Abstract
In this study, the organelle genomes of Polytrichum juniperinum Hedw. and Polytrichum strictum Menzies ex Brid. (Polytrichaceae, Bryophyta) from Antarctica were sequenced and compared with the plastomes of the model moss species Physcomitrella patens Brid. The sizes of the cpDNA in P. juniperinum and P. strictum were estimated to be 55,168 and 20,183 bp, respectively; the sizes of the mtDNA were 88,021 and 58,896 bp, respectively. The genomes are very similar to each other, with the possible loss of petN in the cpDNA, which also showed some gene inversions when compared with the cpDNAs of P. patens Brid. In the mtDNA, it is possible that rps10 was lost. In contrast, Antarctic Polytrichaceae species have nad7 and orf187, without the occurrence of rearrangement events. Phylogenomic analyses of the plastid and mitochondria revealed that the majority-rule tree suggests some differences in the plastids ancestry, however, P. juniperinum and P. strictum were grouped in the same clade in chloroplast, but in mitochondria P. strictum was grouped with Atrichum angustatum (Brid.) Bruch & Schimp. This study helped us understand the evolution of plastomes and chondriosomes in the family Polytrichaceae, and suggest a hybridization event with relation to the mitochondrial data.
1. Introduction
Polytrichum is a cosmopolitan genus with a bipolar distribution [1] (from the Arctic lands to the Antarctic Continent). In the Antarctic, three species have been reported, all of which are confined to the maritime Antarctic: Polytrichum juniperinum Hedw., Polytrichum piliferum Hedw., and Polytrichum strictum Menzies ex Brid [2]. They play an important role in the terrestrial vegetation of this biome as essential constituents in various communities of moss turf subformations as well as fruticose lichens [1]. The phylogenetic relationships of Polytrichales are particularly relevant when considering the evolutionary history of mosses, since the group is probably one of the first lineages to diverge from the common ancestor of all mosses [3,4]. A recent development in plastome sequencing is the use of total genomic DNA as the template for next-generation sequencing [5,6]. The outcome of this new development was huge improvements in our understanding of the phylogenetic relationships among plants, particularly mosses. A previous study had suggested that P. strictum arose from a reticulation event and P. juniperinum is probably its maternal ancestor [4]. However, the phylogenetic position of P. strictum is still unclear. Thus, understanding these relationships is necessary to expand the quantity and quality of phylogenetic molecular data available to evaluate the relationships between P. juniperinum and P. strictum in the Antarctic.
2. Materials and Methods
Gametophyte samples of P. juniperinum (62°12′41.93″ S and 58°55′44.61″ O) and P. strictum (62°12′37.36″ S and 58°57′49.87″ O) were collected from Ardley Island during the austral summer of 2014–2015, Brazilian Antarctic Expedition XXXIII (2014–2015). Part of the samples were incorporated into the Bruno Irgand Herbarium (HBEI) of UNIPAMPA/São Gabriel, under the voucher HBEI 059 and HBEI 060 to P. juniperinum and P. strictum respectively, the remaining samples were used for the analyzes foreseen in the present work. Total genomic DNA was extracted using a modified cetyltrimethylammonium bromide (CTAB) extraction procedure, as described by Shaw [7]. After the DNA extraction, the samples were evaluated with NanoVueTM Plus Spectrophotometer (GE Healthcare, Chicago, IL, USA) and Qubit® 2.0 Fluorometer (Invitrogen, Carlsbad, CA, USA) to ensure the quality and quantity of the samples. This DNA was sheared into fragments averaging approximately 250 bp and then, genome sequencing of the Polytrichaceae DNA samples was performed using the Ion Torrent PGM platform (Life Technologies, Carlsbad, CA, USA). In the same conditions, three genomic-DNA libraries were prepared using the Ion One Touch Template Kit (Life Technologies, Carlsbad, CA, USA) (Table 1). The amplified library was sequenced using Ion PGM™ Hi-Q™ Sequencing Kit within the 318 Chip. The three libraries were concatenate with cat command line and a total of 16,333,496 reads from P. juniperinum and 16,679,733 reads from P. strictum (maximum length, 389 bp and mean length, 170 bp) from single-end type were sequenced (Table 1). The best cut-off values for low quality reads were estimated using the FastQC quality control tool [8]. Then, the reads were filtered for quality using a standard approximating Phred quality score (Q20) in both species set reads with the tool disponible in FASTX Toolkit in the Galaxy platform (https://mississippi.snv.jussieu.fr), decreasing the likelihood of low quality reads in contigs assembly. Assembly of the contigs was performed using the Velvet Assembler for short reads [9] utility cpDNA (NC_005087.1) and mtDNA (NC_007945.1) of Physcomitrella patens as reference, and the best Kmer estimated in 25 (P. juniperinum) and 27 (P. strictum) by Kmergenie [10]. Scaffold assembly for cpDNA and mtDNA was performed using the Scaffold Builder assembler version 2.2 and the chloroplast and mitochondria from P. patens as the reference genomes [11]. Annotation of the chloroplast was performed using web-based Dual Organellar Genome Annotator (DOGMA) [12] and same parameters adjusted (percent identity cut-off for protein-coding genes estimated in 25; percent identity cut-off for RNAs estimated in 25; e-value estimated in 1 × 10−5) and cpGAVAS [13] with e-value estimated in 1 × 10−5 Mitochondrial annotation was performed using Mitofy version 1.3.1 of tRNAscan-SE and version 2.2.28 of National Center for Biotechnology Information-Basic Local Alignment Search Tool (NCBI BLAST) [14]. Annotation of cpDNA and mtDNA genes was manually corrected by comparison with complete chloroplast and mitochondrial genomes of other bryophytes using BLASTn [15]. The species were compared with the reference genomes of P. patens for generating chloroplast and mitochondrial circular maps for coverage visualization, gene content, and presence/absence of genes with Blast Ring Image Generator (BRIG) [16] 0.95. For the phylogenetic analyses, individual alignments were performed for each gene by using Molecular Evolutionary Genetics Analysis Software (MEGA 5.05) [17] and all alignments were concatenated with sequence matrix 1.883 to create a super-alignment. The best model for nucleotide substitution, TN93 model, was established using MEGA 5.05 and adjusted in jModelTest [18] for each gene alignment. The tree was created on the basis of Bayesian statistical analysis with 10,000,000 million Monte Carlo Markov chains to avoid errors in the posterior probability support of the BEAST package [7]. The base frequency was estimated, and the dataset was partitioned (e.g., codon positions) into two partitions (1 + 2), 3 with BEAUti (BEAST package). The majority rule tree was constructed with TreeAnnotator (BEAST package). The support of the nodes was calculated through posterior probability that varies from 0 to 1. Frequency convergence of the trees and 25% burn-in were confirmed with Tracer (BEAST package) and this program was used to estimate when the sampling of the trees was stabilized. The divergence dates were obtained considering the estimated date for bryophyte origin [19].

