Sequence Diversity and Identification of Novel Puroindoline and Grain Softness Protein Alleles in Elymus, Agropyron and Related Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Source of Material
2.2. Germination of Seed Material
2.3. Sequence Analysis
2.4. Construction of Protein Models and Phylogenetic Trees
3. Results
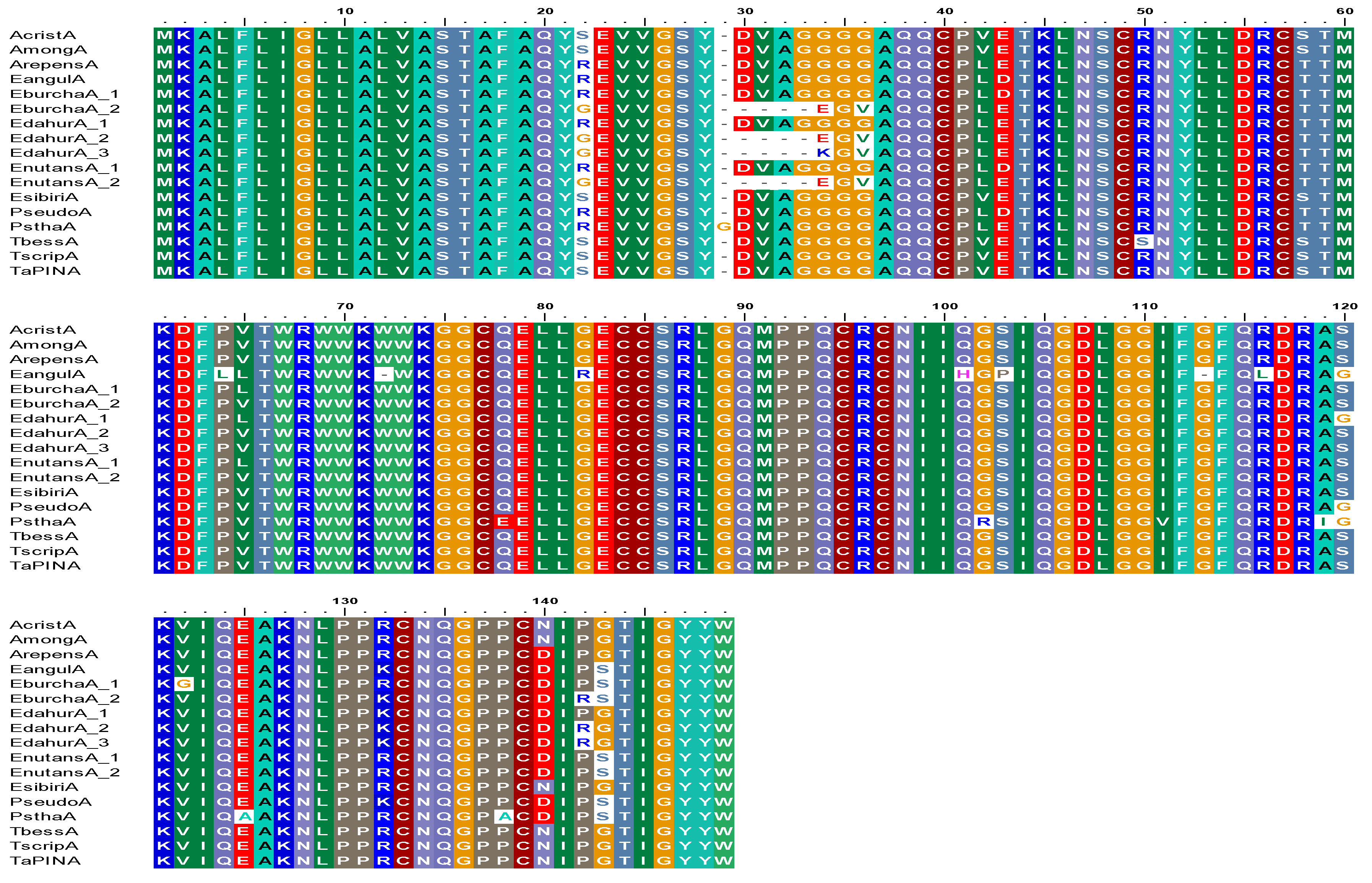
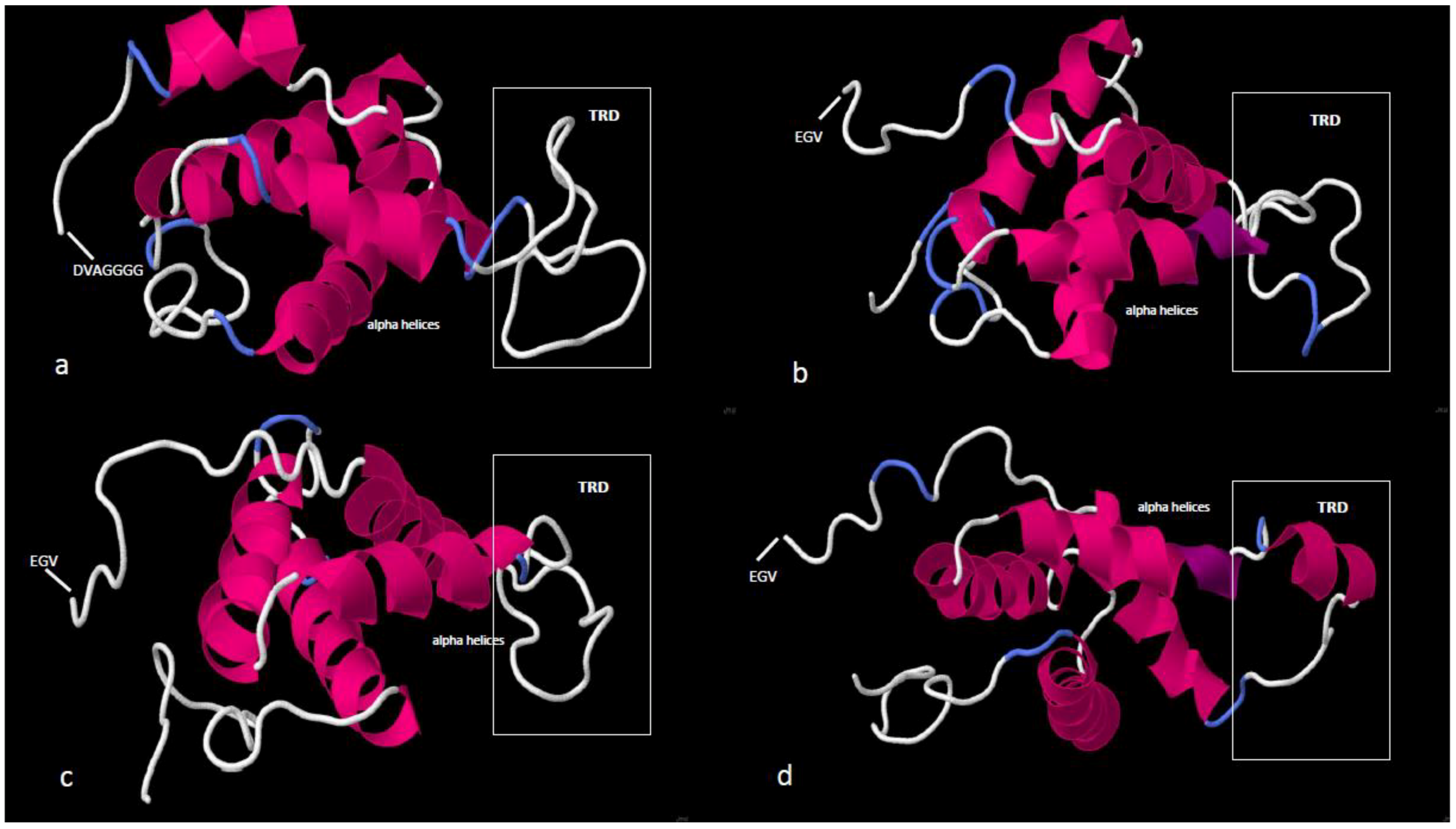
3.1. Sequence Diversity in PINA Proteins
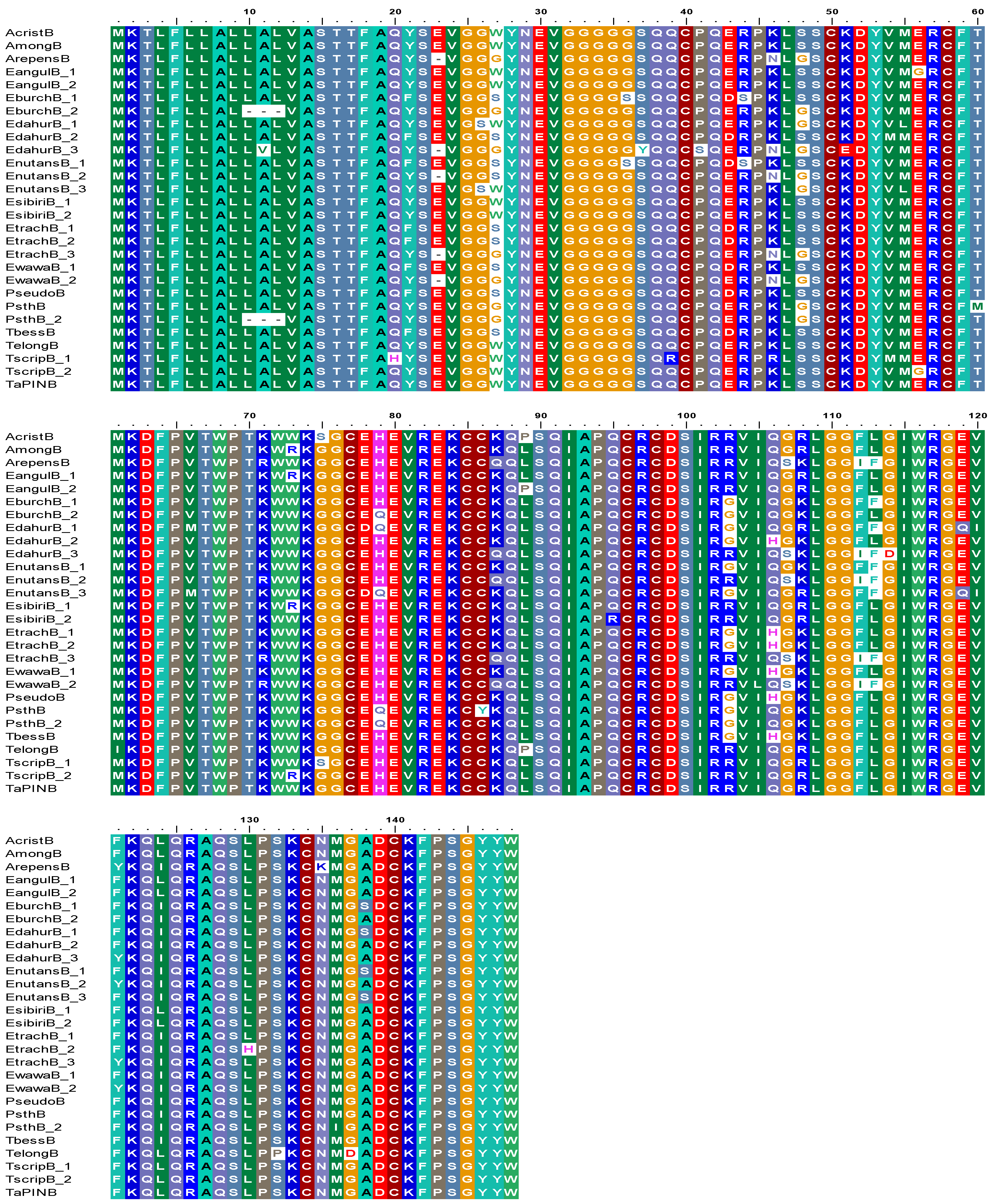
3.2. Sequence Diversity in PINB Proteins
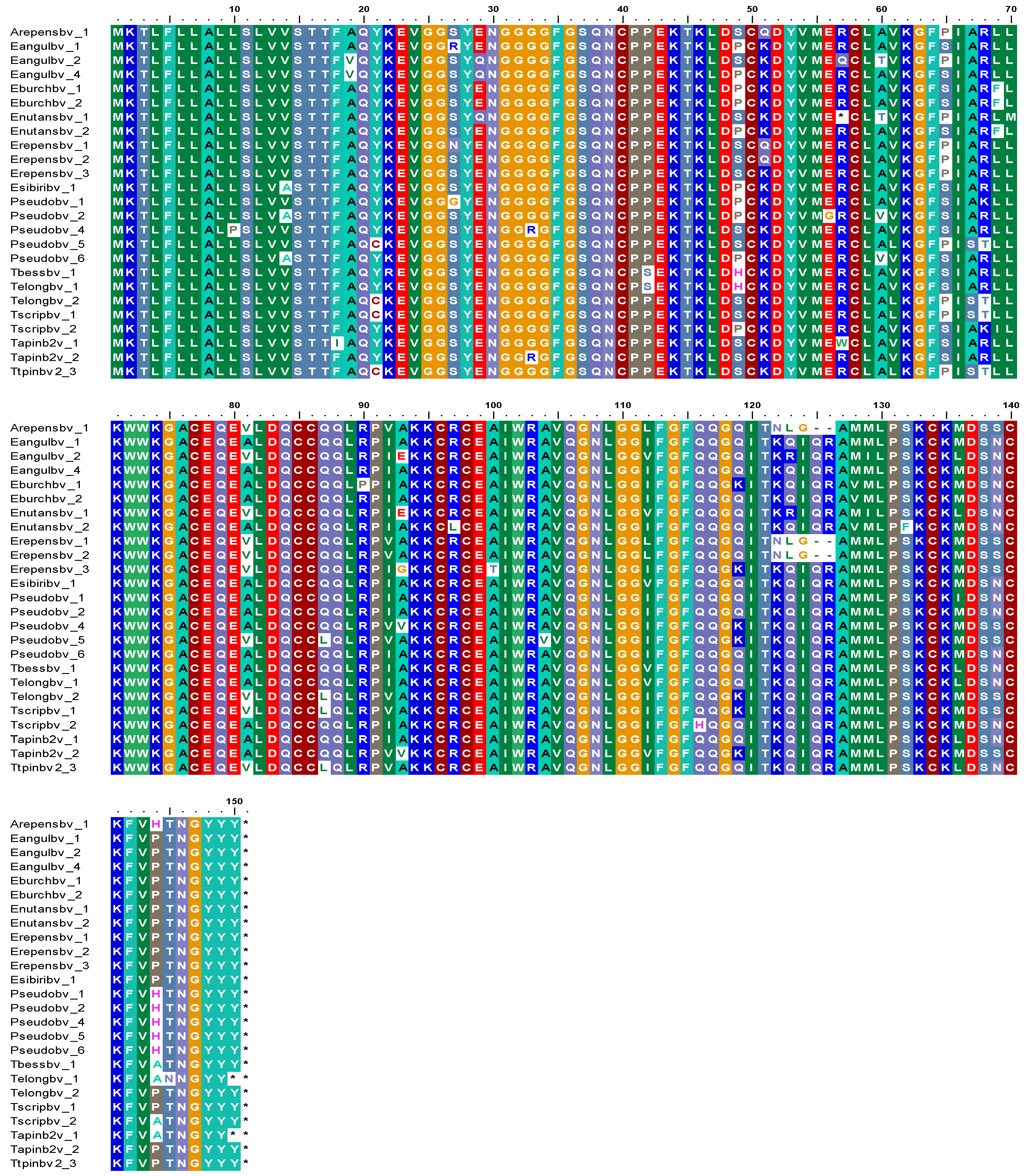
3.3. Sequence Diversity in PINB-2 Proteins
3.4. Gsp-1 Sequences in Elymus Species
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Greenham, T.J.; Altosaar, I. Wheat puroindolines tether to starch granule surfaces in puroindoline-null (Pin-null) plants. J. Cereal Sci. 2018, 79, 286–293. [Google Scholar] [CrossRef]
- Morris, C.F.; Rose, S.P. Wheat. In Cereal Grain Quality; Chapman Hall: London, UK, 1996. [Google Scholar]
- Morris, C.F. Puroindolines: The molecular genetic basis of wheat grain hardness. Plant Mol. Biol. 2002, 48, 633–647. [Google Scholar] [CrossRef] [PubMed]
- Chantret, N.; Salse, J.; Sabot, F.; Rahman, S.; Bellec, A.; Laubin, B.; Dubois, I.; Dossat, C.; Sourdille, P.; Joudrier, P.; et al. Molecular basis of evolutionary events that shaped the hardness locus in diploid and polyploid wheat species (Triticum and Aegilops). Plant Cell 2005, 17, 1033–1045. [Google Scholar] [CrossRef] [PubMed]
- Turner, A.S.; Bradburne, R.P.; Fish, L.; Snape, J.W. New quantitative trait loci influencing grain texture and protein content in bread wheat. J. Cereal Sci. 2004, 40, 51–60. [Google Scholar] [CrossRef]
- Weightman, R.M.; Millar, S.; Alava, J.; Foulkes, M.J.; Fish, L.; Snape, J.W. Effects of drought and the presence of the 1BL/1RS translocation on grain vitreosity, hardness and protein content in winter wheat. J. Cereal Sci. 2008, 47, 457–468. [Google Scholar] [CrossRef]
- Greenwell, P.; Schofield, J.D. A starch granule protein associated with endosperm softness in wheat. Cereal Chem. 1986, 63, 379–380. [Google Scholar]
