A Brief Review of Non-Avian Reptile Environmental DNA (eDNA), with a Case Study of Painted Turtle (Chrysemys picta) eDNA Under Field Conditions
Abstract
1. Introduction
1.1. Environmental DNA Uses and Limitations
1.2. Reptile eDNA
1.3. Painted Turtle eDNA Case Study
2. Materials and Methods
2.1. Experimental Setup and eDNA Collection
2.2. Extraction
2.3. Amplification and Quantification
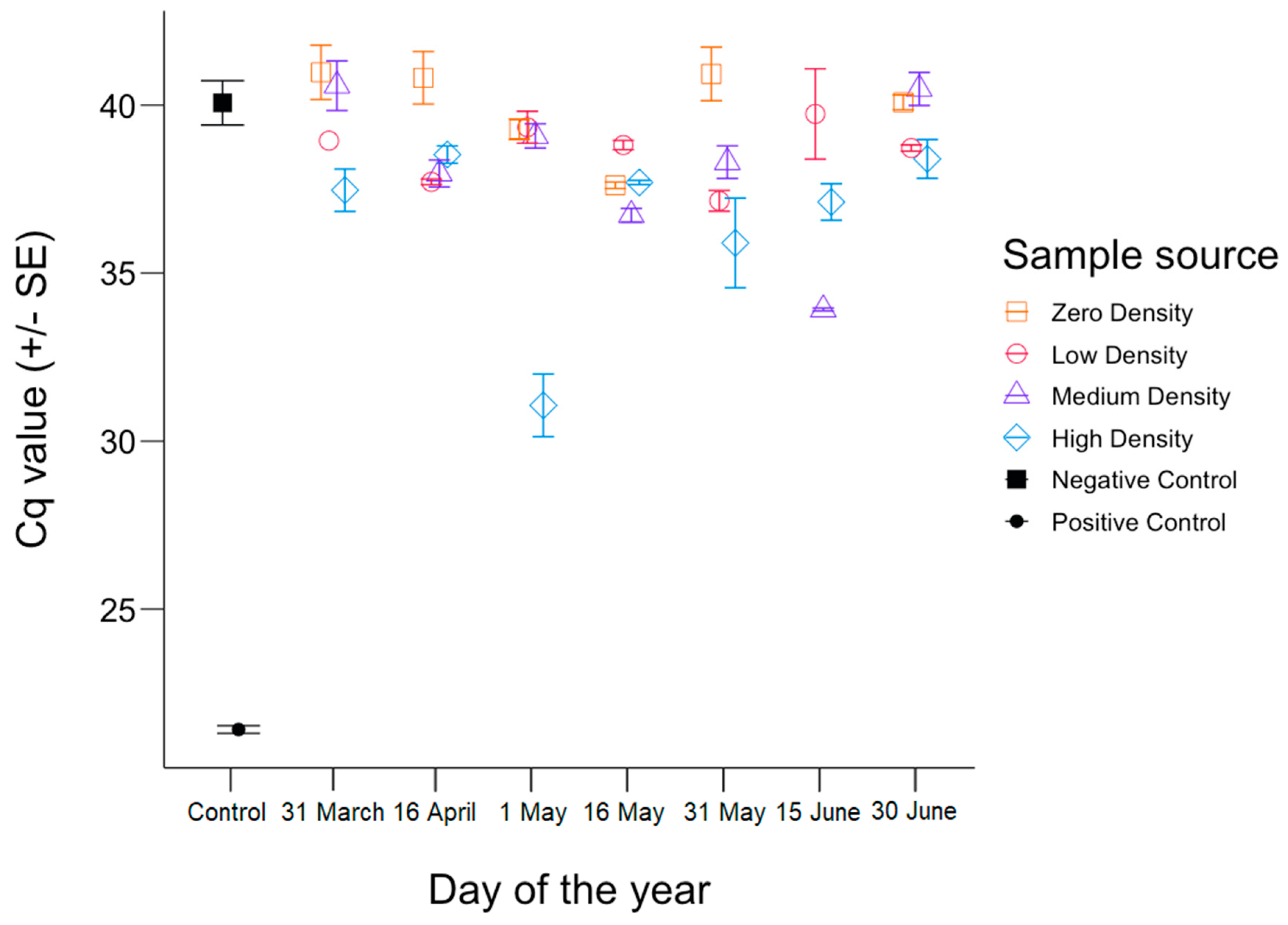
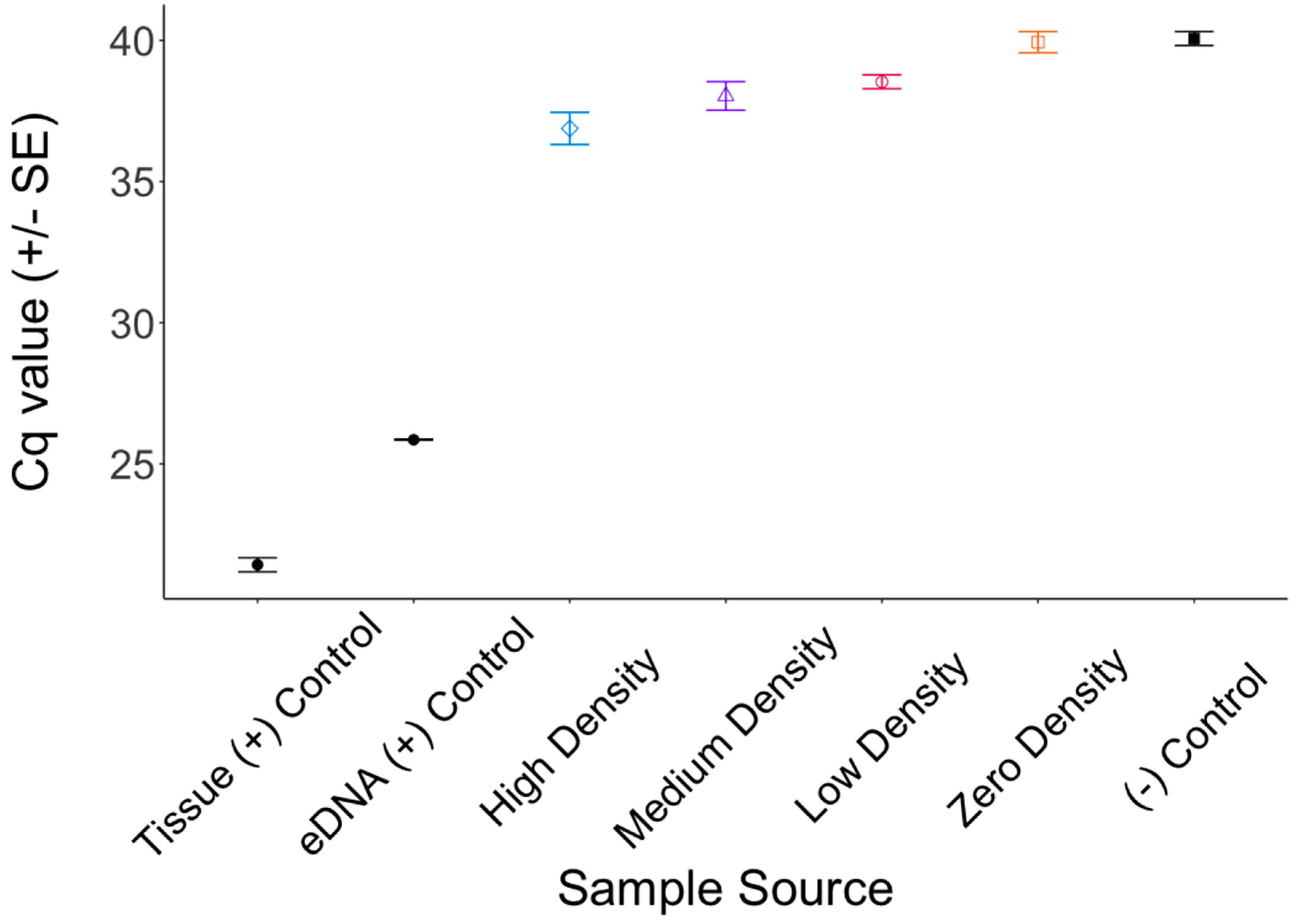
3. Results
4. Discussion
4.1. Inhibition
4.2. The Shedding Hypothesis
4.3. Best Practices
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Caswell, H. Matrix Population Models: Construction, Analysis, and Interpretation; Sinauer Associates: Sunderland, MA, USA, 1989; ISBN 0878930930. [Google Scholar]
- Wells, J.; Richmond, M. Populations, metapopulations and species populations—What are they and who should care? Wildl. Soc. Bull. 1995, 23, 458–462. [Google Scholar]
- Holt, R.D. Bringing the Hutchinsonian niche into the 21st century: Ecological and evolutionary perspectives. Proc. Natl. Acad. Sci. USA 2009, 106, 19659–19665. [Google Scholar] [CrossRef]
- Kendall, D.G. Stochastic processes and population growth. J. R. Stat. Soc. Ser. B 2018, 11, 230–264. [Google Scholar] [CrossRef]
- Sutcliffe, O.L.; Thomas, C.D.; Moss, D. Spatial synchrony and asynchrony in butterfly population dynamics. J. Anim. Ecol. 2006, 65, 85. [Google Scholar] [CrossRef]
- Saether, B.-E.; Bakke, O. Avian life history variation and contribution of demographic traits to the population growth rate. Ecology 2000, 81, 642. [Google Scholar] [CrossRef]
- Frankham, R. Genetic adaptation to captivity in species conservation programs. Mol. Ecol. 2008, 17, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Caswell, H. Matrix models and sensitivity analysis of populations classified by age and stage: A vec-permutation matrix approach. Theor. Ecol. 2012, 5, 403–417. [Google Scholar] [CrossRef]
- Marzluff, J.M.; McGowan, K.J.; Donnelly, R.; Knight, R.L. Causes and Consequences of Expanding American Crow Populations; Marzluff, J.M., Bowman, R., Donnelly, R., Eds.; Springer: Boston, MA, USA, 2001; ISBN 9978-1-4613-5600-4, 978-1-4615-1531-9. [Google Scholar]
- Mills, C.E. Jellyfish blooms: Are populations increasing globally in response to changing ocean conditions? Hydrobiologia 2001, 451, 55–68. [Google Scholar] [CrossRef]
- Dobson, F.S.; Kjelgaard, J.D. The influence of food resources on population dynamics in Columbian ground squirrels. Can. J. Zool. 1985, 63, 2095–2104. [Google Scholar] [CrossRef]
- Gargan, L.M.; Morato, T.; Pham, C.K.; Finarelli, J.A.; Carlsson, J.E.L.; Carlsson, J. Development of a sensitive detection method to survey pelagic biodiversity using eDNA and quantitative PCR: A case study of devil ray at seamounts. Mar. Biol. 2017, 164, 112. [Google Scholar] [CrossRef]
- Tucker, A.J.; Chadderton, W.L.; Jerde, C.L.; Renshaw, M.A.; Uy, K.; Gantz, C.; Mahon, A.R.; Bowen, A.; Strakosh, T.; Bossenbroek, J.M.; et al. A sensitive environmental DNA (eDNA) assay leads to new insights on Ruffe (Gymnocephalus cernua) spread in North America. Biol. Invasions 2016, 18, 3205–3222. [Google Scholar] [CrossRef]
- Cilleros, K.; Valentini, A.; Allard, L.; Dejean, T.; Etienne, R.; Grenouillet, G.; Iribar, A.; Taberlet, P.; Vigouroux, R.; Brosse, S. Unlocking biodiversity and conservation studies in high-diversity environments using environmental DNA (eDNA): A test with Guianese freshwater fishes. Mol. Ecol. Resour. 2019, 19, 27–46. [Google Scholar] [CrossRef] [PubMed]
- Ficetola, G.F.; Miaud, C.; Pompanon, F.O.; Taberlet, P. Species detection using environmental DNA from water samples. Biol. Lett 2008, 4, 423–425. [Google Scholar] [CrossRef] [PubMed]
- Dejean, T.; Valentini, A.; Miquel, C.; Taberlet, P.; Bellemain, E.; Miaud, C. Improved detection of an alien invasive species through environmental DNA barcoding: The example of the American bullfrog Lithobates catesbeianus. J. Appl. Ecol. 2012, 49, 953–959. [Google Scholar] [CrossRef]
- Secondi, J.; Dejean, T.; Valentini, A.; Audebaud, B.; Miaud, C. Detection of a global aquatic invasive amphibian, Xenopus laevis, using environmental DNA. Amphib. Reptil. 2016, 37, 131–136. [Google Scholar] [CrossRef]
- Ardura, A.; Zaiko, A.; Martinez, J.L.; Samulioviene, A.; Semenova, A.; Garcia-Vazquez, E. eDNA and specific primers for early detection of invasive species—A case study on the bivalve Rangia cuneata, currently spreading in Europe. Mar. Environ. Res. 2015, 112, 48–55. [Google Scholar] [CrossRef]