Table 1.
Selected statistics for chloroplast and mitochondria genomes of Polytrichum species.
3. Results
3.1. Sequence Data
A set of 16,333,496 single-end reads of Polytrichum juniperinum and 16,679,733 single-end reads of Polytrichum strictum was generated with a mean length of 170 bp from a 3/3 run on an Ion Torrent sequencer in PGM plattform. Read quality was satisfactory, with a low ratio of duplicates (2%). The representation of the two plant cell genomes in the data were as follows: 103,776 reads were mapped to the chloroplast genome of P. juniperinum with a coverage level of 10083, and 32,931 reads to the mitochondria of P. juniperinum with coverage level of 2988; 58,624 reads to the chloroplast of P. strictum with a coverage level of 1890, and 29,620 reads to the mitochondria genome of P. strictum with a coverage level of 26,564. Details from reads and libraries can be visualized in Table 1. Physcomitrella patens (Funariaceae) was chosen as a reference species for the assembly of the genome of chloroplasts and mitochondria due mainly to the availability of complete and updated genomic data, as well as to its relatively proximate phylogenetic position with the Polytrichaceae family. The obvious choice as a reference would be of a genome from a same family species, but the lack of complete genomic data for species closer to Polytrichum makes this strategy impracticable. Atrichum angustatum (Brid.) Bruch & Schimp. (Polytrichaceae) has its mitochondrial genome sequenced, but we can not include it only in the assembly of the mitochondrial genome, this could raise misinterpretations about our inference between presence/absence genes in Polytrichum and other species of this family.
3.2. Genomic Organization and Gene Content
After assembly, the Polytrichum juniperinum plastid genome (cpDNA) obtained was 55,168 bp in length and had a G + C content of 44.9%, including 51 putative coding genes, 31 tRNAs, and 4 rRNAs. Furthermore, the cpDNA revealed 19 putative protein-coding genes related to photosynthesis, such as putative photosystem I and II proteins. The Polytrichum strictum cpDNA assembly generated a size genome of 20,183 bp with a G + C content of 46.8% (similar to P. juniperinum). The cpDNA of P. strictum also showed 44 putative coding genes, 14 tRNAs, and 4 rRNAs, and 18 putative protein-coding genes related to photosynthesis, such as putative photosystem I and II proteins.
Our data suggests a possible absence of rpoA in the cpDNA P. juniperinum and P. strictum (Table 2). petN was absent in Syntrichia ruralis (Hedw.) F. Weber & D. Mohr [20] and Tetraphis pellucida Hedw. [21] and was probably translocated to the nucleus in P. juniperinum and P. strictum. The BLAST analyses showed that ycf66 had 90% identity among the Antarctic Polytrichaceae species. In P. juniperinum, this gene presented 100% identity with the homologous gene in Sanionia uncinata (Hedw.) Loeske, and in P. strictum, 94.3% identity with the homologous gene in S. ruralis. Some gene regions were found to have an identity lower than 90% when compared with the reference, for example, psaB, trnV, and ndhD (Figure 1). We also observed one inversion event in ndhA and ycf2 between Physcomitrella patens and both Polytrichum samples from Antartica (Figure 1) and (Figure 2 and Figure 3).

Table 2.
Gene content of cpDNA and mtDNA from algae, bryophytes, and higher plants.
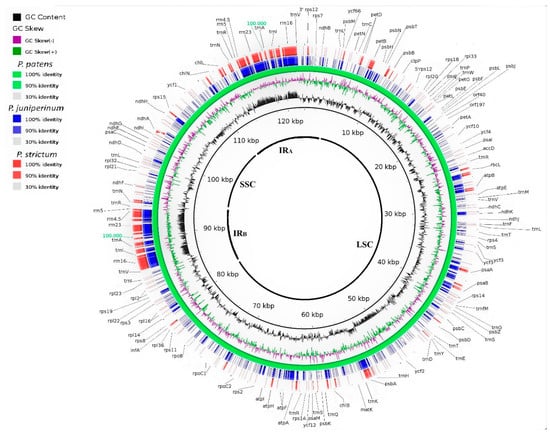
Figure 1.
Blast Ring Image Generator output image of the chloroplast genome comparing Physcomitrella patens and Antarctic Polytrichum species. The internal ring represents the P. patens chloroplast genome (green). BLAST match of Polytrichum juniperinum and Polytrichum strictum are in blue and red gradient, respectively. The legend showing color gradient for percentage similarity between the reference and Polytrichum species. The innermost rings show the GC skew (purple/green) and GC content (black). The highlighted blocks show the inversions observed between P. strictum and P. juniperinum with the reference genome. The inverted repeat region (IRs), long single copy section (LSC), and short single copy section (SSC) are indicated.
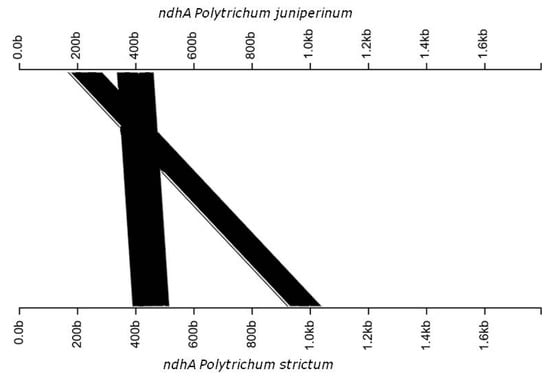
Figure 2.
Linear model of ndhA gene inversion in Polytrichum strictum.
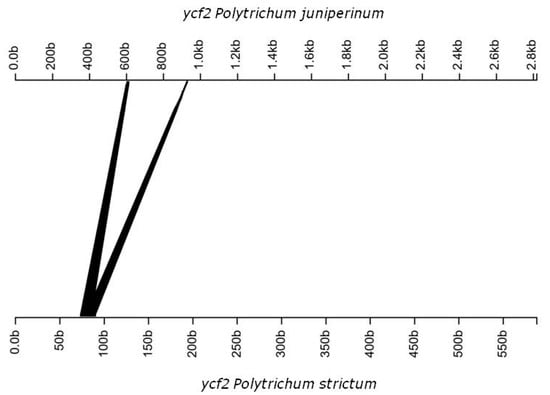
Figure 3.
Linear model of ycf2 gene inversion in Polytrichum juniperinum.