- Ali, I.; Saradar, Z.; Rasheed, A.; Mahmood, T. Molecular characterization of the puroindoline-a and b alleles in synthetic hexaploid wheats and in in silico functional and structural insights into Pina-D1. J. Theor. Biol. 2015, 376, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Beecher, B.; Bettge, A.; Smidansky, E.; Giroux, M.J. Expression of wild-type pinB sequence in transgenic wheat complements a hard phenotype. Theor. Appl. Genet. 2002, 105, 870–877. [Google Scholar] [PubMed]
- Martin, J.M.; Meyer, F.D.; Smidasky, E.D.; Wangugi, H.; Blechl, A.E.; Giroux, M.J. Complementation of the pina (null) allele with the wild type Pina sequence restores a soft phenotype in transgenic wheat. Theor. Appl. Genet. 2006, 113, 1563–1570. [Google Scholar] [CrossRef] [PubMed]
- Heinze, K.; Kiszonas, A.M.; Murray, J.C.; Morris, C.F.; Lullien-Pellerin, V. Puroindoline genes introduced into durum wheat reduce milling energy and change milling behaviour similar to soft common wheats. J. Cereal Sci. 2016, 71, 183–189. [Google Scholar] [CrossRef]
- Xia, L.; Geng, H.; Chen, X.; He, Z.; Lillemo, M.; Morris, C.F. Silencing of puroindoline a alters the kernel texture in transgenic bread wheat. J. Cereal Sci. 2008, 47, 331–338. [Google Scholar] [CrossRef]
- Gasparis, S.; Orczyk, W.; Zalewski, W.; Nadolska-Orcyzk, A. The RNA-mediated silencing of one the Pin genes in allohexaploid wheat simultaneously decreases the expression of the other and increases grain hardness. J. Exp. Bot. 2001, 62, 4025–4036. [Google Scholar] [CrossRef] [PubMed]
- Jolly, C.J.; Glenn, G.M.; Rahman, S. GSP-1 genes are linked to the grain hardness locus (Ha) on wheat chromosome 5D. Proc. Natl. Acad. Sci. USA 1996, 93, 2408–2413. [Google Scholar] [CrossRef] [PubMed]
- Tranquilli, G.; Heaton, J.; Chicaiza, O.; Dubcovsky, J. Substitutions and deletions of genes related to grain hardness in wheat and their effect on grain texture. Crop Sci. 2002, 42, 1812–1817. [Google Scholar] [CrossRef]
- Wilkinson, M.D.; Tosi, P.; Lovegrove, A.; Corol, D.I.; Ward, J.L.; Palmer, R.; Powers, S.; Passmore, D.; Webster, G.; Marcus, S.E.; et al. The Gsp-1 genes encode the wheat arabinogalactan peptide. J. Cereal Sci. 2017, 74, 155–164. [Google Scholar] [CrossRef]
- Turner, M.; Mukai, Y.; Leroy, P.; Charaf, B.; Appels, R.; Rahman, S. The Ha locus of wheat: identification of a polymorphic region for tracing grain harness in crosses. Genome 1999, 42, 1242–1250. [Google Scholar] [CrossRef] [PubMed]
