CrossRef]
- Collard, B.C.Y.; Mackill, D.J. Marker-assisted selection: An approach for precision plant breeding in the twenty-first century. Philos. Trans. R. Soc. B 2007, 363, 557–572. [Google Scholar] [CrossRef]
- Grover, A.; Sharma, P.C. Development and use of molecular markers: Past and present. Crit. Rev. Biotechnol. 2016, 36, 290–302. [Google Scholar] [CrossRef]
- Nogueira, A.M.; Ferreira, A.; Ferreira, M.F.S. Transferability of Microsatellites from Psidium guajava to Eugenia, Myrciaria, Campomanesia, and Syzygium Species (Myrtaceae). Plant Mol. Biol. Rep. 2016, 34, 249–256. [Google Scholar] [CrossRef]
- Erayman, M.; Ilhan, E.; Güzel, Y.; Eren, A. Transferability of SSR markers from distantly related legumes to Glycyrrhiza species. Turk. J. Agric. For. 2014, 38, 32–38. [Google Scholar] [CrossRef]
- Miranda, E.A.G.C.; Boaventura-Novaes, C.R.D.; Braga, R.S.; Reis, E.F.; Pinto, J.F.N.; Telles, M.P.C. Validation of EST-derived microsatellite markers for two Cerrado-endemic Campomanesia (Myrtaceae) species. Genet. Mol. Res. 2016, 15, 16. [Google Scholar] [CrossRef] [PubMed]
- De Paula, L.F.A.; Leal, B.S.S.; Rexroth, J.; Porembski, S.; Palma-Silva, C. Transferability of microsatellite loci to Vellozia plicata (Velloziaceae), a widespread species on Brazilian inselbergs. Braz. J. Bot. 2017, 40, 1071–1075. [Google Scholar] [CrossRef]
- Camacho, L.M.D.; Schatzer, C.A.F.; Alves-Pereira, A.; Zucchi, M.I.; Carvalho, M.A.M.; Gaspar, M. Development, characterization and cross-amplification of microsatellite markers for Chrysolaena obovata, an important Asteraceae from Brazilian Cerrado. J. Genet. 2017, 96, 47–53. [Google Scholar] [CrossRef]
- Wright, S. The interpretation of population structure by F-statistics with special regard to systems of Mating. Evolution 1965, 19, 395–420. [Google Scholar] [CrossRef]
- Wright, S. Evolution and the Genetics of Populations. The Theory of Gene Frequencies. v.2; University of Chicago Press: Chicago, IL, USA, 1969; p. 512. [Google Scholar]
- Diniz-Filho, J.A.F.; Telles, M.P.C.; Bonatto, S.L.; Eizirik, E.; Freitas, T.R.O.; Marco Junior, P.; Santos, F.R.; Sole-Cava, A.; Soares, T.N. Mapping the evolutionary twiligh zone: Molecular markers, populations and geography. J. Biogeogr. 2008, 35, 753–763. [Google Scholar] [CrossRef]
- Gao, Y.; Yin, S.; Yang, H.; Wu, L.; Yan, Y. Genetic diversity and Phylgenetic relationships of seven Amorphophallus species in southwestern China revealed by chloroplast DNA sequences. Mitochondrial DNA 2018, 29, 679–686. [Google Scholar] [CrossRef]
- Resende-Moreira, L.C.; Vasconcelos, P.N.; Souto, A.P.; Menezes, A.P.A.; Lemos-Filho, J.P.; Lovato, M.B. East-west divergence in central Brazilian Cerrado revealed by cpDNA sequences of a bird-dispersed tree species. Biochem. Syst. Ecol. 2017, 70, 247–253. [Google Scholar] [CrossRef]
- Zhao, X.L.; Gao, X.F.; Zhu, Z.M.; Gao, Y.D.; Xu, B. The demographic response of a deciduous shrub (the Indigofera bungeana complex, Fabaceae) to the Pleistocene climate changes in East Asia. Sci. Rep. 2017, 7, 697. [Google Scholar] [CrossRef]
- Hartl, D.L.; Clark, A.G. Princípios de Genética de Populações, 4th ed.; Artmed Editora: Porto Alegre, Brazil, 2010. [Google Scholar]
- Guo, W.; Ng, W.L.; Wu, H.; Li, W.; Zhang, L.; Qiao, S.; Yang, X.; Shi, X.; Huang, Y. Chloroplast Phylgeography of a widely distributed mangrove species, Excoecaria agallocha, in the Indo-West Pacific region. Hydrobiologia 2017, 807, 333–347. [Google Scholar] [CrossRef]
- Qiong, L.A.; Zhang, W.; Wang Hao Zeng, L.; Birks, H.J.B.; Zhong, Y. Testing the effect of the Himalayan mountains as a physical barrier to gene flow in Hippophae tibetana Schlect. (Elaeagnaceae). PLoS ONE 2017, 12, e0172948. [Google Scholar] [CrossRef]