- Carim, K.J.; Christianson, K.R.; McKelvey, K.M.; Pate, W.M.; Silver, D.B.; Johnson, B.M.; Galloway, B.T.; Young, M.K.; Schwartz, M.K. Environmental DNA marker development with sparse biological information: A case study on opossum shrimp (Mysis diluviana). PLoS ONE 2016, 11, e0161664. [Google Scholar] [CrossRef]
- Dougherty, M.M.; Larson, E.R.; Renshaw, M.A.; Gantz, C.A.; Egan, S.P.; Erickson, D.M.; Lodge, D.M. Environmental DNA (eDNA) detects the invasive rusty crayfish Orconectes rusticus at low abundances. J. Appl. Ecol. 2016, 53, 722–732. [Google Scholar] [CrossRef]
- Forsström, T.; Vasemägi, A. Can environmental DNA (eDNA) be used for detection and monitoring of introduced crab species in the Baltic Sea? Mar. Pollut. Bull. 2016, 109, 350–355. [Google Scholar] [CrossRef]
- Larson, E.R.; Renshaw, M.A.; Gantz, C.A.; Umek, J.; Chandra, S.; Lodge, D.M.; Egan, S.P. Environmental DNA (eDNA) detects the invasive crayfishes Orconectes rusticus and Pacifastacus leniusculus in large lakes of North America. Hydrobiologia 2017, 800, 173–185. [Google Scholar] [CrossRef]
- Piaggio, A.J.; Engeman, R.M.; Hopken, M.W.; Humphrey, J.S.; Keacher, K.L.; Bruce, W.E.; Avery, M.L. Detecting an elusive invasive species: A diagnostic PCR to detect Burmese python in Florida waters and an assessment of persistence of environmental DNA. Mol. Ecol. Resour. 2014, 14, 374–380. [Google Scholar] [CrossRef]
- Davy, C.M.; Kidd, A.G.; Wilson, C.C. Development and validation of environmental DNA (eDNA) markers for detection of freshwater turtles. PLoS ONE 2015, 10, e0130965. [Google Scholar] [CrossRef]
- Klymus, K.E.; Marshall, N.T.; Stepien, C.A. Environmental DNA (eDNA) metabarcoding assays to detect invasive invertebrate species in the Great Lakes. PLoS ONE 2017, 12, e0177643. [Google Scholar] [CrossRef]
- Goldberg, C.S.; Sepulveda, A.; Ray, A.; Baumgardt, J.; Waits, L.P. Environmental DNA as a new method for early detection of New Zealand mudsnails (Potamopyrgus antipodarum). Freshw. Sci. 2013, 32, 792–800. [Google Scholar] [CrossRef]
- Xia, Z.; Zhan, A.; Gao, Y.; Zhang, L.; Haffner, G.D.; MacIsaac, H.J. Early detection of a highly invasive bivalve based on environmental DNA (eDNA). Biol. Invasions 2018, 20, 437–447. [Google Scholar] [CrossRef]
- Thomsen, P.F.; Kielgast, J.; Iversen, L.L.; Wiuf, C.; Rasmussen, M.; Gilbert, M.T.P.; Orlando, L.; Willerslev, E. Monitoring endangered freshwater biodiversity using environmental DNA. Mol. Ecol. 2012, 21, 2565–2573. [Google Scholar] [CrossRef] [PubMed]
- Rees, H.C.; Baker, C.A.; Gardner, D.S.; Maddison, B.C.; Gough, K.C. The detection of great crested newts year round via environmental DNA analysis. BMC Res. Notes 2017, 10, 327. [Google Scholar] [CrossRef]
- Schmelzle, M.C.; Kinziger, A.P. Using occupancy modelling to compare environmental DNA to traditional field methods for regional-scale monitoring of an endangered aquatic species. Mol. Ecol. Resour. 2016, 16, 895–908. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, K.; Doi, H.; Tanaka, K.; Kawai, T.; Negishi, J.N. Using environmental DNA to detect an endangered crayfish Cambaroides japonicus in streams. Conserv. Genet. Resour. 2016, 8, 231–234. [Google Scholar] [CrossRef]
- Piggott, M.P. An environmental DNA assay for detecting Macquarie perch, Macquaria australasica. Conserv. Genet. Resour. 2017, 9, 257–259. [Google Scholar] [CrossRef]
- Brozio, S.; Manson, C.; Gourevitch, E.; Burns, T.J.; Greener, M.S.; Downie, J.R.; Hoskisson, P.A. Development and application of an eDNA method to detect the critically endangered Trinidad golden tree frog (Phytotriades auratus) in bromeliad Phytotelmata. PLoS ONE 2017, 12, e0170619. [Google Scholar] [CrossRef]
- Laramie, M.B.; Pilliod, D.S.; Goldberg, C.S. Characterizing the distribution of an endangered salmonid using environmental DNA analysis. Biol. Conserv. 2015, 183, 29–37. [Google Scholar] [CrossRef]
- Olson, Z.H.; Briggler, J.T.; Williams, R.N. An eDNA approach to detect eastern hellbenders (Cryptobranchus a. alleganiensis) using samples of water. Wildl. Res. 2012, 39, 629–636. [Google Scholar] [CrossRef]
- Parsons, K.M.; Everett, M.; Dahlheim, M.; Park, L. Water, water everywhere: Environmental DNA can unlock population structure in elusive marine species. R. Soc. Open Sci. 2018, 5, 180537. [Google Scholar] [CrossRef]