- Gollan, P.; Smith, K.; Bhave, M. Gsp-1 genes comprise a multigene family in wheat that exhibits a unique combination of sequence diversity yet conservation. J. Cereal Sci. 2007, 45, 184–198. [Google Scholar] [CrossRef]
- Wilkinson, M.D.; Castells-Brooke, N.; Shewry, P.R. Diversity of sequences encoded by the Gsp-1 genes in wheat and other grass species. J. Cereal Sci. 2013, 57, 1–9. [Google Scholar] [CrossRef]
- Wilkinson, M.; Wan, Y.; Tosi, P.; Leverington, M.; Snape, J.; Mitchell, R.A.C.; Shewry, P.R. Identification and genetic mapping of variant forms of puroindoline b expressed in developing wheat grain. J. Cereal Sci. 2008, 48, 722–728. [Google Scholar] [CrossRef]
- Chen, F.; Beecher, B.; Morris, C.F. Physical mapping and a new variant of Puroindoline b-2 genes in wheat. Theor. Appl. Genet. 2010, 120, 745–751. [Google Scholar] [CrossRef] [PubMed]
- Geng, H.; Beecher, B.S.; He, Z.; Kiszonas, A.M.; Morris, C.F. Prevalence of Puroindoline D1 and Puroindoline b-2 variants in U.S. Pacific Northwest wheat breeding germplasm pools, and their association with kernel texture. Theor. Appl. Genet. 2012, 124, 1259–1269. [Google Scholar] [CrossRef] [PubMed]
- Geng, H.; Beecher, B.S.; Pumphrey, M.; He, Z.; Morris, C.F. Segregation analysis indicates that Puroindoline b-2 variants 2 and 3 are allelic in Triticum aestivum and that a revision to Puroindolineb-2 gene symbolization is indicated. J. Cereal Sci. 2013, 57, 61–66. [Google Scholar] [CrossRef]
- Chen, F.; Xu, H.X.; Zhang, F.Y.; Xia, X.C.; He, Z.H.; Wang, D.W.; Dong, Z.D.; Zhan, K.H.; Cheng, X.Y.; Cui, D.Q. Physical mapping of puroindoline b-2 genes and molecular characterization of a novel variant in durum wheat (Triticum turgidum L.). Mol. Biol. 2011, 28, 153–161. [Google Scholar] [CrossRef]
- Chen, F.; Zhang, F.Y.; Cheng, X.Y.; Morris, C.F.; Xu, H.X.; Dong, Z.D.; Zhan, K.H.; Cui, D.Q. Association of Puroindoline b-B2 variants with grain traits, yield components and flag leaf size in bread wheat (Triticum aestivum L.) varieties of the Yellow and Huai valleys of China. J. Cereal Sci. 2010, 52, 247–253. [Google Scholar] [CrossRef]