With these results we have so far, it is possible, but not confirmed yet, that the genes psaJ, psaM, atpE, rpl36, and rps14 may be absent in the two assembled genomes, but they have been reported in P. patens [22]. Other genes were analyzed separately, as they were not found with the tools used for annotation in this study. For example, psaM and ccsA cpDNA genes in P. juniperinum and psaI, rpl23, rpl32, rps7, ycf4, ccsA, matK, and certain tRNAs in P. strictum were found only with BLAST [15] by using the total genome of Polytrichaceae and a lower e-value (10−5). It is possible that these regions were not sequenced and therefore not included in the percentage of genome coverage or these genes probably have a high degree of rearrangement (deletions, tandem duplications, and inversions) and substitutions. Scaffold Builder assembler version 2.2 is not effective when the sequences have a high degree of rearrangements [23], and the sequences of these genes and sequences of the genome have at least 80% identity. These genes were not accounted for during our analysis, and further studies on the presence/absence of these genes in Polytrichaceae mosses are required.
The P. juniperinum mitochondrial genome (mtDNA) has a total of 88,021 bp and 41.4% GC content. In total, this genome contains 67 genes, including 2 rRNA genes (1 rnl and 1 rns), 19 tRNAs, 3 rRNAs, 3 open reading frames (ORFs; ORF533, ORF622, and ORF187), and 12 protein-coding genes related to mitochondrial oxidative metabolism. Among these, 12 ribosomal proteins (4 rpl and 8 rps) with absence of rps10. The P. strictum mtDNA has a total of 58,896 bp and 41.1% GC content. The genome contains a total of 62 genes, including 2 rRNA genes as in P. juniperinum (1 rnl and 1 rns), 19 tRNAs, 3 rRNAs, 3 ORFs (ORF533, ORF622, and ORF187), 13 protein-coding genes related to mitochondrial oxidative metabolism, and 13 ribosomal proteins (4 rpl and 9 rps). The rps10 gene that encoded a protein from the 40S subunit of the ribosome was not found in Antarctic Polytrichaceae, apparently that region was lost before the split of mosses lineage since this absence is also evident in Tetraphidaceae [21] and Funariaceae [24]. The nad7 pseudogene in Marchantia polymorpha and ORF187 is frequently observed in the mtDNA of M. polymorpha [25] and contradictorily does not occur in Tetraphis pellucida but seems to be present in the Polytrichum species studied (Table 2). The BLAST analyses showed that nad7 and ORF187 has 55.3% and 43.8% of similarity, respectively, in both species studied by us. The nad7 gene from P. juniperinum showed 98% identity with its homologue in Sanionia uncinata, and P. strictum showed 97.4% identity with its homologue in Atrichum angustatum. ORF187 in P. juniperinum showed 96.8% identity with ORF187 in Marchantia paleacea, and P. strictum showed 98.6% identity with its homologue in A. angustatum. This suggests that, in relation to the analysis of ycf66, nad7, and ORF187, for both Polytrichum samples, seems that these genes evolved independently, presenting significant mismatch and gaps in the sequence between the homologues in the different species. This can occur due to different types of RNA editing events during the evolution in the two species. Other potential causes of these differences include a gene duplication event leading paralogue genes in the Polytrichum species, although we did not find obvious signatures for these affirmations in our data. However, complementary analyses on the substitution rates and the type of selection pressure on each gene reported are necessary to verify the hypothesis of different type RNA editing event or paralogy between the homologues. The genes rps19, nad3, sdh3, atp8, and atp9, so far, were not found in the mtDNA of P. strictum, similar to the cpDNA.
With respect to mtDNA it is evident that P. juniperinum and P. strictum share many blocks with P. patens (Funaraceae) (Figure 4). Rearrangements were not observed in the mitochondrial genome.
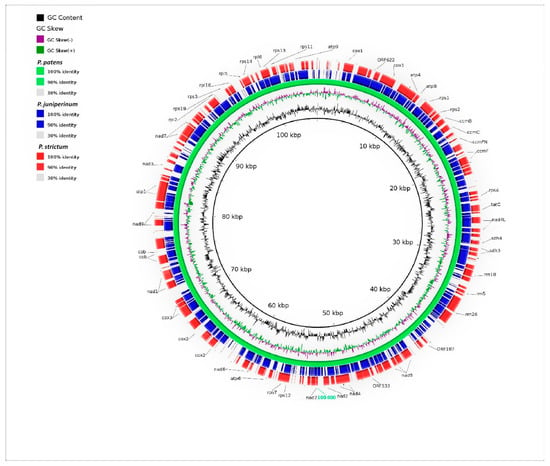
Figure 4.
Blast Ring Image Generator output image of the draft mitochondrial genome. The internal rings represent the Physcomitrella patens genome (green). Basic Local Alignment Search Tool match of Polytrichum juniperinum and Polytrichum strictum are in blue and red gradient, respectively. The legend showing color gradient for percentage similarity between the reference and Polytrichum species. The referred mitochondrial genes are indicated around the map. The innermost rings show the GC skew (purple/green) and GC content (black).
The differences in the gene content of the cpDNAs and mtDNAs of the three classes of bryophytes, including the representatives of the family Polytrichaceae and seed plants, are summarized in Table 2. In the chloroplast and mitochondrial genomes, the two algal lineages diverge with respect to the content of the preserved genes and those that have been lost; this shows how the algal lineage varies with respect to both size and gene content because of the various rearrangements that occurred during evolution [26]. Marchantia polymorpha with its large-sized cpDNA [27] and mtDNA [28] genomes remains with some unknown genes compared with other species, including many ORFs predicted as possible genes, as example the ORF187 that is shared with Polytrichum but not T. pellucida. Marchantia polymorpha share the lack of petN in the cpDNA with some mosses, exceptly with P. patens. This species has the mitochondrial nad7 as a pseudogene; Anthoceros formosae Steph [29] has maturase K and rps15 as pseudogenes, characterizing these two pseudogenes in cpDNA of Anthocerotophyta [30]. In addition, nad7 from T. pellucida is considered a pseudogene [8]. The mosses share practically the same lack of gene among their representatives; only T. pellucida shows a lack of rps10 and ORF187 in the mtDNA. Seed plants usually have gene contents that apparently do not differ substantially, and previous studies showed that gene loss in plastids is associated with an increase in parasitism [31,32].