- Adams, C.I.M.; Knapp, M.; Gemmell, N.J.; Jeunen, G.-J.; Bunce, M.; Lamare, M.D.; Taylor, H.R. Beyond biodiversity: Can environmental DNA (eDNA) cut it as a population genetics tool? Genes 2019, 10, 192. [Google Scholar] [CrossRef]
- Witmer, G.W. Wildlife population monitoring: Some practical considerations. Wildl. Res. 2005, 32, 259. [Google Scholar] [CrossRef]
- Hoffmann, C.; Schubert, G.; Calvignac-Spencer, S. Aquatic biodiversity assessment for the lazy. Mol. Ecol. 2016, 25, 846–848. [Google Scholar] [CrossRef]
- Jerde, C.L.; Mahon, A.R.; Chadderton, W.L.; Lodge, D.M. “Sight-unseen” detection of rare aquatic species using environmental DNA. Conserv. Lett. 2011, 4, 150–157. [Google Scholar] [CrossRef]
- Hinlo, R.; Furlan, E.; Suitor, L.; Gleeson, D. Environmental DNA monitoring and management of invasive fish: Comparison of eDNA and fyke netting. Manag. Biol. Invasions 2017, 8, 89–100. [Google Scholar] [CrossRef]
- Evans, N.T.; Shirey, P.D.; Wieringa, J.G.; Mahon, A.R.; Lamberti, G.A. Comparative cost and effort of fish distribution detection via environmental DNA analysis and electrofishing. Fisheries 2017, 42, 90–99. [Google Scholar] [CrossRef]
- Hebert, P.; Penton, E.H.; Burns, J.M.; Janzen, D.H.; Hallwachs, W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc. Natl. Acad. Sci. USA 2004, 101, 14812–14817. [Google Scholar] [CrossRef]
- Fletcher, L.M.; Zaiko, A.; Atalah, J.; Richter, I.; Dufour, C.M.; Pochon, X.; Wood, S.A.; Hopkins, G.A. Bilge water as a vector for the spread of marine pests: A morphological, metabarcoding and experimental assessment. Biol. Invasions 2017, 19, 2851–2867. [Google Scholar] [CrossRef]
- Ardura, A.; Zaiko, A.; Martinez, J.L.; Samuiloviene, A.; Borrell, Y.; Garcia-Vazquez, E. Environmental DNA evidence of transfer of North Sea molluscs across tropical waters through ballast water. J. Molluscan Stud. 2015, 81, 495–501. [Google Scholar] [CrossRef]
- Pielou, E.C. The measurement of diversity in different types of biological collections. J. Theor. Biol. 1966, 13, 131–144. [Google Scholar] [CrossRef]
- Eichmiller, J.J.; Bajer, P.G.; Sorensen, P.W. The relationship between the distribution of common carp and their environmental DNA in a small lake. PLoS ONE 2014, 9, e112611. [Google Scholar] [CrossRef]
- Doi, H.; Uchii, K.; Takahara, T.; Matsuhashi, S.; Yamanaka, H.; Minamoto, T. Use of droplet digital PCR for estimation of fish abundance and biomass in environmental DNA surveys. PLoS ONE 2015, 10, e0122763. [Google Scholar] [CrossRef] [PubMed]
- Uthicke, S.; Lamare, M.; Doyle, J.R. eDNA detection of corallivorous seastar (Acanthaster cf. solaris) outbreaks on the Great Barrier Reef using digital droplet PCR. Coral Reefs 2018, 37, 1229–1239. [Google Scholar] [CrossRef]
- Sassoubre, L.M.; Yamahara, K.M.; Gardner, L.D.; Block, B.A.; Boehm, A.B. Quantification of environmental DNA (eDNA) shedding and decay rates for three marine fish. Environ. Sci. Technol. 2016, 50, 10456–10464. [Google Scholar] [CrossRef] [PubMed]
- Iversen, L.L.; Kielgast, J.; Sand-Jensen, K. Monitoring of animal abundance by environmental DNA—An increasingly obscure perspective: A reply to Klymus et al., 2015. Biol. Conserv. 2015, 192, 479–482. [Google Scholar] [CrossRef]
- Lacoursière-Roussel, A.; Rosabal, M.; Bernatchez, L. Estimating fish abundance and biomass from eDNA concentrations: Variability among capture methods and environmental conditions. Mol. Ecol. Resour. 2016, 16, 1401–1414. [Google Scholar] [CrossRef] [PubMed]
- Bista, I.; Carvalho, G.R.; Tang, M.; Walsh, K.; Zhou, X.; Hajibabaei, M.; Shokralla, S.; Seymour, M.; Bradley, D.; Liu, S.; et al. Performance of amplicon and shotgun sequencing for accurate biomass estimation in invertebrate community samples. Mol. Ecol. Resour. 2018, 18, 1020–1034. [Google Scholar] [CrossRef] [PubMed]
- Klymus, K.E.; Richter, C.A.; Chapman, D.C.; Paukert, C. Quantification of eDNA shedding rates from invasive bighead carp Hypophthalmichthys nobilis and silver carp Hypophthalmichthys molitrix. Biol. Conserv. 2015, 183, 77–84. [Google Scholar] [CrossRef]
- Stewart, K.A. Understanding the effects of biotic and abiotic factors on sources of aquatic environmental DNA. Biodivers. Conserv. 2019, 28, 1–19. [Google Scholar] [CrossRef]
- Seymour, M.; Durance, I.; Cosby, B.J.; Ransom-Jones, E.; Deiner, K.; Ormerod, S.J.; Colbourne, J.K.; Wilgar, G.; Carvalho, G.R.; de Bruyn, M.; et al. Acidity promotes degradation of multi-species environmental DNA in lotic mesocosms. Commun. Biol. 2018, 1, 4. [Google Scholar] [CrossRef]