- Feng, X.; Liu, J.; Chiang, Y.C.; Gong, X. Investigating the genetic diversity, population differentiation and population dynamics of Cycas segmentifida (Cycadaceae) endemic to Southwest China by multiple molecular markers. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef]
- Wei, S.; Yang, W.; Wang, X.; Hou, Y. High genetic diversity in an endangered medicinal plant, Saussurea involucrata (Saussurea, Asteraceae), in western Tianshan Mountains, China. Conserv. Genet. 2017, 18, 1435–1447. [Google Scholar] [CrossRef]
- Chang, Y.-J.; Cao, Y.-F.; Zhang, J.-M.; Tian, L.-M.; Dong, X.-G.; Zhang, Y.; Qi, D.; Zhang, X.-S. Study on chloroplast DNA diversity of cultivated and wild pears (Pyrus L.) in Northern China. Tree Genet. Genomes 2017, 13, 44. [Google Scholar] [CrossRef]
- Goodwin, S.; McPherson, J.D.; McCombie, W.R. Coming of age: Ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016, 17, 333. [Google Scholar] [CrossRef]
- Van Dijk El Auger, H.; Jaszczyszyn, Y.; Thermes, C. Ten years of next-generation sequencing technology. Trends Genet. 2014, 30, 418–426. [Google Scholar] [CrossRef] [PubMed]
- Souza, U.J.B.D.; Telles, M.P.D.C.; Diniz-Filho, J.A.F. Tendências da literatura científica sobre genética de populações de plantas do Cerrado. Hoehnea 2016, 43, 461–477. [Google Scholar] [CrossRef][Green Version]
- Aguilar, R.; Quesada, M.; Ashworth, L.; Herrerias-Diego, Y.; Lobo, J. Genetic consequences of habitat fragmentation in plant populations: Susceptible signals in plant traits and methodological approaches. Mol. Ecol. 2008, 17, 5177–5188. [Google Scholar] [CrossRef] [PubMed]
- Ballesteros-Mejia, L.; Lima, N.E.; Lima-Ribeiro, M.S.; Collevatti, R.G. Pollination mode and mating system explain patterns in genetic differentiation in neotropical plants. PLoS ONE 2016, 11, e0158660. [Google Scholar] [CrossRef]
- Hamrick, J.L.; Godt, M.J.W. Allozyme divcrsity in plant species. Plant Population Genetics, Breeding, and Genetic Resources. Sinauer, Sunder-land, MA Jolls, Cl & Chenier, TC. Gynodioccy in Silene vulgaris (Caryophyllaceae): Progeny success, experimental design, and maternal effects. Am. J. Bot. 1989, 76, 1360–1367. [Google Scholar]
- Hamrick, J.L.; Godt, M.J.W. Effects of life history traits on genetic diversity in plant species. Philos. Trans. R. Soc. Lond. B 1996, 351, 1291–1298. [Google Scholar] [CrossRef]
- Aguinagalde, I.; Hampe, A.; Mohanty, A.; Martín, J.P.; Duminil, J.; Petit, R.J. Effects of life-history traits and species distribution on genetic structure at maternally inherited markers in European trees and shrubs. J. Biogeogr. 2005, 32, 329–339. [Google Scholar] [CrossRef]
- Duminil, J.; Fineschi, S.; Hampe, A.; Jordano, P.; Salvini, D.; Vendramin, G.G.; Petit, R.J. Can population genetic structure be predicted from life-history traits? Am. Nat. 2007, 169, 662–672. [Google Scholar] [CrossRef]
- Garland, T., Jr. Rate tests for phenotypic evolution using phylogenetically independent contrasts. Am. Nat. 1992, 140, 509–519. [Google Scholar] [CrossRef] [PubMed]
- Garland, T., Jr.; Harvey, P.H.; Ives, A.R. Procedures for the analysis of comparative data using phylogenetically independent contrasts. Syst. Biol. 1992, 41, 18–32. [Google Scholar] [CrossRef]
- IUCN [Internet]. The IUCN Red List of Threatened Species. Version 2017-3. 2018. Available online: http://www.iucnredlist.org (accessed on 12 July 2018).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017; Available online: https://www.R-project.org/ (accessed on 31 January 2018).