- Ramalingam, A.; Palombo, E.A.; Bhave, M. The Pinb-2 genes in wheat comprise a multigene family with sequence diversity and important variants. J. Cereal Sci. 2012, 56, 171–180. [Google Scholar] [CrossRef]
- Giroux, M.J.; Kim, K.H.; Hogg, A.C.; Martin, J.M.; Beecher, B. The Puroindoline b-2 variants are expressed at low levels relative to the Puroindoline D1 genes in wheat seeds. Crop Sci. 2013, 53, 833–841. [Google Scholar] [CrossRef]
- Gautier, M.F.; Cosson, P.; Guirao, A.; Alary, R.; Joudrier, P. Puroindoline genes are highly conserved in diploid ancestor wheats and related species but absent in tetraploid Triticum species. Plant Sci. 2000, 153, 81–91. [Google Scholar] [CrossRef]
- Chen, M.; Wilkinson, M.; Tosi, P.; He, G.; Shewry, P.R. Novel puroindoline and grain softness protein alleles in Aegilops species with the C, D, S, M and U genomes. Theor. Appl. Genet. 2005, 111, 1159–1166. [Google Scholar] [CrossRef] [PubMed]
- Massa, A.N.; Morris, C.F. Molecular evolution of the puroindoline-a, puroindoline-b, and grain softness protein-1 genes in the tribe Triticeae. J. Mol. Evol. 2006, 63, 526–536. [Google Scholar] [CrossRef] [PubMed]
- Bhave, M.; Morris, C. Molecular genetics of puroindolines and related genes: Allelic diversity in wheat and other grasses. Plant Mol. Biol. 2008, 66, 205–219. [Google Scholar] [CrossRef] [PubMed]
- Linc, G.; Gaál, E.; Molnár, I.; Icso, D.; Badaeva, E.; Molnár-Láng, M. Molecular cytogenetic (FISH) and genome analysis of diploid wheatgrasses and their phylogenetic relationship. PLoS ONE 2017, 12, e0173623. [Google Scholar] [CrossRef] [PubMed]
- McMillan, E.; Sun, G. Genetic relationships of tetraploid Elymus species and their genomic donor species inferred from polymerase chain reaction-restriction length polymorphism analysis of chloroplast gene regions. Theor. Appl. Genet. 2004, 108, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.R.C. Wild Crop Relatives: Genomic and Breeding Resources. In Agropyron and Psathyrostachys; Springer: Berlin/Heidelberg Germany, 2011. [Google Scholar]
- Gazza, L.; Galassi, E.; Ciccoritti, R.; Cacciatori, P.; Pogna, N.E. Qualitative traits of perennial wheat lines derived from different Thinopyrum species. Genet. Res. Crop Evol. 2016, 63, 209–219. [Google Scholar] [CrossRef]