- Collins, R.A.; Wangensteen, O.S.; O’Gorman, E.J.; Mariani, S.; Sims, D.W.; Genner, M.J. Persistence of environmental DNA in marine systems. Commun. Biol. 2018, 1, 185. [Google Scholar] [CrossRef]
- Takahara, T.; Minamoto, T.; Doi, H. Effects of sample processing on the detection rate of environmental DNA from the Common Carp (Cyprinus carpio). Biol. Conserv. 2015, 183, 64–69. [Google Scholar] [CrossRef]
- Wilcox, T.M.; McKelvey, K.S.; Young, M.K.; Sepulveda, A.J.; Shepard, B.B.; Jane, S.F.; Whiteley, A.R.; Lowe, W.H.; Schwartz, M.K. Understanding environmental DNA detection probabilities: A case study using a stream-dwelling char Salvelinus fontinalis. Biol. Conserv. 2016, 194, 209–216. [Google Scholar] [CrossRef]
- Darling, J.A.; Mahon, A.R. From molecules to management: Adopting DNA-based methods for monitoring biological invasions in aquatic environments. Environ. Res. 2011, 111, 978–988. [Google Scholar] [CrossRef] [PubMed]
- Ficetola, G.F.; Pansu, J.; Bonin, A.; Coissac, E.; Giguet-Covex, C.; De Barba, M.; Gielly, L.; Lopes, C.M.; Boyer, F.; Pompanon, F.; et al. Replication levels, false presences and the estimation of the presence/absence from eDNA metabarcoding data. Mol. Ecol. Resour. 2015, 15, 543–556. [Google Scholar] [CrossRef] [PubMed]
- Lahoz-Monfort, J.J.; Guillera-Arroita, G.; Tingley, R. Statistical approaches to account for false-positive errors in environmental DNA samples. Mol. Ecol. Resour. 2016, 16, 673–685. [Google Scholar] [CrossRef]
- Muha, T.P.; Rodríguez-Rey, M.; Rolla, M.; Tricarico, E. Using Environmental DNA to improve species distribution models for freshwater invaders. Front. Ecol. Evol. 2017, 5, 158. [Google Scholar] [CrossRef]
- Willoughby, J.R.; Wijayawardena, B.K.; Sundaram, M.; Swihart, R.K.; DeWoody, J.A. The importance of including imperfect detection models in eDNA experimental design. Mol. Ecol. Resour. 2016, 16, 837–844. [Google Scholar] [CrossRef]
- Mackenzie, D.I.; Nichols, J.D.; Hines, J.E.; Knutson, M.G.; Franklin, A.B.; Knu, M.G.; Franklin, A.B. Estimating site occupancy, colonization, and local extinction when a species is detected imperfectly. Ecology 2003, 84, 2200–2207. [Google Scholar] [CrossRef]
- Hunter, M.E.; Oyler-McCance, S.J.; Dorazio, R.M.; Fike, J.A.; Smith, B.J.; Hunter, C.T.; Reed, R.N.; Hart, K.M. Environmental DNA (eDNA) sampling improves occurrence and detection estimates of invasive Burmese pythons. PLoS ONE 2015, 10, e0121655. [Google Scholar] [CrossRef] [PubMed]
- Böhm, M.; Collen, B.; Baillie, J.E.M.; Bowles, P.; Chanson, J.; Cox, N.; Hammerson, G.; Hoffmann, M.; Livingstone, S.R.; et al. The conservation status of the world’s reptiles. Biol. Conserv. 2013, 157, 372–385. [Google Scholar] [CrossRef]
- Van Dijk, P.P.; Iverson, J.; Rhodin, A.; Shaffer, B.; Bour, R. Turtles of the World, 7th Edition: Annotated checklist of taxonomy, synonymy, distribution with maps, and conservation status. Chelonian Res. Monogr. 2014, 5, 329–479. [Google Scholar] [CrossRef]
- Egeter, B.; Peixoto, S.; Brito, J.C.; Jarman, S.; Puppo, P.; Velo-Antón, G. Challenges for assessing vertebrate diversity in turbid Saharan water-bodies using environmental DNA. Genome 2018, 61, 807–814. [Google Scholar] [CrossRef]
- Lacoursière-Roussel, A.; Dubois, Y.; Normandeau, E.; Bernatchez, L.; Adamowicz, S. Improving herpetological surveys in eastern North America using the environmental DNA method. Genome 2016, 59, 991–1007. [Google Scholar] [CrossRef]
- Hart, K.M.; Cherkiss, M.S.; Smith, B.J.; Mazzotti, F.J.; Fujisaki, I.; Snow, R.W.; Dorcas, M.E. Home range, habitat use, and movement patterns of non-native Burmese pythons in Everglades National Park, Florida, USA. Anim. Biotelemetry 2015, 3, 8. [Google Scholar] [CrossRef]
- Kucherenko, A.; Herman, J.E.; III, E.M.E.; Urakawa, H. Terrestrial snake environmental DNA accumulation and degradation dynamics and its environmental application. Herpetologica 2018, 74, 38–49. [Google Scholar] [CrossRef]
- Baker, S.J.; Niemiller, M.L.; Stites, A.J.; Ash, K.T.; Davis, M.A.; Dreslik, M.J.; Phillips, C.A. Evaluation of environmental DNA to detect Sistrurus catenatus and Ophidiomyces ophiodiicola in crayfish burrows. Conserv. Genet. Resour. 2018, 1–3. [Google Scholar] [CrossRef]
- Halstead, B.J.; Wood, D.A.; Bowen, L.; Waters, S.C.; Vandergast, A.G.; Ersan, J.S.; Skalos, S.M.; Casazza, M.L. An Evaluation of the Efficacy of Using Environmental DNA (eDNA) to Detect Giant Gartersnakes (Thamnophis gigas); USGS Open-File Reports; U.S. Geological Survey: Reston, VA, USA, 2017; 41p.