3.3. Phylogenetic Analysis: Chloroplast and Mitochondria
The phylogenetic analysis of the chloroplast included all assembly sequences from both species genomes. The plastid genomes of eight species, four species from the division Marchantiophyta, two species from Anthocerophyta, and the two Polytrichum species, were included in the analysis. Marchantia polymorpha L. were included as the outgroup, due to be the sister group of all land plants groups, as mosses and hornworts [33,34]. The majority-rule tree constructed with the plastid genomes showed branching into three clades, which is consistent with the taxonomic classification of the division Bryophyta sensu lato and Marchantia polymorpha as the sister group (Figure 4). Peristomate mosses [35] were resolved as a clade supported by 0.99 Bayesian posterior probability (pP), with the representatives of Sphagnum as the basal lineage. A sister relationship between P. juniperinum and P. strictum received high support (pP = 1). Furthermore, these species were grouped with T. pellucida, indicating Polytrichum as an apparent basal lineage; thus, the grouping characterizes the nematodontous mosses [8,36], supported by a pP of 1. This positioning is corroborated by other studies [37,38,39,40,41]. However, other authors have reported Tetraphidopsida as the basal group for Polytrichopsida [26]. The remaining species of the division Marchantiophyta formed a clade with little support (Pp = 0.60); however, this result is consistent with the phylogenomic study of Qiu et al [34]. Finally, Anthocerophyta species Nothoceros aenigmaticus J.C. Villarreal & K.D. McFarland and Anthoceros angustus Steph. formed a supported group (Pp = 0.93), and this is consistent with the findings of Qiu et al [34]. The tree topology showed no conflicting clade, corroborating the results of other authors [40,41,42].
The phylogenetic relationship, resulting from the majority rule tree inferred from assembly of mitochondrial genome from Polytrichum species selected for the present study—including 31 moss species, 2 hornwort species, and 4 liverwort species—are shown in Figure 5. The same outgroup species used for chloroplast phylogenomic analysis was chosen for mithochondrial phylogenomics (Marchantia polymorpha). In the present study, the analysed species were placed in three separate clades, one corresponding to Bryophyta; one, Marchantiophyta; and one mixed with Anthocerophyta and the moss species Ptychomnion cygnisetum (Müll. Hal.) Kindb. The two representative species of Antarctic Polytrichum are closely related moss species, forming a supported clade (Pp = 1) but not grouped in the same branch. The other Polytrichaceae species included in the present analysis, Atrichum angustatum, was grouped with P. strictum to form a close clade. The topology of the tree not according to that proposed by Liu et al. [38,43,44].
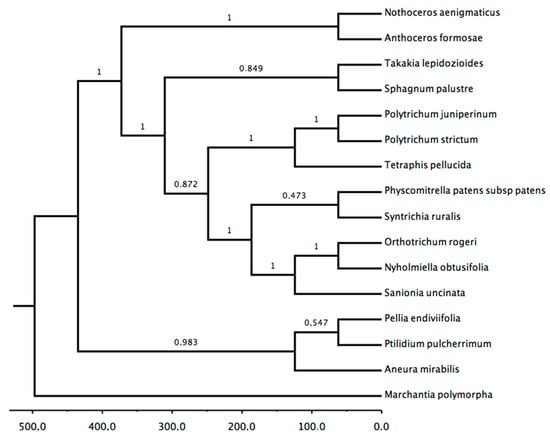
Figure 5.
Maximum clade credibility tree created using Bayesian analysis of the chloroplast gene dataset. The robustness of each node is represented by a posterior probability value (Pp) that varies between 0 and 1 and was obtained after 10,000,000 Monte Carlo Markov chains (MCMC). The tree was re-root using M. polymorpha as outgroup, due to be the sister group of all land plants groups. Time scale root age estimated in 497 ma [19].
4. Discussion
The chloroplast genome of P. juniperinum and P. strictum had a smaller cpDNA than those of some moss, liverwort, and hornwort species deposited in NCBI database [32,45]. The smaller size of Polytrichum genomes, obtained until now in the present study, might reflect the life history [46], evolutionary affiliation [47], and geographical distribution [48] of the species. The latter is more important here, as the Antarctic is geographically isolated continent and their hardly conditions contributing to a selective force [49] that can affects genome size variation, but further studies, including samples from other climatic region become necessary to assess whether those conditions are really affecting the size of the genomes of these species. However, this small size does not interfere with the G + C content, which is high. The plastid genomes of the closest Polytrichum species have a GC percentage of 28% to 33% [21,22] and those of seed plants range between 34% and 40% [50]. Cai et al., observed high G + C contents in the chloroplast coding regions, and certain regions had higher percentages than others, such as the IR region with its four genes with high levels of G and C [51]. Thus, the distribution of GC content in the chloroplast is unequal, and, perhaps, the higher content presented by the Polytrichaceae species refers to the significant number of coding regions scaffolded. The mitochondria of Polytrichaceae are smaller size than those of other mosses and similar to the mtDNA of Buxbaumia aphylla Hedw.; the size of the mitochondrial genome of mosses is between 100,000 and 141,000 bp, and Anthocerophyta species have the largest mitochondrial genome (209,482 bp). However, we must take into account that our results refer to only one draft of mt genomes, which further studies can contribute to whole picture about the size of these genomes. Despite the small size of the mitochondrial genome, % GC is consistent with the mitochondrial GC content of other moss species [21,24,52].
The gene content of cpDNA in the two Polytrichaceae genomes was similar to that of Tetraphis pellucida, however, for Polytrichaceae species, so far, no maturase K have been found in their genome; further studies based in overall Polytrichum species both from Antarctica and other regions are required to evaluate this loss. The rpoA gene seems to have been lost in Polytrichaceae. The absence of this gene had been reported in T. pellucida as well as in all arthrodontous groups [53,54]. Goffinet et al. [55] showed that rpoA does not seem to have been lost in Polytrichum pallidisetum Funck. However, we did not identify this gene, and it is possible that this gene has been lost or translocated in the Antarctic Polytrichaceae species, but only complementary data could confirm or not this hypothesis. According to Sheveleva et al. [56], the presence of rpoA gene is quite variable from species to species. The membrane thylakoid gene ycf66 is absent in A. formosae [30,57,58] but remains more stable in Polytrichaceae than in ferns [59]. Only two species of bryophytes are currently known to lack the petN gene [20,21], part of the photosynthetic cytochrome b6lf complex in the chloroplast, and it is possible, according Oliver et al., that another nuclear-encoded gene product performs the same function as a subunit of the complex [20].
The overall gene content of the mitochondrial genome from the two Polytrichaceae species is close to that observed in P. patens (Figure 4). The rps10 gene seems be absent in the Polytrichaceae mitochondrial genome but is present in P. patens. Adams et al., reported that rps10 has been frequently lost (26 times) and transferred to the nucleus of 277 diverse angiosperms, and they suggest that the gene loss is a frequent event [60]. The mitochondrial genes that seemed to remain in the Polytrichaceae species are nad7 and ORF187; these were identified initially in M. polymorpha [28] and later in P. patens mtDNA [24]. Absence of the nad7 mitochondrial gene in the Nicotiana sylvestris Speg. CMSII mutant caused an abnormal phenotype, poor growth, and male sterility [61]. In the Antarctic Polytrichaceae species, the nad7 gene perhaps has a key role in sustaining the phenotype.