- Posada, D. jModelTest: Phylogenetic Model Averaging. Mol. Biol. Evol. 2008, 25, 1253–1256. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.G.L.; Taboada, R.; Doallo, D. Posada jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.M.; Teslenko, P.; van der Mark, D.L.; Ayres, A.; Darling, S.; Hohna, B.; Larget, L.; Liu, M.A.; Suchard, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Escobar, J.S.; Scornavacca, C.; Cenci, A.; Guilhaunmon, C.; Santoni, S.; Douzery, E.J.; Ranwez, V.; Glemin, S.; David, J. Multigenic phylogeny and analysis of tree incongruences in Triticeae (Poaceae). BMC Evol. Biol. 2011, 11, 181. [Google Scholar] [CrossRef] [PubMed]
- Gautier, M.F.; Aleman, M.E.; Guirao, A.; Marion, D.; Joudrier, P. Triticum aestivum puroindolines, two basic cysteine-rich seed proteins: cDNA sequence analysis and development gene expression. Plant Mol. Biol. 1994, 25, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Pouch, M.; Vaculova, K.; Milotova, J.; Stehno, Z.; Curn, V. Molecular and phylogenetic study of puroindoline genes in wild and cultivated wheat species with different ploidy levels. 2018; in press. [Google Scholar]
- Terasawa, Y.; Rahman, S.M.; Takata, K.; Ikeda, T.M. Distribution of Hordoindoline genes in the genus Hordeum. Theor. Appl. Gene. 2012, 124, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Shaaf, S.; Sharma, R.; Baloch, F.S.; Badaeva, E.D.; Knupffer, H.; Kilian, B.; Ozkan, H. The grain Hardness locus characterized in a diverse wheat panel (Triticum aestivum L.) adapted to the central part of the Fertile Crescent: genetic diversity, haplotype structure, and phylogeny. Mol. Genet. Genom. 2016, 291, 1259–1275. [Google Scholar] [CrossRef] [PubMed]
- Lillemo, M.; Simeone, M.C.; Morris, C.F. Analysis of puroindoline a and b sequences from Triticum aestivum cv. ‘Penawawa’ and related diploid taxa. Euphytica 2002, 126, 321–331. [Google Scholar] [CrossRef]
- Yan, X.; Wang, Y.-Q.; Zhao, J.-X.; Wu, J.; Chen, X.-H.; Cheng, X.-N. Isolation and sequence analysis on the grain hardness genes in Psathyrostachys huashanica. 2018; in press. [Google Scholar]
- Terasawa, Y.; Takata, K.; Anai, T.; Ikeda, T.M. Identification and distribution of Puroindoline b-2 variant gene homologs in Hordeum. Genetica 2013, 141, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, M.D.; Shewry, P.R. Screening of Aegilops species for Puroindoline b variants. 2018; in press. [Google Scholar]