- Halstead, B.J.; Wylie, G.D.; Casazza, M.L. Habitat suitability and conservation of the Giant Gartersnake (Thamnophis gigas) in the Sacramento Valley of California. Copeia 2010, 2010, 591–599. [Google Scholar] [CrossRef]
- Cannon, M.V.; Hester, J.; Shalkhauser, A.; Chan, E.R.; Logue, K.; Small, S.T.; Serre, D. In silico assessment of primers for eDNA studies using PrimerTree and application to characterize the biodiversity surrounding the Cuyahoga River. Sci. Rep. 2016, 6, 22908. [Google Scholar] [CrossRef]
- de Souza, L.S.; Godwin, J.C.; Renshaw, M.A.; Larson, E. Environmental DNA (eDNA) detection probability is influenced by seasonal activity of organisms. PLoS ONE 2016, 11, e0165273. [Google Scholar] [CrossRef]
- Feist, S.M.; Jones, R.L.; Copley, J.L.; Pearson, L.S.; Berry, G.A.; Qualls, C.P. Development and validation of an environmental DNA method for detection of the Alligator Snapping Turtle (Macrochelys temminckii). Chelonian Conserv. Biol. 2018, 17, 271–279. [Google Scholar] [CrossRef]
- Kelly, R.P.; Port, J.A.; Yamahara, K.M.; Crowder, L.B. Using environmental DNA to census marine fishes in a large mesocosm. PLoS ONE 2014, 9, e86175. [Google Scholar] [CrossRef]
- Kundu, S.; Kumar, V.; Tyagi, K.; Chandra, K. Environmental DNA (eDNA) testing for detection of freshwater turtles in a temple pond. Herpetol. Notes 2018, 11, 369–371. [Google Scholar]
- Raemy, M.; Ursenbacher, S. Detection of the European pond turtle (Emys orbicularis) by environmental DNA: Is eDNA adequate for reptiles? Amphibia-Reptilia 2018, 39, 135–143. [Google Scholar] [CrossRef]
- Wilson, J.-J.; Sing, K.-W.; Chen, P.-N.; Zieritz, A. Tracking the southern river terrapin (Batagur affinis) through environmental DNA: Prospects and challenges. Mitochondrial DNA Part A 2017, 29, 862–866. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, J.F.M.; Coelho, M.T.P.; Varela, S.; Diniz-Filho, J.A.F. Invasion risk of the pond slider turtle is underestimated when niche expansion occurs. Freshw. Biol. 2016, 61, 1119–1127. [Google Scholar] [CrossRef]
- Fonseca, V.G. Pitfalls in relative abundance estimation using eDNA metabarcoding. Mol. Ecol. Resour. 2018, 18, 923–926. [Google Scholar] [CrossRef]
- Ernst, C.H.; Lovich, J.E. Turtles of the United States and Canada, 2nd ed.; JHU Press: Baltimore, MD, USA, 2009; ISBN 0801891213. [Google Scholar]
- Iverson, J.B. Biomass in turtle populations: A neglected subject. Oecologia 1982, 55, 69–76. [Google Scholar] [CrossRef]
- Congdon, J.D.; Greene, J.L.; Gibbons, J.W. Biomass of freshwater turtles: A geographic comparison. Am. Midl. Nat. 1986, 115, 165–173. [Google Scholar] [CrossRef]
- Ernst, C.H. Population dynamics and activity cycles of Chrysemys picta in southeastern Pennsylvania. J. Herpetol. 1971, 5, 151–160. [Google Scholar] [CrossRef]
- Dunker, K.J.; Sepulveda, A.J.; Massengill, R.L.; Olsen, J.B.; Russ, O.L.; Wenburg, J.K.; Antonovich, A. Potential of Environmental DNA to Evaluate Northern Pike (Esox lucius) Eradication Efforts: An Experimental Test and Case Study. PLoS ONE 2016, 11, e0162277. [Google Scholar] [CrossRef] [PubMed]
- Tsuji, S.; Yamanaka, H.; Minamoto, T. Effects of water pH and proteinase K treatment on the yield of environmental DNA from water samples. Limnology 2017, 18, 1–7. [Google Scholar] [CrossRef]
- Deiner, K.; Walser, J.C.; Mächler, E.; Altermatt, F. Choice of capture and extraction methods affect detection of freshwater biodiversity from environmental DNA. Biol. Conserv. 2015, 183, 53–63. [Google Scholar] [CrossRef]
- Taberlet, P.; Griffin, S.; Goossens, B.; Questiau, S.; Manceau, V.; Escaravage, N.; Waits, L.P.; Bouvet, J. Reliable genotyping of samples with very low DNA quantities using PCR. Nucleic Acids Res. 1996, 24, 3189–3194. [Google Scholar] [CrossRef]
- Olds, B.P.; Jerde, C.L.; Renshaw, M.A.; Li, Y.; Evans, N.T.; Turner, C.R.; Deiner, K.; Mahon, A.R.; Brueseke, M.A.; Shirey, P.D.; et al. Estimating species richness using environmental DNA. Ecol. Evol. 2016, 6, 4214–4226. [Google Scholar] [CrossRef]
- Sow, F.B.; Gallup, J.M.; Sacco, R.E.; Ackermann, M.R. Laser capture microdissection revisited as a tool for transcriptomic analysis: Application of an excel-based qPCR preparation software (PREXCEL-Q). Int. J. Biomed. Sci. 2009, 5, 105–124. [Google Scholar] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- McKee, A.M.; Spear, S.F.; Pierson, T.W. The effect of dilution and the use of a post-extraction nucleic acid purification column on the accuracy, precision, and inhibition of environmental DNA samples. Biol. Conserv. 2015, 183, 70–76. [Google Scholar] [CrossRef]
- Jonckheere, A.R. A distribution-free k-sample test against ordered alternatives. Biometrika 1954, 41, 133. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2013; Available online: http://www.R-project.org/ (accessed on 24 February 2019).