- APG—Angiosperm Phylogeny Group IV. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linean Soc. 2016, 181, 1–20. [Google Scholar] [CrossRef]
- Gastauer, M.; Meira-Neto, J.A.A. Updated angiosperm family tree for analyzing phylogenetic diversity and community structure. Acta Bot. Bras. 2017, 31, 191–198. [Google Scholar] [CrossRef]
- Ackerly, D.D. Taxon sampling, correlated evolution, and independent contrasts. Evolution 2000, 54, 1480–1492. [Google Scholar] [CrossRef]
- Guénard, G.; Legendre, P.; Peres-Neto, P. Phylogenetic eigenvector maps: A framework to model and predict species traits. Methods Ecol. Evol. 2013, 4, 1120–1131. [Google Scholar] [CrossRef]
- Guénard, G. A Phylogenetic Modelling Tutorial Using Phylogenetic Eigenvector Maps (PEM) as Implemented in R Package MPSEM (0:3-2). 2017. Available online: https://cran.r-project.org/web/packages/MPSEM/vignettes/MPSEM.pdf (accessed on 15 November 2017).
- Jakob, S.S.; Blattner, F.R. A chloroplast genealogy of Hordeum (Poaceae): Long-term persisting haplotypes, incomplete lineage sorting, regional extinction, and the consequences for Phylgenetic inference. Mol. Biol. Evol. 2006, 23, 1602–1612. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Abbott, R.J.; Zheng, W.E.I.; Chen, P.; Wang, Y.; Liu, J. History and evolution of alpine plants endemic to the Qinghai-Tibetan Plateau: Aconitum gymnandrum (Ranunculaceae). Mol. Ecol. 2009, 18, 709–721. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Chiang, T.Y.; George, M.; Liu, J.Q.; Abbott, R.J. Phylgeography of the Qinghai-Tibetan Plateau endemic Juniperus przewalskii (Cupressaceae) inferred from chloroplast DNA sequence variation. Mol. Ecol. 2005, 14, 3513–3524. [Google Scholar] [CrossRef] [PubMed]
- Anderson, L.L.; Hu, F.S.; Nelson, D.M.; Petit, R.J.; Paige, K.N. Ice-age endurance: DNA evidence of a white spruce refugium in Alaska. Proc. Natl. Acad. Sci. USA 2006, 103, 12447–12450. [Google Scholar] [CrossRef]
- Mullis, K.B. The unusual origin of the polymerase chain reaction. Sci. Am. 1990, 262, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Tarbelet, P.; Gielly, L.; Bouvet, J. Universal primers for amplification of three noncoding regions of chloroplast DNA. Plant Mol. Biol. 1991, 17, 1105–1109. [Google Scholar] [CrossRef]
- Demesure, B.; Sodzi, N.; Petit, R.J. A set of universal primers for amplification of polymorphic non-coding regions of mitochondrial and chloroplast DNA in plants. Mol. Ecol. 1995, 4, 129–134. [Google Scholar] [CrossRef]
- Dumolin-Lapegue, S.; Pemonge, M.-H.; Petit, R.J. An enlarged set of consensus primers for the study of organelle DNA in plants. Mol. Ecol. 1997, 6, 393–397. [Google Scholar] [CrossRef] [PubMed]