Some genes seem to have been lost multiple times in the chloroplast and mitochondrial genomes during evolution [50], and other genes appear to be present or absent only in particular clades. For example, diverse genes are lacking in mosses and liverworts, such as rps16; however, the gene is present in hornworts and some vascular plants. The gene psaM is absent in three polypoid ferns (Adiantum capillus-veneris L., Cheilanthes lindheimeri Hook., and Pteridium aquilinum (L.) Kuhn.) as well as two Selaginella plastomes, and most of the seed plant plastomes. Seed plant plastomes as well as two Selaginella plastomes lack rpl21. In angiosperms, most of the gene transfer to the nuclear genome affects the subunits of the ribosomal proteins as rps and rpl [60]. In contrast, some genes remain present, such as the plastid gene ycf66, that seems to be an independent loss in multiple clades of land plants, including hornworts, ferns, and seed plants [62]. Gene transfer is a continuous event in plant evolution, and this is promoted possibly by high-frequency translocation of gene-rich organelle DNA into the nucleus and the relatively rare, or entirely absent, transfer of DNA encoding complete genes from the nucleus to the organelles [63].
The chloroplast and mitochondrial genomes of P. juniperinum and P. strictum are identical with respect to overall gene content and structure, as shown in the maps; however, P. strictum genomes show lower degree of sinteny with the reference. We observed variations in the chloroplast genomes, for example, inversions. Inversions represent a type of rearrangement, and one gene inversions were observed in the cpDNA of P. juniperinum (Figure 3), which is not shared with P. patens/P. strictum, and one inversion between the cpDNAs of P. strictum (Figure 2) and P. patens/P. juniperinum. The gene content and gene arrangement of the chloroplast are highly conserved in land plants [64]. Large inversions and other chloroplast genome rearrangements are relatively uncommon among land plants [65], but small inversions are common and widespread in the plant plastid genomes and have been reported in a variety of plants, including bryophytes [66,67,68]. Generally, such small inversions provide a rather interesting phylogenetic marker between species, but also a vision of the relationships among groups. These inversions seem restricted to the species and do not characterize the genera. The mitochondrial genome shows conservation between Antarctic Polytrichum species. Previous studies have shown that the structural evolution of the mitochondrial genome is highly conservative not only within each individual lineage but also across mosses; however, this is most evident when compared with more distant orders within the large group Bryophyta that shows some rearrangements that are very conserved [50]. The occurrence of some rearrangements was observed between Marchantia polymorpha mtDNA and P. patens, as these species diverged more than 375 million years ago [69].
Over the last few decades, single-gene phylogenetic analyses have served as powerful tools for reconstructing the evolutionary history of every major lineage of life on Earth [70]. Indeed, with next-generation sequencing technologies, complete plastome sequences are now being fastest generated [71,72,73]. We sought to analysis the phylogenetic positions of P. juniperinum and P. strictum by using the plastid and mitochondrial gene data of representative moss families, hornworts, and liverworts deposited in GenBank. We wanted to form a hypothesis on the origin of the P. strictum, since there is oodles debate on its origin and definition as a species or variant of P. juniperinum. Polytrichum strictum has morphological characteristics similar to those of P. juniperinum [74,75,76,77] and it differs from P. juniperinum in that it occurs in habitats in the north, such as wetlands (North America), and has, among other morphological characteristics, a remarkable coverage of white rhizoids [78]. Bell and Hyvönen conducted a study on the phylogeny of mosses of the class Polytrichopsida and proposed that the origin of P. strictum (samples used were from Chile and Finland) could be from a cross-linking event [4]. For these authors, P. strictum could be the product of hybridization between the P. juniperinum lineage (sample used from Finland) and a basal lineage of another Polytrichaceae representative. According to the topology presented by the study, the samples of P. strictum were grouped into the same branch, and P. juniperinum appears in a sister branch of P. strictum, suggesting this species as maternal ancestor of P. strictum. In the present study, despite the low number of samples studied, ours results corroborating this ancestry. Because of the lack of well-supported resolution for the positions of Polytrichum hyperboreum R. Br. and P. piliferum, one of these species or a related extinct taxon could easily be the paternal progenitor. In another study on Polytrichales, molecular and morphological data suggested the grouping of P. juniperinum and P. piliferum [40].
The phylogenetic analysis of the partial data from the chloroplast presented a substantial support branch (Figure 5). The Bayesian analysis was selected, as it would be more effective for a large amount of data used in the phylogenetic analysis. The nematodontous mosses are grouped in the same clade, suggesting that P. strictum is a sister of P. juniperinum and T. pellucida appears outside of these grouping. This topology is consistent with that reported by several authors who study nematodontous mosses [21] and that of Cox et al., with respect to the earliest divergence from Tetraphidales and Polytrichales [79]. The Bryopsida clade comprising Nyholmiella obtusifolia (Brid.) Holmen & E. Warncke, Orthotrichum rogeri Brid, Syntrichia ruralis, S. uncinata, and Takakia lepidozioides S. Hatt. & Inoue was supported; however, some authors have proposed different topologies for the class Bryopsida [79,80,81,82]. Although a smaller number of chloroplast genomes than mitochondrial genomes are available for moss species, the phylogenetic positioning of some branches could be reconstructed with a larger amount of data. Whole-genome phylogeny has been shown to be congruent with respect to most of the topologies already inferred for bryophytes, especially when compared to the approaches where only some regions—such as psaA, atpH, atpI, chlL, rbcL, and rpl16 for chloroplast and atp1, ccmB, cob, nad3, nad4, rpl5, rpl6, rps1, and rps11 for mitochondria—are used for the resolution of evolutionary relations [83,84].