- Mächler, E.; Deiner, K.; Spahn, F.; Altermatt, F. Fishing in the water: Effect of sampled water volume on environmental DNA-based detection of macroinvertebrates. Environ. Sci. Technol. 2016, 50, 305–312. [Google Scholar] [CrossRef]
- Furlan, E.M.; Gleeson, D.; Hardy, C.M.; Duncan, R.P. A framework for estimating the sensitivity of eDNA surveys. Mol. Ecol. Resour. 2016, 16, 641–654. [Google Scholar] [CrossRef] [PubMed]
- Nathan, L.M.; Simmons, M.; Wegleitner, B.J.; Jerde, C.L.; Mahon, A.R. Quantifying environmental DNA signals for aquatic invasive species across multiple detection platforms. Environ. Sci. Technol. 2014, 48, 12800–12806. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Paparini, A.; Monis, P.; Ryan, U. Comparison of next-generation droplet digital PCR (ddPCR) with quantitative PCR (qPCR) for enumeration of Cryptosporidium oocysts in faecal samples. Int. J. Parasitol. 2014, 44, 1105–1113. [Google Scholar] [CrossRef]
- Rački, N.; Dreo, T.; Gutierrez-Aguirre, I.; Blejec, A.; Ravnikar, M. Reverse transcriptase droplet digital PCR shows high resilience to PCR inhibitors from plant, soil and water samples. Plant Methods 2014, 10, 42. [Google Scholar] [CrossRef] [PubMed]
- Barnes, M.A.; Turner, C.R.; Jerde, C.L.; Renshaw, M.A.; Chadderton, W.L.; Lodge, D.M. Environmental conditions influence eDNA persistence in aquatic systems. Environ. Sci. Technol. 2014, 48, 1819–1827. [Google Scholar] [CrossRef] [PubMed]
- Renshaw, M.A.; Olds, B.P.; Jerde, C.L.; Mcveigh, M.M.; Lodge, D.M. The room temperature preservation of filtered environmental DNA samples and assimilation into a phenol-chloroform-isoamyl alcohol DNA extraction. Mol. Ecol. Resour. 2015, 15, 168–176. [Google Scholar] [CrossRef]
- Wegleitner, B.J.; Jerde, C.L.; Tucker, A.; Chadderton, W.L.; Mahon, A.R. Long duration, room temperature preservation of filtered eDNA samples. Conserv. Genet. Resour. 2015, 7, 789–791. [Google Scholar] [CrossRef]
- Pierson, T.W.; McKee, A.M.; Spear, S.F.; Maerz, J.C.; Camp, C.D.; Glenn, T.C. Detection of an enigmatic Plethodontid salamander using environmental DNA. Copeia 2016, 104, 78–82. [Google Scholar] [CrossRef]
- Wilson, I.G. Inhibition and facilitation of nucleic acid amplification. Appl. Environ. Microbiol. 1997, 63, 3741–3751. [Google Scholar]
- Schrader, C.; Schielke, A.; Ellerbroek, L.; Johne, R. PCR inhibitors—Occurrence, properties and removal. J. Appl. Microbiol. 2012, 113, 1014–1026. [Google Scholar] [CrossRef]
- Li, J.; Lawson Handley, L.-J.; Read, D.S.; Hänfling, B. The effect of filtration method on the efficiency of environmental DNA capture and quantification via metabarcoding. Mol. Ecol. Resour. 2018, 18, 1102–1114. [Google Scholar] [CrossRef]
- Weldon, P.J.; Demeter, B.J.; Rosscoe, R. A survey of shed skin-eating (Dermatophagy) in amphibians and reptiles. J. Herpetol. 1993, 27, 219. [Google Scholar] [CrossRef]
- Ernst, C.H. Growth of the painted turtle, Chrysemys picta, in Southeastern Pennsylvania. Herpetologica 1971, 27, 135–141. [Google Scholar]
- Parmenter, R.R. Digestive turnover rates in freshwater turtles: The influence of temperature and body size. Comp. Biochem. Physiol. Part A Physiol. 1981, 70, 235–238. [Google Scholar] [CrossRef]
- Northmore, D.; Granda, A. Ocular dimensions and schematic eyes of freshwater and sea turtles. Vis. Neurosci. 1991, 7, 627–635. [Google Scholar] [CrossRef] [PubMed]
- Sansom, B.J.; Sassoubre, L.M. Environmental DNA (eDNA) Shedding and Decay Rates to Model Freshwater Mussel eDNA Transport in a River. Environ. Sci. Technol. 2017, 51, 14244–14253. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, C.S.; Turner, C.R.; Deiner, K.; Klymus, K.E.; Thomsen, P.F.; Murphy, M.A.; Spear, S.F.; McKee, A.; Oyler-McCance, S.J.; Cornman, R.S.; et al. Critical considerations for the application of environmental DNA methods to detect aquatic species. Methods Ecol. Evol. 2016, 7, 1299–1307. [Google Scholar] [CrossRef]
- Taberlet, P.; Bonin, A.; Zinger, L.; Coissac, E. Environmental DNA: For Biodiversity Research and Monitoring; Oxford University Press: Oxford, UK, 2018; ISBN 0198767226. [Google Scholar]
- Deiner, K.; Bik, H.M.; Mächler, E.; Seymour, M.; Lacoursière-Roussel, A.; Altermatt, F.; Creer, S.; Bista, I.; Lodge, D.M.; de Vere, N.; et al. Environmental DNA metabarcoding: Transforming how we survey animal and plant communities. Mol. Ecol. 2017, 26, 5872–5895. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.K.; Meyer, M.J.; Mcphee, C.; Gaston, J.R.; Venesky, M.D.; Case, B.F. Seasonal and diel signature of eastern hellbender environmental DNA. J. Wildl. Manag. 2018, 82, 217–225. [Google Scholar] [CrossRef]