- Schulman, A.H. Molecular markers to assess genetic diversity. Euphytica 2007, 158, 313–321. [Google Scholar] [CrossRef]
- Shaw, J.; Lickey, E.B.; Beck, J.T.; Farmer, S.B.; Liu, W.; Miller, J.; Siripun, K.C.; Winder, C.T.; Schilling, E.E.; Small, R.L. The tortoise and the hare II: Relative utility of 21 noncoding chloroplast DNA sequences for Phylgenetic analysis. Am. J. Bot. 2005, 92, 142–166. [Google Scholar] [CrossRef]
- Mardis, E.R. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008, 24, 133–141. [Google Scholar] [CrossRef]
- Shendure, J.; Ji, H. Next-generation DNA sequencing. Nat. Biotechnol. 2008, 26, 1135. [Google Scholar] [CrossRef]
- Wu, J.; Xiao, J.; Zhang, R.; Yu, J. DNA sequencing leads to genomics progress in China. Sci. China Life Sci. 2011, 54, 290–292. [Google Scholar] [CrossRef][Green Version]
- Cyranoski, D. The Sequencing Superpower. Nature 2016, 534, 462. [Google Scholar] [CrossRef]
- De Almeida, E.C.E.; Guimarães, J.A. Brazil’s growing production of scientific articles-how are we doing with review articles and other qualitative indicators? Scientometrics 2013, 97, 287–315. [Google Scholar] [CrossRef]
- Bellard, C.; Leclerc, C.; Leroy, B.; Bakkenes, M.; Veloz, S.; Thuiller, W.; Courchamp, F. Vulnerability of biodiversity hotspots to global change. Glob. Ecol. Biogeogr. 2014, 23, 1376–1386. [Google Scholar] [CrossRef]
- Miranda, R.; Garcia-Carpinteiro, E. Overcitation and overrepresentation of review papers in the most cited papers. J. Informetr. 2018, 12, 1015–1030. [Google Scholar] [CrossRef]
- Wang, J. Citation time window choice for research impact evaluation. Scientometrics 2013, 94, 851–872. [Google Scholar] [CrossRef]
- Nakamura, A.T.; Longhi-Wagner, H.M.; Scatena, V.L. Desenvolvimento de óvulo, fruto e semente de espécies de Poaceae (Poales). Rev. Bras. Bot. 2009, 165–176. [Google Scholar] [CrossRef]
- Xiang, Q.-P.; Wei, R.; Shao, Y.-Z.; Yang, Z.-Y.; Wang, X.-Q.; Zhang, X.-C. Phylgenetic relationships, possible ancient hybridization, and biogeographic history of Abies (Pinaceae) based on data from nuclear, plastid, and mitochondrial genomes. Mol. Phylgenet. Evol. 2015, 82, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Potokina, E.K.; Kiseleva, A.A.; Nikolaeva, M.A.; Ivanov, A.S.; Ulianich, O.S.; Potokin, A.F. Analysis of the polymorphism of organelle DNA to elucidate the Phylgeography of Norway spruce in the East European Plain. Russ. J. Genet. Appl. Res. 2015, 5, 430–439. [Google Scholar] [CrossRef]