Analysis of the mitochondrial genes showed a high branch support for most nodes and little support in some branches, such as the node of Racomitrium and Codriophorus but even so agrees partial with Sawicki et al., that grouping species from Grimniaceae family [85]. The same occurred with the node for Climacium americanum Brid., S. uncinata, Hypnum imponens Hedw. with Orthotrichaceae. Polytrichaceae representatives appear to form a clade, but Atrichum angustatum appears inside this clade, close to P. strictum. Although A. angustatum is a more basal species in Polytrichaceae phylogeny [4], here, the species seems to be placed incorrectly in the mitochondrial tree. However, this result can generate two interpretations, the first involving a possible misunderstanding due to the reference genome used in assembling the plastids sequenced for the Polytrichum species used since the reference was based on the genome of P. patens; this may induce the resulting genomes to be more similar to the genomes of P. patens than A. angustatum. Particularly, recent literature gives assurances of mtDNA conservation among the moss lineage, which lowers the expectations of the results obtained from an assembly error [25,43,52]. A second interpretation would be that mtDNA ancestry is distinct for Polytrichum juniperinum and Polytrichum strictum, consequently the resolution of this relationship will remain unclear until more genomic data are generated for inclusion in this analysis, although we have obtained good coverage of the mitochondrial genome
Our mitochondrial phylogenomic tree (Figure 6) does not match the reconstructed plastid tree (Figure 5) presenting another topology. Differential inheritance of organelles in the same cytoplasm can break the typically expected linkage equilibrium between the chloroplast and mitochondrion [86,87,88] and if this happens, then phylogenetic reconstructions of these two organellar genomes can conflict. The uniparental inheritance in Rhizomnium moss both for chloroplast as mitochondria genomes has been previously reported [89]. In contrast to higher plants, there have been few studies on organellar inheritance in bryophytes [90]; however, the maternal inheritance of the chloroplast in mosses has been reported [89,91]. Therefore, the stasis on the mitochondria genome evolution in mosses should also be taken into account. Liu et al., reports the stasis for mt genomes in mosses, suggesting that the mt genome structure remained virtually frozen for 350 My [29], which may contribute to the distinct topology in the phylogeny obtained from mitochondrial genome data. Although the various gene losses or the psedogenizations identified in the mitochondrial genomes of mosses [43] may also influence these ambiguous topologies, such as the approximation of Ptychmonium cygnisetum and hornworts in our phylogenomic analysis for mt genomic data. A previous study reported that incongruence can be caused by a very small number of characters that are in conflict with other sources of data and excluding part of the data would be warranted only if we knew a priori which part of our data is unreliable [40]. Potential incongruence between chloroplast DNA and mitochondrial DNA markers has been reported [92,93].
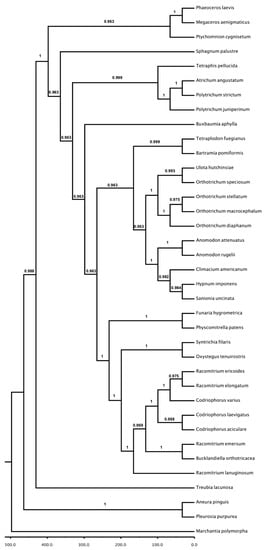
Figure 6.
Maximum clade credibility tree obtained using Bayesian analysis of the mitochondrial gene dataset. The robustness of each node is represented by the posterior probability value obtained after 10,000,000 Monte Carlo Markov chains (MCMC). The tree was reroot using M. polymorpha as outgroup due to be the sister group of all land plants groups. Time scale root age estimated in 497 ma [19].
Molecular phylogenies derived from plastidial, mitochondrial, and nuclear plant genomes can provide insight into the evolutionary history of plant groups influenced by reticulation events [94]. In this study, the chloroplast phylogeny suggests that P. juniperinum is a sister species of P. strictum, at least from Antarctic samples studied; however, the mitochondrial phylogeny suggests P. juniperinum as a maternal ancestor for P. strictum, as well as in study reported by Bell and Hyvönen for both Polytrichum species diverse world regions [4]. This can indicate that the structure of mitochondria remained virtually frozen in Antarctic Polytrichum. However, to confirm the suggested hypothesis, it is necessary include the other all Polytrichum species samples collected from different locations to study the distribution of the species. Currently, constructing a phylogeny for a group of poorly studied organisms requires substantial research. This study present contributes to a preliminary understanding of plastomes and chondriosomes evolution in the family Polytrichaceae, and so far, reveals prelude information that allows the distinction between P. juniperinum and P. strictum from a molecular scenery.
5. Nucleotide Sequence Accession Numbers
This draft genome BioProject has been deposited at GenBank under accession number SUB2397616. The genome accession numbers are KY795004, KY795005, KY795006, and KY795007 from Polytrichum juniperinum cpDNA and mtDNA and Polytrichum strictum cpDNA and mtDNA, respectively.
Author Contributions
K.E.J.d.F carried out experiments in both Polytrichum juniperinum and Polytrichum strictum, carried out the DNA extraction for sequencing analysis, performed the initial genome assembly, annotation of the partial genome assembled and wrote the manuscript with assistance from the co-authors. G.F.M carried out the initial bioinformatic analysis. E.R.P.C. carried out the initial phylogemic analysis. L.F.W.R. designed and carried out the Next Generation Sequencing. A.B.P. co-directed the project and carried. F.C.V. conceived and co-directed the project, designed and carried out the sample collection, the evolutionary analysis, and genome assembly.
Funding
This study was supported by the Brazilian Antarctic Program through the National Council for Research and Development—CNPq (process no. 574018/2008); Research Foundation of the State of Rio de Janeiro—FAPERJ (process E-26/170.023/2008); Ministry of Environment—MMA, Ministry of Science and Technology—MCT; and Interministerial Commission for Sea Resources—CIRM, whom the authors thank for the licenses to collect in Antarctica, as well as the financial and logistic support required to perform this study.