- Bhave, M.; Morris, C. Molecular genetics of puroindolines and related genes: Regulation of expression, membrane binding properties and applications. Plant Molec. Biol. 2008, 66, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Li, H.; Cui, D. Discovery, distribution and diversity of Puroindoline-D1 genes in bread wheat from five countries (Triticum aestivum L.). Plant Biol. 2013, 13, 125. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Sajjad, M.; Wang, J.; Yang, W.; Sun, J.; Li, X.; Zhang, A.; Liu, D. Diversity, distribution of Puroindoline genes and their effect on kernel hardness in a diverse panel of Chinese wheat germplasm. BMC Plant Biol. 2017, 17, 158. [Google Scholar] [CrossRef] [PubMed]
- Boehm, J.D.; Ibba, M.I.; Kiszonas, A.M.; See, D.R.; Skinner, D.Z.; Morris, C.F. Genetic analysis of kernel texture (grain hardness) in a hard-red spring wheat (Triticum aestivum L.) bi-parental population. J. Cereal Sci. 2018, 79, 57–65. [Google Scholar] [CrossRef]
- Cuesta, S.; Guzman, C.; Alvarez, J.B. Allelic diversity and molecular characterization of puroindoline genes in five diploid species of the Aegilops genus. J. Exp. Bot. 2013, 64, 5133–5143. [Google Scholar] [CrossRef] [PubMed]
- Bihan, T.L.; Blochet, J.E.; Desormeaux, A.; Marion, D.; Pezolet, M. Determination of the secondary structure and conformation of puroindolines by infrared and Raman spectroscopy. Biochemistry 1996, 35, 12712–12722. [Google Scholar] [CrossRef]
- Shewry, P.R.; Halford, N.G. Cereal seed storage proteins: Structures, properties and role in grain utilization. J. Exp. Bot. 2012, 53, 947–958. [Google Scholar] [CrossRef]
- Elmorjani, K.; Geneix, N.; Dagalarrondo, M.; Branlard, G.; Didier, M. Wheat grain softness protein (Gsp-1) is a puroindoline-like protein that displays a specific post-translational maturation and does not interact with lipids. J. Cereal Sci. 2013, 58, 117–122. [Google Scholar] [CrossRef]
- Strobl, S.M.; Mühlhahn, P.; Bernstein, R.; Wiltscheck, R.; Maskos, K.; Wunderlich, M.; Huber, R.; Glockshuber, R.; Holak, T.A. Determination of the three-dimensional structure of the bifunctional alpha-amylase/trypsin inhibitor from ragi seeds by NMR spectroscopy. Biochemistry 1995, 34, 8281–8293. [Google Scholar] [CrossRef]