- Potter, K.M.; Hipkins, V.D.; Mahalovich, M.F.; Means, R.E. Nuclear genetic variation across the range of ponderosa pine (Pinus ponderosa): Phylgeographic, taxonomic and conservation implications. Tree Genet. Genomes 2015, 11, 38. [Google Scholar] [CrossRef]
- Yang, Y.-X.; Wang, M.-L.; Zhu, J.; Yan, M.-Y.; Li, Z.-H. Nucleotide polymorphism and Phylgeographic history of an endangered conifer species Pinus bungeana. Biochem. Syst. Ecol. 2015, 64, 89–96. [Google Scholar] [CrossRef]
- Faghir, M.B.; Attar, F.; Farazmand, A.; Kazempour, S. Phylgeny of the genus Potentilla (Rosaceae) in Iran based on nrDNA ITS and cpDNA trnL-F sequences with a focus on leaf and style characters’ evolution. Turk. J. Bot. 2014, 38, 417–429. [Google Scholar] [CrossRef]
- Fu, P.-C.; Gao, Q.-B.; Zhang, F.-Q.; Xing, R.; Khan, G.; Wang, J.-L.; Liu, H.-R.; Chen, S.-L. Responses of plants to changes in Qinghai–Tibetan Plateau and glaciations: Evidence from Phylgeography of a Sibiraea (Rosaceae) complex. Biochem. Syst. Ecol. 2016, 65, 72–82. [Google Scholar] [CrossRef]
- Khan, G.; Zhang, F.; Gao, Q.; Fu, P.; Zhang, Y.; Chen, S. Spiroides shrubs on Qinghai-Tibetan Plateau: Multilocus Phylgeography and palaeodistributional reconstruction of Spiraea alpina and S. Mongolica (Rosaceae). Mol. Phylgenet. Evol. 2018, 123, 137–148. [Google Scholar] [CrossRef]
- Lamoreux, J.; Akçakaya, H.R.; Bennun, L.; Collar, N.J.; Boitani, L.; Brackett, D.; Bräutigam, A.; Brooks, T.M.; Da Fonseca, G.A.B.; Mittermeier, R.A.; et al. Value of the IUCN Red List. Trends Ecol. Evolut. 2003, 18, 214–215. [Google Scholar] [CrossRef]
- Wilson, J.B.; Peet, R.K.; Dengler, J.; Pärtel, M. Plant species richness: The world records. J. Veg. Sci. 2012, 23, 796–802. [Google Scholar] [CrossRef]
- Christenhusz, M.J.M.; Byng, J.W. The number of known plants species in the world and its annual increase. Phytotaxa 2016, 261, 201–217. [Google Scholar] [CrossRef]
- The Plant List Version 1.1. 2013. Available online: http://www.theplantlist.org/ (accessed on 24 October 2018).
- Darrah, S.E.; Bland, L.M.; Bachman, S.P.; Clubbe, C.P.; Trias-Blasi, A. Using coarse-scale species distribution data to predict extinction risk in plants. Divers. Distrib. 2017, 23, 435–447. [Google Scholar] [CrossRef]
- IUCN Standards and Petitions Subcommittee. Guidelines for Using the IUCN Red List Categories and Criteria. Version 11 (February 2014). Prepared by the Standards and Petitions Working Group of the IUCN SSC Biodiversity Assessments Subcommittee; IUCN Standards and Petitions Subcommittee. 2014. Available online: http://www.iucnredlist.org/documents/RedListGuidelines.pdf (accessed on 10 March 2014).