- Spear, S.F.; Groves, J.D.; Williams, L.A.; Waits, L.P. Using environmental DNA methods to improve detectability in a hellbender (Cryptobranchus alleganiensis) monitoring program. Biol. Conserv. 2015, 183, 38–45. [Google Scholar] [CrossRef]
- O’Donnell, J.L.; Kelly, R.P.; Shelton, A.O.; Samhouri, J.F.; Lowell, N.C.; Williams, G.D. Spatial distribution of environmental DNA in a nearshore marine habitat. PeerJ 2017, 5, e3044. [Google Scholar] [CrossRef]
- Deiner, K.; Altermatt, F. Transport distance of invertebrate environmental DNA in a natural river. PLoS ONE 2014, 9, e88786. [Google Scholar] [CrossRef]
- Jane, S.F.; Wilcox, T.M.; Mckelvey, K.S.; Young, M.K.; Schwartz, M.K.; Lowe, W.H.; Letcher, B.H.; Whiteley, A.R. Distance, flow and PCR inhibition: eDNA dynamics in two headwater streams. Mol. Ecol. Resour. 2015, 15, 216–227. [Google Scholar] [CrossRef]
- Nukazawa, K.; Hamasuna, Y.; Suzuki, Y. Simulating the advection and degradation of the environmental DNA of common carp along a river. Environ. Sci. Technol. 2018, 52, 10562–10570. [Google Scholar] [CrossRef]
- Wilson, C.C.; Wozney, K.M.; Smith, C.M. Recognizing false positives: Synthetic oligonucleotide controls for environmental DNA surveillance. Methods Ecol. Evol. 2015, 7, 23–29. [Google Scholar] [CrossRef]
- Wilcox, T.M.; McKelvey, K.S.; Young, M.K.; Jane, S.F.; Lowe, W.H.; Whiteley, A.R.; Schwartz, M.K. Robust detection of rare species using environmental DNA: The importance of primer specificity. PLoS ONE 2013, 8, e59520. [Google Scholar] [CrossRef] [PubMed]
- Turner, C.R.; Barnes, M.A.; Xu, C.C.Y.; Jones, S.E.; Jerde, C.L.; Lodge, D.M. Particle size distribution and optimal capture of aqueous macrobial eDNA. Methods Ecol. Evol. 2014, 5, 676–684. [Google Scholar] [CrossRef]
- Hinlo, R.; Gleeson, D.; Lintermans, M.; Furlan, E. Methods to maximise recovery of environmental DNA from water samples. PLoS ONE 2017, 12, e0179251. [Google Scholar] [CrossRef]
- Williams, K.E.; Huyvaert, K.P.; Piaggio, A.J. Clearing muddied waters: Capture of environmental DNA from turbid waters. PLoS ONE 2017, 12, e0179282. [Google Scholar] [CrossRef]
- Beans, C. Core Concept: Environmental DNA helps researchers track pythons and other stealthy creatures. Proc. Natl. Acad. Sci. USA 2018, 115, 8843–8845. [Google Scholar] [CrossRef] [PubMed]
- Levi, T.; Allen, J.M.; Bell, D.; Joyce, J.; Russell, J.R.; Tallmon, D.A.; Vulstek, S.C.; Yang, C.; Yu, D.W. Environmental DNA for the enumeration and management of Pacific salmon. Mol. Ecol. Resour. 2018, 394445. [Google Scholar] [CrossRef] [PubMed]
- Rees, H.C.; Bishop, K.; Middleditch, D.J.; Patmore, J.R.M.; Maddison, B.C.; Gough, K.C. The application of eDNA for monitoring of the great crested newt in the UK. Ecol. Evol. 2014, 4, 4023–4032. [Google Scholar] [CrossRef] [PubMed]
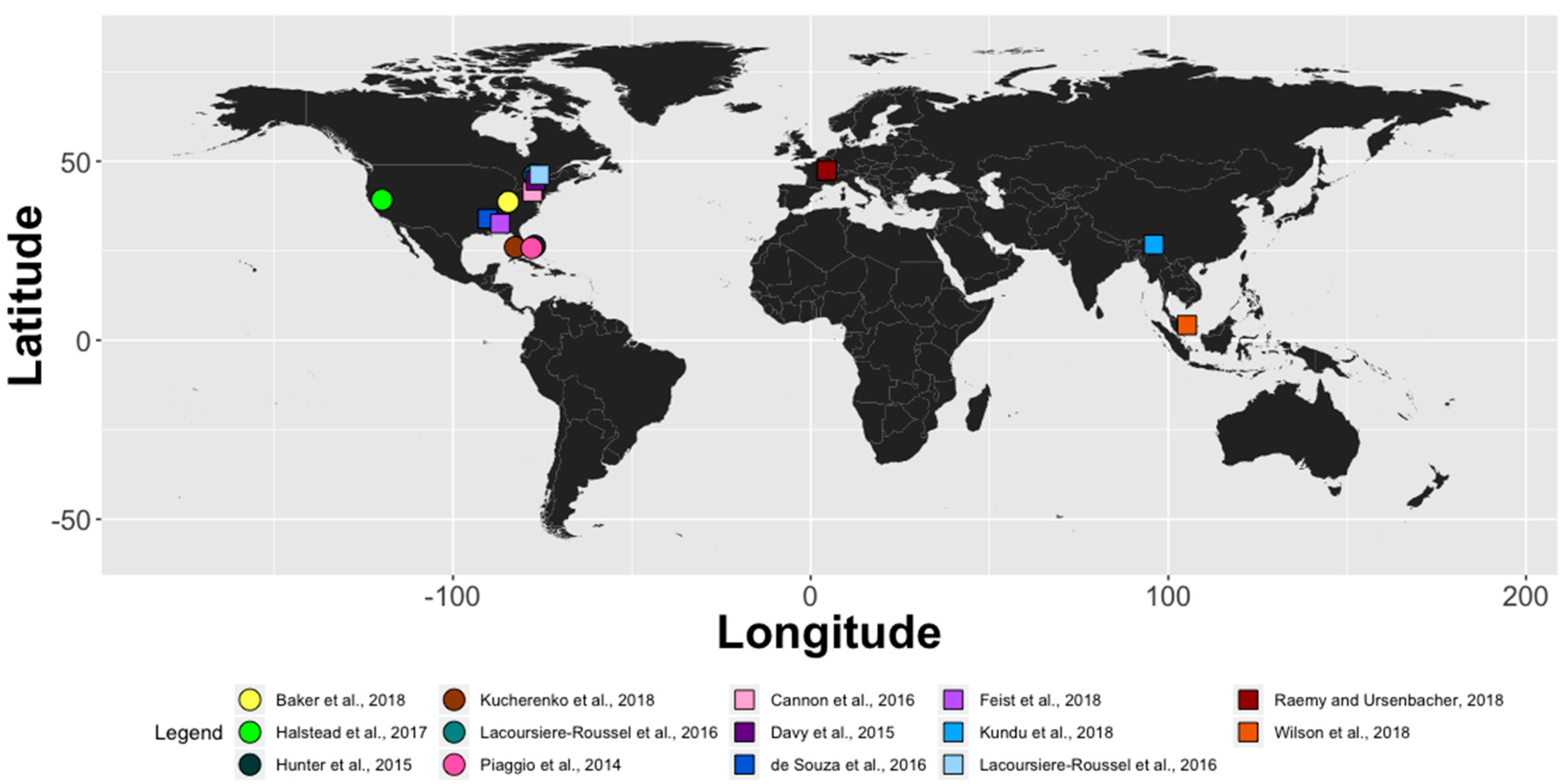
| Study | Order | Species | Country | Laboratory | Field | Consistent Field Detection? | Water Collection Details | Assay Type and Molecular Marker |
|---|---|---|---|---|---|---|---|---|
| Detection? | Detection? | |||||||
| Baker et al., 2018 [73] | Squamata | Sistrurus catenatus | U.S.A. | - | Yes | 2/100 samples amplified with S. catenatus. | 50 mL of water filtered with a 1.4 µM cellulose acetate filter; DNA extracted with the Qiagen DNeasy Blood and Tissue kit | qPCR Taqman assay for COI |
| Cannon et al., 2016 [76] | Testudines | Terrapene carolina | U.S.A. | - | Yes | 2/91 samples amplified from universal amphibian primers. | 50 mL of water centrifuged; DNA extracted with the Qiagen DNeasy Blood and Tissue Kit | Metabarcoded with CytB, Illumina MiSeq 2 × 250. |
| Davy et al., 2015 [24] | Testudines | Trachemys scripta | Canada | Yes | Yes | Yes, all PCR replicates of a field sample for T. scripta in a local pond. Other turtles not test for in a field setup. | 1 L of water filtered through 1.2 µm Whatman GF/C glass microfiber filters; DNA extracted with the Qiagen DNeasy Blood and Tissue Kit | qPCR SYBR (Intercalating dye) assay for COI |
| de Souza et al., 2016 [77] | Testudines | Sternotherus depressus | U.S.A. | Yes | Yes | Yes, four water samples required in the warm season and 14 water samples required in the cold season for a 95% detection probability. | 1 L of water filtered through a 0.80 µm cellulose nitrate filter; DNA extracted with the Qiagen DNeasy Blood and Tissue Kit. | qPCR EvaGreen (Intercalating dye) for 16S rDNA |