Acknowledgments
The authors would like to thank Paulo Eduardo Aguiar Saraiva Camara and Micheline Carvalho-Silva for the review of the text and ideas presented in this paper. The first author thanks the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for the scholarship granted during the accomplishment of the present research.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ochyra, R. The Moss Flora of King George Island Antarctic; Polish Academy of Sciences, W. Szafer Institute of Botany: Cracow, Poland, 1998. [Google Scholar]
- Greene, S.W.; Greene, D.M.; Brown, P.D.; Pacey, J.M. Antarctic Moss Flora: 1. The Genera Andreaea, Pohlia, Polytrichum, Psilopilum and Sarconeurum, 64th ed.; British Antartic Survey: London, UK, 1970. [Google Scholar]
- Mishler, B.D.; Churchill, S.P. A Cladistic Approach to the Phylogeny of the “Bryophytes”. Brittonia 1984, 36, 406–424. [Google Scholar] [CrossRef]
- Bell, N.E.; Hyvönen, J.P. Phylogeny of the moss class Polytrichopsida (BRYOPHYTA): Generic level structure and incongruent gene trees. Mol. Phylogenet. Evol. 2010, 55, 381–398. [Google Scholar] [CrossRef] [PubMed]
- Nock, C.; Waters, D.L.; Edwards, M.A.; Bowen, S.G.; Rice, N.; Cordeiro, G.M.; Henry, R.J. Chloroplast genome sequences from total DNA for exploring plant relationships. Plant Biotechnol. J. 2010, 9, 328–333. [Google Scholar] [CrossRef] [PubMed]
- Atherton, R.A.; McComish, B.J.; Shepherd, L.D.; Berry, L.A.; Albert, N.W.; Lockhart, P.J. Whole genome sequencing of enriched chloroplast DNA using the Illumina GAII platform. Plant Methods 2010, 6, 22. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.J.; Suchard, M.A.; Xie, D.; Rambaut, A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 2012, 29, 1969–1973. [Google Scholar] [CrossRef] [PubMed]
- Leggett, R.M.; Ramirez-Gonzalez, R.H.; Clavijo, B.; Waite, D.; Davey, R.P. Sequencing quality assessment tools to enable data-driven informatics for high throughput genomics. Front. Genet. 2013, 4, 288. [Google Scholar] [CrossRef] [PubMed]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008, 18, 821–829. [Google Scholar] [CrossRef] [PubMed]
- Chikhi, R.; Medvedev, P. Informed and automated k-mer size selection for genome assembly. Bioinformatics 2014, 30, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Benson, D.A.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Wheeler, D.L. GenBank. Nucleic Acids Res. 2005, 33, 34–38. [Google Scholar] [CrossRef] [PubMed]
- Wyman, S.K.; Jansen, R.K.; Boore, J.L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 2004, 20, 3252–3255. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Shi, L.; Zhu, Y.; Chen, H.; Zhang, J.; Lin, X.; Guan, X. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genom. 2012, 13, 715. [Google Scholar] [CrossRef] [PubMed]
- Alverson, A.J.; Wei, X.; Rice, D.W.; Stern, D.B.; Barry, K.; Palmer, J.D. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). Mol. Biol. Evol. 2010, 27, 1436–1448. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acid Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Alikhan, N.F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetic Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Posada, D. JModelTest: Phylogenetic model averaging. Mol. Biol. Evol. 2008, 25, 1253–1256. [Google Scholar] [CrossRef] [PubMed]
- Magallón, S.; Hilu, K.W. The Timetree of Life; Hedges, S.B., Kumar, S., Eds.; Oxford University Press: Oxford, UK, 2009. [Google Scholar]
- Oliver, M.J.; Murdock, A.G.; Mishler, B.D.; Kuehl, J.V.; Boore, J.L.; Mandoli, D.F.; Everett, K.D.; Wolf, P.G.; Duffy, A.M.; Karol, K.G. Chloroplast genome sequence of the moss Tortula ruralis: Gene content, polymorphism, and structural arrangement relative to other green plant chloroplast genomes. BMC Genom. 2010, 11, 143. [Google Scholar] [CrossRef] [PubMed]
- Bell, N.E.; Boore, J.L.; Mishler, B.D.; Hyvönen, J. Organellar genomes of the four-toothed moss. Tetraphis pellucida. BMC Genom. 2014, 15, 383. [Google Scholar]
- Sugiura, C.; Kobayashi, Y.; Aoki, S.; Sugita, C.; Sugita, M. Complete chloroplast DNA sequence of the moss Physcomitrella patens: Evidence for the loss and relocation of rpoA from the chloroplast to the nucleus. Nucleic Acids Res. 2003, 31, 5324–5331. [Google Scholar] [CrossRef] [PubMed]
- Silva, G.G.; Dutilh, B.E.; Matthews, T.D.; Elkins, K.; Schmieder, R.; Dinsdale, E.A.; Edwards, R.A. Combining de novo and reference-guided assembly with scaffold builder. Source Code Biol. Med. 2013, 8, 23. [Google Scholar] [CrossRef] [PubMed]
- Terasawa, K.; Odahara, M.; Kabeya, Y.; Kikugawa, T.; Sekine, Y.; Fujiwara, M.; Sato, N. The mitochondrial genome of the moss Physcomitrella patens sheds new light on mitochondrial evolution in land plants. Mol. Biol. Evol. 2007, 24, 699–709. [Google Scholar] [CrossRef] [PubMed]
- Oda, K.; Yamato, K.; Ohta, E.; Nakamura, Y.; Takemura, M.; Nozato, N.; Akashi, K.; Kanegae, T.; Ogura, Y.; Kohchi, T.; et al. Gene organization deduced from the complete sequence of liverwort Marchantia polymorpha mitochondrial DNA: A primitive form of plant mitochondrial genome. J. Mol. Biol. 1992, 223, 1–7. [Google Scholar] [CrossRef]
- Graham, L.E. Green algae to land plants: An evolutionary transition. J. Plant Res. 1996, 109, 241–251. [Google Scholar] [CrossRef]
- Ohyama, K.; Fukuzawa, H.; Kohchi, T.; Shirai, H.; Sano, T.; Sano, S.; Umesono, K.; Shiki, Y.; Takeuchi, M.; Chang, Z.; et al. Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA. Nature 1986, 322, 572–574. [Google Scholar] [CrossRef]
- Ohyama, K. Chloroplast and mitochondrial genomes from a liverwort, Marchantia polymorpha—Gene organization and molecular evolution. Biosci. Biotechnol. Biochem. 1996, 60, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Medina, R.; Goffinet, B. 350 My of mitochondrial genomes stasis in mosses, an early land plant lineage. Mol. Biol. Evol. 2014, 31, 2586–2591. [Google Scholar] [CrossRef] [PubMed]
- Kugita, M.; Kaneko, A.; Yamamoto, Y.; Takeya, Y.; Matsumoto, T.; Yoshinaga, K. The complete nucleotide sequence of the hornwort (Anthoceros formosae) chloroplast genome: Insight into the earliest land plants. Nucleic Acids Res. 2003, 31, 716–721. [Google Scholar] [CrossRef] [PubMed]