- Frankham, R. Genetics and extinction. Biol. Cons. 2005, 126, 131–140. [Google Scholar] [CrossRef]
- Spielman, D.; Brook, B.W.; Frankham, R. Most species are not driven to extinction before genetic factors impact them. Proc. Natl. Acad. Sci. USA 2004, 101, 15261–15264. [Google Scholar] [CrossRef]
- Falk, E.D.D.A.; Falk, D.A.; Holsinger, K.E.; Holsinger, K.E. Genetics and Conservation of Rare Plants; Oxford University Press on Demand: Oxford, UK, 1991. [Google Scholar]
- Meirmans, P.G.; Hedrick, P.W. Assessing population structure: FST and related measures. Mol. Ecol. Resour. 2010, 11, 5–18. [Google Scholar] [CrossRef]
- Harvey, P.H.; Pagel, M.D. The Comparative Method in Evolutionary Biology; Oxford University Press: Oxford, UK, 1991. [Google Scholar]
- Bennett, P.M.; Owens, I.P.F. Evolutionary Ecology of Birds: Life Histories, Mating Systems, and Extinction; Oxford University Press: Oxford, UK, 2002. [Google Scholar]
- Tanksley, S.D.; McCouch, R. Seed banks and molecular maps: Unlocking genetic potential from the wild. Science 1997, 277, 1063–1066. [Google Scholar] [CrossRef]
- Doebley, J.F.; Gaut, B.S.; Smith, B.D. The Molecular Genetics of Crop Domestication. Cell 2006, 127, 1309–1321. [Google Scholar] [CrossRef]
- Reif, J.C.; Zhang, P.; Dreisigacker, S.; Warburton, M.L.; van Ginkel, M.; Hoisington, D.; Bohn, M.; Melchinger, A.E. Wheat genetic diversity trends during domestication and breeding. Theor. Appl. Genet. 2005, 110, 859–864. [Google Scholar] [CrossRef]
- Hollingsworth, P.M.; Dawson IKGoodall-Copestake, W.P.; Richardson, J.E.; Weber, J.C.; Sotelo Montes, C.; Pennington, R.T. Do farmers reduce genetic diversity when they domesticate tropical trees? A case study from Amazonia. Mol. Ecol. 2005, 14, 497–501. [Google Scholar] [CrossRef]
- De Oliveira, F.A.; Vigna, B.B.Z.; Silva, C.C.; Favero, A.P.; Matta, F.P.; Azevedo, A.L.S.; Souza, A.P. Apomixis-related genes identified from a coexpression network in Paspalum notatum, a Neotropical grass. bioRxiv 2018, 369280. [Google Scholar] [CrossRef]
- Radhakrishna, A.; Dwivedi, K.K.; Srivastava, M.K.; Roy, A.K.; Malaviya, D.R.; Kaushal, P. Transcriptomic data of pre-meiotic stage of floret development in apomictic and sexual types of guinea grass (Panicum maximum Jacq.). Data Brief 2018, 18, 590–593. [Google Scholar] [CrossRef]
- Kaushal, P.; Dwivedi, K.K.; Radhakrishna, A.; Saxena, S.; Paul, S.; Srivastava, M.K.; Baig, M.J.; Roy, A.K.; Malaviya, D.R. Ploidy dependent expression of apomixis and its components in guinea grass (Panicum maximum Jacq.). Euphytica 2018, 214, 152. [Google Scholar] [CrossRef]
- Davidson, A.M.; Jennions, M.; Nicotra, A.B. Do invasive species show higher phenotypic plasticity than native species and, if so, is it adaptive? A meta-analysis. Ecol. Lett. 2011, 14, 419–431. [Google Scholar] [CrossRef]