| Species and Genome | Pina Gene 1 | Pinb Gene 2 | Pinb-2 Gene 3 | Gsp-1 Gene 4 | Summary of Sequence Information |
|---|---|---|---|---|---|
| Agropyron cristatum (2x)-P | +1 (3) | +1 (6) | - | +5 (14 *) | 100% identity P33432 1; 99% identity AHC54604 2; [19] 4 |
| Agropyron mongolicum (2x)-P | +1 (6) | +1 (6) | - | +9 (22 *) | 100% identity P33432 1; 100% identity ALI87022 2; [19] 4 |
| Thinopyrum bessarabicum (2x)-Eb or J | +1 (6) | +1 (4) | +1 (3) | +2 (10 *) | 99% identity P33432 1; 97% identity AER62841 2; 98% identity AFB35605 3; [19] |
| Thinopyrum elongatum (2x)-J | - | +1-(6) | +2-(14) | +2-(11 *) | 98% identity AHC54604 2; 98% identity AFB35605 3 & 100% identity CCH14734 3; [19] 4 |
| Thinopyrum scripeum (2x)-E or Je | +1 (4) | +1 (6) | +2 (7) | +1 (2) | 100% identity P33432 1; 97% identity CAH10199 2; 97% identity AFB35605 3 & 100% identity CCH14734 3; 95% identity XP_020170444 4 |
| Pseudoroegneria spicata (2x)-St | +1 (7) | +1 (9) | +2 (21) | +3 (11 *) | 97% identity AER62831 1; 97% identity AER62841 2; 93% identity CAQ16391 3 & 99% identity AFB35608 3; [19] 4 |
| Psathyrostachys juncea (2x)-Ns | +1 (8) | +2 (4) | - | +4 (9 *) | 99% identity AER62828 1; 97–99% identity AER62849 2; [19] 4 |
| Elymus burchan-buddae (4x)-StY | +2 (9) | +2 (2) | +1 (4) | +1 (8) | 98% identity AER62831 1 & Novel Pin a sequence GenBank accession number LT669797; & 100% AER62849 2 & Novel Pin b sequence Genbank accession number LR025202 2; 97% identity AFB35605 3; AGP1 detected [19] 4 |
| Elymus sibiricus (4x)-StH | +1 (6) | +2 (7) | +1 (2) | +1 (2) | 100% identity P33432 1; 100% identity ALI87022 2 & 99% Q10464 2; 97% identity AFB35605 3; AGP3 detected [19] |
| Elymus trachycaulus subsp. subsecundus (4x)-StH | - | +3 (10) | - | +2 (11) | 97% identity AER62841 2 and 98% identity BAK64220 2; AGP3 and AGP13 [19] 4 |
| Elymus wawawaiensis (4x)-StH | - | +2 (5) | - | +3 (9) | 97% identity AER62841 2 & 98% identity BAK64220 2; AGP3 and AGP13 detected [19] 4; Novel Gsp-1 sequence GenBank accession number LT669800 4 |