| Feist et al., 2018 [78] | Testudines | Macrochelys temminckii | U.S.A. | Yes | Yes | 2/3 to 1/6 replications amplified in the field when amplification occurred. | Between 850 and 1500 mL of water filtered through two 1.5 µm microfiber glass filters; DNA extracted with the DNeasy Blood and Tissue Kit | qPCR Taqman assay for CR of mtDNA |
| Halstead et al., 2017 [74] | Squamata | Thamnophis gigas | U.S.A. | Yes, limited. | No | No, no samples amplified. | 1 L of water filtered with a 0.45 µm nitrocellulose filter; DNA extracted with the MoBio PowerWater DNA isolation kit. | qPCR Taqman assay for ND4 |
| Hunter et al., 2015 [66] | Squamata | Python bivittatus | U.S.A. | Yes | Yes | Yes, 58% positive technical replicate detection near radio-tracked snakes | 950 mL of water filtered with a 0.45 µm cellulose nitrate filter and extracted with the PowerWater DNA Isolation Kit. | qPCR Taqman assay for ND4 |
| Kelly et al., 2014 [79] | Testudines | Chelonia mydas | U.S.A. | No—metabarcoding, yes—species-specific PCR | - | - | 1 L of water filtered with a 0.22 µm durapore membrane filter; DNA extracted with either the DNeasy Blood and Tissue kit or the PowerSoil DNA Isolation Kit | Metabarcoded with 12S rRNA gene or conventional PCR targeting CR of mtDNA |
| Kucherenko et al., 2018 [72] | Squamata | Pantherophis guttatus, Python bivittatus | U.S.A. | Yes | Yes | 66.7% successful detection rate. | 0.5 g of soil extracted with a phenol-chloroform protocol | Conventional PCR with a CytB primer |
| Kundu et al., 2018 [80] | Testudines | Nilssonia nigricans, Nilssonia gangetica, Chitra indica | India | - | Yes | No information given on how many of the 10 replicates were successful. | 15–20 mL sodium acetate precipitation; DNA extracted with the QIAamp Tissue Extraction Kit | Conventional PCR with a COI primer |
| Lacoursiere-Roussel et al., 2016 [70] | Testudines, Squamata | Chelydra serpentina, Glyptemys insculpta, Nerodia sipedon, Lampropeltis triangulum, Storeria occipitomaculata | Canada | Yes | Yes | Yes, targeted qPCR detected wood turtle in 9/9 locations. eDNA metabarcoding detected two turtle species in 3/9 locations, but 4/9 metabarcoding locations did not detect wood turtle otherwise detected with qPCR methodology. Snake species were found in 3/9 locations. | 2 L of water filtered through a 1.2 µm glass microfiber filter (Whatman GF/C); DNA extracted with the QIAshredder and DNeasy Blood and Tissue Kit | qPCR Taqman Assay targeting the COI gene for the Wood Turtle; Metabarcoding using CytB and COI for non-turtle species. |
| Piaggio et al., 2014 [23] | Squamata | Python bivittatus | U.S.A. | Yes | Yes | Yes, 5/5 field sites with known presence amplified. | 15 mL sodium acetate precipitation; DNA extracted with the QIAamp DNA Micro Kit. | Conventional PCR for CytB |
| Raemy and Ursenbacher, 2018 [81] | Testudines | Emys orbicularis | Switzerland | Yes | Yes | 3/6 to 6/6 replications amplified in the field when amplification occurred. | 90 mL of water filtered through a 0.22 µm filter; DNA extracted with the QIAamp Tissue Extraction Kit | qPCR SYBR (Intercalating dye) assay for CytB |
| Wilson et al., 2018 [82] | Testudines | Batagur affinis | Malaysia | Yes | Yes | Yes, with live individuals within a 1 km vicinity of turtle presence. | 250 mL of water filtered through a 0.45 µm cellulose nitrate filter; DNA extracted with the NucleoSpin Tissue Kit | Conventional PCR with a CytB primer |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adams, C.I.M.; Hoekstra, L.A.; Muell, M.R.; Janzen, F.J. A Brief Review of Non-Avian Reptile Environmental DNA (eDNA), with a Case Study of Painted Turtle (Chrysemys picta) eDNA Under Field Conditions. Diversity 2019, 11, 50. https://doi.org/10.3390/d11040050
Adams CIM, Hoekstra LA, Muell MR, Janzen FJ. A Brief Review of Non-Avian Reptile Environmental DNA (eDNA), with a Case Study of Painted Turtle (Chrysemys picta) eDNA Under Field Conditions. Diversity. 2019; 11(4):50. https://doi.org/10.3390/d11040050
Chicago/Turabian StyleAdams, Clare I. M., Luke A. Hoekstra, Morgan R. Muell, and Fredric J. Janzen. 2019. "A Brief Review of Non-Avian Reptile Environmental DNA (eDNA), with a Case Study of Painted Turtle (Chrysemys picta) eDNA Under Field Conditions" Diversity 11, no. 4: 50. https://doi.org/10.3390/d11040050
APA StyleAdams, C. I. M., Hoekstra, L. A., Muell, M. R., & Janzen, F. J. (2019). A Brief Review of Non-Avian Reptile Environmental DNA (eDNA), with a Case Study of Painted Turtle (Chrysemys picta) eDNA Under Field Conditions. Diversity, 11(4), 50. https://doi.org/10.3390/d11040050