| Variables | k | p | h | p | π | p | FST | p |
|---|---|---|---|---|---|---|---|---|
| Dist | 0.015 | 0.025 | 4 × 10−7 | 0.990 | 2 × 10−4 | 0.789 | 0.037 | 0.058 |
| Phyl | 0.829 | 2 × 10−16 | 0.799 | 2 × 10−16 | 0.877 | 2 × 10−16 | 0.600 | 4 × 10−8 |
| LF | 0.015 | 0.781 | 0.022 | 0.650 | 0.020 | 0.728 | 0.064 | 0.293 |
| Hab | 3 × 10−5 | 0.994 | 0.004 | 0.587 | 0.007 | 0.373 | 0.069 | 0.035 |
| IUCN | 0.006 | 0.841 | 0.011 | 0.674 | 0.011 | 0.703 | 0.011 | 0.784 |
| Dist and Phyl | 0.832 | 2 × 10−16 | 0.826 | 2 × 10−16 | 0.881 | 2 × 10−16 | 0.616 | 2 × 10−8 |
| LF and Dist | 0.028 | 0.445 | 0.022 | 0.743 | 0.020 | 0.805 | 0.127 | 0.052 |
| Hab and Dist | 0.020 | 0.092 | 0.004 | 0.7564 | 0.008 | 0.558 | 0.115 | 0.010 |
| Dist and IUCN | 0.023 | 0.283 | 0.012 | 0.783 | 0.012 | 0.790 | 0.043 | 0.389 |
| LF and Phyl | 0.878 | 2 × 10−16 | 0.861 | 2 × 10−16 | 0.899 | 2 × 10−16 | 0.668 | 1 × 10−8 |
| Hab and Phyl | 0.858 | 2 × 10−16 | 0.807 | 2 × 10−16 | 0.880 | 2 × 10−16 | 0.651 | 3 × 10−9 |
| Hab and IUCN | 0.006 | 0.955 | 0.017 | 0.714 | 0.019 | 0.658 | 0.085 | 0.145 |
| IUCN and Phyl | 0.838 | 2 × 10−16 | 0.803 | 2 × 10−16 | 0.918 | 2 × 10−16 | 0.655 | 6 × 10−9 |
| LF and IUCN | 0.020 | 0.921 | 0.034 | 0.767 | 0.031 | 0.829 | 0.072 | 0.556 |
| LF and Hab | 0.015 | 0.901 | 0.027 | 0.710 | 0.030 | 0.630 | 0.132 | 0.076 |
| Dist and IUCN and Phyl | 0.866 | 2 × 10−16 | 0.811 | 2 × 10−16 | 0.924 | 2 × 10−16 | 0.714 | 1 × 10−10 |
| Dist and IUCN and LF | 0.003 | 0.667 | 0.034 | 0.827 | 0.032 | 0.870 | 0.132 | 0.175 |
| Hab and Dist and Phyl | 0.863 | 2 × 10−16 | 0.083 | 2 × 10−16 | 0.886 | 2 × 10−16 | 0.710 | 3 × 10−11 |
| Hab and Dist and IUCN | 0.028 | 0.339 | 0.018 | 0.775 | 0.019 | 0.753 | 0.123 | 0.062 |
| IUCN and Phyl and LF | 0.894 | 2 × 10−16 | 0.864 | 2 × 10−16 | 0.942 | 2 × 10−16 | 0.695 | 1 × 10−8 |
| IUCN and Phyl and Hab | 0.866 | 2 × 10−16 | 0.811 | 2 × 10−16 | 0.924 | 2 × 10−16 | 0.714 | 1 × 10−10 |
| LF and Hab and Dist | 0.030 | 0.546 | 0.027 | 0.780 | 0.031 | 0.697 | 0.214 | 0.005 |
| Phyl and LF and Dist | 0.883 | 2 × 10−16 | 0.899 | 2 × 10−16 | 0.901 | 2 × 10−16 | 0.696 | 2 × 10−9 |
| LF and Hab and Phyl | 0.907 | 2 × 10−16 | 0.870 | 2 × 10−16 | 0.904 | 2 × 10−16 | 0.717 | 7 × 10−10 |
| LF and Hab and IUCN | 0.021 | 0.968 | 0.040 | 0.785 | 0.043 | 0.746 | 0.142 | 0.188 |
| Dist and IUCN and Hab and Phyl | 0.872 | 2 × 10−16 | 0.845 | 2 × 10−16 | 0.934 | 2 ×10−16 | 0.746 | 1 × 10−11 |
| LF and Dist and Hab and Phyl | 0.920 | 2 × 10−16 | 0.923 | 2 × 10−16 | 0.908 | 2 × 10−16 | 0.781 | 0.023 |
| LF and Dist and IUCN and Phyl | 0.901 | 2 × 10−16 | 0.906 | 2 × 10−16 | 0.945 | 2 × 10−16 | 0.709 | 1 × 10−8 |