| Elymus angulatus (6x)-StYH | +1 (3) | +2 (5) | +2 (4) | +1 (6) | 92% identity AER62832 1 (two stop codons present); 100% identity AHC54604 & 99% identity ALI87022 2; 97% identity AFB35605 3 & 97% BAM43258 3; AGP1 detected [19] stop codon present 4 |
| Elymus dahuricus subsp. excelsus (6x)-StYH | +2 (8) | +3 (12) | - | +1 (2) | 97% identity AER62831 1 & Novel Pin a sequence GenBank accession number LT669797; sequence ID SHD75392 1; 98% identity BAK64220 2 & Novel Pin b sequence GenBank accession number LT669796 2; AGP1 detected [19] 4 |
| Elymus nutans (6x)-StYH | +2 (8) | +3 (6) | +1 (6) | +2 (8) | 99% identity AER62831 1 & Novel Pin a sequence GenBank accession number LT669797 1; 91% identity CAC33506 2; 98% identity BAK64220 2 & 2 Novel Pin b sequence GenBank accession number LT669796 2 & LS991245 2; 93% identity AFB35607 3; AGP1 detected [19]; Novel Gsp-1 sequence GenBank accession number LT669799 4 |
| Agropyron repens (6x)-StStH | +1 (6) | +1 (6) | +1 (8) | +1 (4 *) | 98% identity AER62831 1; 98% identity BAK64220 2; Novel Pinb-2 sequence GenBank accession number LT669798 3; [19] 4 |
| Elymus repens (6x)-StStH | +(not sequenced) | +(not sequenced) | +2 (9) | +2 (3 *) | 95% identity AFB35607 3 & Novel Pinb-2 sequence GenBank accession number LT669798 3; [19] 4 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wilkinson, M.D.; King, R.; Grimaldi, R. Sequence Diversity and Identification of Novel Puroindoline and Grain Softness Protein Alleles in Elymus, Agropyron and Related Species. Diversity 2018, 10, 114. https://doi.org/10.3390/d10040114
Wilkinson MD, King R, Grimaldi R. Sequence Diversity and Identification of Novel Puroindoline and Grain Softness Protein Alleles in Elymus, Agropyron and Related Species. Diversity. 2018; 10(4):114. https://doi.org/10.3390/d10040114
Chicago/Turabian StyleWilkinson, Mark D., Robert King, and Roberta Grimaldi. 2018. "Sequence Diversity and Identification of Novel Puroindoline and Grain Softness Protein Alleles in Elymus, Agropyron and Related Species" Diversity 10, no. 4: 114. https://doi.org/10.3390/d10040114
APA StyleWilkinson, M. D., King, R., & Grimaldi, R. (2018). Sequence Diversity and Identification of Novel Puroindoline and Grain Softness Protein Alleles in Elymus, Agropyron and Related Species. Diversity, 10(4), 114. https://doi.org/10.3390/d10040114