| LF and Hab and Dist and IUCN | 0.038 | 0.731 | 0.040 | 0.826 | 0.043 | 0.799 | 0.221 | 1 × 10−12 |
| LF and IUCN and Hab and Phyl | 0.921 | 2 × 10−16 | 0.873 | 2 × 10−16 | 0.948 | 2 × 10−16 | 0.750 | 3 × 10−10 |
| Dist and IUCN and Hab and Phyl and LF | 0.936 | 2 × 10−16 | 0.931 | 2 × 10−16 | 0.956 | 2 × 10−16 | 0.789 | 9 × 10−12 |
| Residual | 0.063 | 0.069 | 0.043 | 0.059 |
| Variables | k | p | h | p | π | p | FST | p |
|---|---|---|---|---|---|---|---|---|
| LC | 0.001 | 0.777 | 0.030 | 0.324 | 0.002 | 0.736 | 0.015 | 0.483 |
| F | 0.001 | 0.814 | 0.090 | 0.046 | 1 × 10−4 | 0.986 | 0.002 | 0.895 |
| P | 0.003 | 0.785 | 5 × 10−4 | 0.852 | 0.003 | 0.665 | 0.003 | 0.830 |
| Phyl | 0.829 | 2 × 10−16 | 0.690 | 1 × 10−4 | 0.904 | 2 × 10−16 | 0.868 | 7 × 10−15 |
| LC and F | 0.002 | 0.950 | 0.112 | 0.122 | 0.002 | 0.942 | 0.017 | 0.796 |
| LC and Phyl | 0.830 | 2 × 10−16 | 0.703 | 2 × 10−4 | 0.913 | 2 × 10−16 | 0.899 | 2 × 10−16 |
| LC and P | 0.006 | 0.858 | 0.036 | 0.688 | 0.006 | 0.781 | 0.024 | 0.677 |
| F and Phyl | 0.832 | 2 × 10−16 | 0.762 | 9 × 10−6 | 0.905 | 2 × 10−16 | 0.874 | 2 × 10−14 |
| F and P | 0.009 | 0.686 | 0.099 | 0.171 | 0.003 | 0.908 | 0.012 | 0.887 |
| P and Phyl | 0.833 | 2 × 10−16 | 0.769 | 5 × 10−6 | 0.914 | 2 × 10−16 | 0.875 | 1 × 10−14 |
| LC and F and P | 0.011 | 0.834 | 0.119 | 0.266 | 0.007 | 0.935 | 0.029 | 0.840 |
| LC and F and Phyl | 0.832 | 2 × 10−16 | 0.769 | 2 × 10−5 | 0.914 | 2 × 10−16 | 0.913 | 2 × 10−16 |
| LC and P and Phyl | 0.834 | 2 × 10−16 | 0.810 | 1 × 10−6 | 0.922 | 2 × 10−16 | 0.916 | 2 × 10−16 |
| F and P and Phyl | 0.834 | 2 × 10−16 | 0.900 | 5 × 10−11 | 0.915 | 2 × 10−16 | 0.881 | 6 × 10−14 |
| LC and F and P and Phyl | 0.835 | 2 × 10−16 | 0.928 | 3 × 10−12 | 0.924 | 2 × 10−16 | 0.925 | 2 × 10−16 |
| Resíduo | 0.063 | 0.072 | 0.075 | 0.074 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carvalho, Y.G.S.; Vitorino, L.C.; Souza, U.J.B.d.; Bessa, L.A. Recent Trends in Research on the Genetic Diversity of Plants: Implications for Conservation. Diversity 2019, 11, 62. https://doi.org/10.3390/d11040062
Carvalho YGS, Vitorino LC, Souza UJBd, Bessa LA. Recent Trends in Research on the Genetic Diversity of Plants: Implications for Conservation. Diversity. 2019; 11(4):62. https://doi.org/10.3390/d11040062
Chicago/Turabian StyleCarvalho, Yasmin G. S., Luciana C. Vitorino, Ueric J. B. de Souza, and Layara A. Bessa. 2019. "Recent Trends in Research on the Genetic Diversity of Plants: Implications for Conservation" Diversity 11, no. 4: 62. https://doi.org/10.3390/d11040062
APA StyleCarvalho, Y. G. S., Vitorino, L. C., Souza, U. J. B. d., & Bessa, L. A. (2019). Recent Trends in Research on the Genetic Diversity of Plants: Implications for Conservation. Diversity, 11(4), 62. https://doi.org/10.3390/d11040062