- Lockhart, J. Plastid genes that were lost along the road to parasitism. Plant Cell 2013, 25, 3636. [Google Scholar] [CrossRef] [PubMed]
- Daniell, H.; Chase, C.D. Molecular Biology and Biotechnology of Plant Organelles: Chloroplasts and Mitochondria; Springer Science & Business Media: Berlin, Germany, 2007. [Google Scholar]
- Knoop, V. Looking for sense in the nonsense: A short review of non-coding organellar DNA elucidating the phylogeny of bryophytes. Trop. Bryol. 2010, 31, 51–60. [Google Scholar]
- Qiu, Y.L.; Li, L.; Wang, B.; Chen, Z.; Knoop, V.; Groth-Malonek, M.; Dombrovska, O.; Lee, J.; Kent, L.; Rest, J.; et al. The deepest divergences in land plants inferred from phylogenomic evidence. Proc. Natl. Acad. Sci. USA 2006, 103, 15511–15516. [Google Scholar] [CrossRef] [PubMed]
- Edwards, S.R. New Manual of Bryology; The Hattori Botanical Laboratory: Nichican, Japan, 1984. [Google Scholar]
- Magombo, Z.L. The phylogeny of basal peristomate mosses: Evidence from cpDNA, and implications for peristome evolution. Syst. Bot. 2003, 28, 24–38. [Google Scholar]
- Volkmar, U.; Knoop, V. Introducing intron locus cox1i624 for phylogenetic analysis in bryophytes: On the issue of Takakia as sister genus to all other extant mosses. J. Mol. Evol. 2010, 70, 506–518. [Google Scholar] [CrossRef] [PubMed]
- Ligrone, R.; Ducket, J.G. Morphology versus molecules in moss phylogeny: New insights (or controversies) from placental and vascular anatomy in Oedipodium griffithianum. Plant Syst. Evol. 2011, 296, 275–282. [Google Scholar] [CrossRef]
- Cox, C.J.; Goffinet, B.; Shaw, J.; Boles, S.B. Phylogenetic relationships among the mosses based on heterogeneous bayesian analysis of multiple genes from multiple genomic compartments. Syst. Bot. 2004, 29, 234–250. [Google Scholar] [CrossRef]
- Hyvönen, J.; Koskinen, S.; Smith, M.G.L.; Hedderson, T.A.; Stenroos, S. Phylogeny of the Polytrichales (Bryophyta) based on simultaneous analysis of molecular and morphological data. Mol. Phylogenet. Evol. 2004, 31, 915–928. [Google Scholar] [CrossRef] [PubMed]
- Newton, A.E.; Cox, C.J.; Duckett, J.G.; Wheeler, J.A.; Goffinet, B.; Hedderson, T.A.J.; Mishler, B.D. Evolution of the major moss lineages: Phylogenetic analyses based on multiple gene sequences and morphology. Bryologist 2000, 103, 187–211. [Google Scholar] [CrossRef]
- Goffinet, B.; Buck, W.R. Monographs in Systematic Botany from the Missouri Botanical Garden. In Molecular Systematics of Bryophytes; Goffinet, B., Hollowell, V., Magill, R., Eds.; Missouri Botanical Garden Press: St. Louis, MO, USA, 2004; pp. 205–239. [Google Scholar]
- Liu, Y.; Cox, C.J.; Wang, W.; Goffinet, B. Mitochondrial phylogenomics of early land plants: Mitigating the effects of saturation, compositional heterogeneity, and codon-usage bias. Syst. Biol. 2014, 63, 862–878. [Google Scholar] [CrossRef] [PubMed]
- Daniell, H.; Lin, C.S.; Yu, M.; Chang, W.J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef] [PubMed]
- Lane, N.; Martin, W. The energetics of genome complexity. Nature 2010, 467, 929–934. [Google Scholar] [CrossRef] [PubMed]
- Leitch, I.J.; Chase, M.W.; Bennett, M.D. Phylogenetic analysis of DNA c-values provides evidence for a small ancestral genome size in flowering plants. Ann. Bot. 1998, 82, 85–94. [Google Scholar] [CrossRef]
- Vesely, P.; Bures, P.; Smarda, P.; Pavlicek, T. Genome size and DNA base composition of geophytes: The mirror of phenology and ecology? Ann. Bot. 2011, 109, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.-M.; Wang, J.; Feng, L.; Liu, S.; Pang, H.; Qi, L.; Li, J.; Sun, W.Y.; Qiao, W.; Zhang, L.; et al. Inferring the evolutionary mechanism of the chloroplast genome size by comparing whole-chloroplast genome sequences in seed plants. Sci. Rep. 2017, 7, 1555. [Google Scholar]
- Jansen, R.K.; Ruhlman, T.A. Genomics of Chloroplasts and Mitochondria; Bock, R., Knoop, V., Eds.; Springer: New York, NY, USA, 2012; pp. 103–126. [Google Scholar]
- Cai, Z.; Guisinger, M.; Kim, H.G.; Ruck, E.; Blazier, J.C.; McMurtry, V.; Kuehl, J.V.; Boore, J.; Jansen, R.K. Extensive reorganization of the plastid genome of Trifolium subterraneum (Fabaceae) is associated with numerous repeated sequences and novel DNA insertions. J. Mol. Evol. 2008, 67, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xue, J.Y.; Wang, B.; Li, L.; Qiu, Y.L. The mitochondrial genomes of the early land plants Treubia lacunosa and Anomodon rugelii: Dynamic and conservative evolution. PLoS ONE 2011, 6, e25836. [Google Scholar] [CrossRef] [PubMed]
- Goffinet, B.; Cox, C.J. Phylogenetic relationships among basal-most arthrodontous mosses with special emphasis on the evolutionary significance of the Funariineae. Bryologist 2000, 103, 212–223. [Google Scholar] [CrossRef]
- Cox, C.J.; Hedderson, T.A.J. Phylogenetic relationships among the ciliate arthrodontous mosses: Evidence from chloroplast and nuclear DNA sequences. Plant Syst. Evol. 1999, 215, 119–139. [Google Scholar] [CrossRef]
- Goffinet, B.; Wickett, N.J.; Shaw, A.J.; Cox, C.J. Phylogenetic significance of the rpoA loss in the chloroplast genome of mosses. Taxon 2005, 54, 353–360. [Google Scholar] [CrossRef]
- Sheveleva, E.V.; Giordani, N.V.; Hallick, R.B. Identification and comparative analysis of the chloroplast α-subunit gene of DNA-dependent RNA polymerase from seven Euglena species. Nucleic Acids Res. 2002, 30, 1247–1254. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Nakamura, Y.; Kaneko, T.; Asamizu, E.; Tabata, S. Complete structure of the chloroplast genome of Arabidopsis thaliana. DNA Res. 1999, 6, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Wakasugi, T.; Nagai, T.; Kapoor, M.; Sugita, M.; Ito, M.; Ito, S.; Tsudzuki, J.; Nakashima, K.; Tsudzuki, T.; Suzuki, Y.; et al. Complete nucleotide sequence of the chloroplast genome from the green alga Chlorella vulgaris: The existence of genes possibly involved in chloroplast division. Proc. Natl. Acad. Sci. USA 1997, 94, 5967–5972. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Zhou, Y.; Wang, Z.W.; Su, Y.J.; Wang, T. Evolution of the rpoB-psbZ region in fern plastid genomes: Notable structural rearrangements and highly variable intergenic spacers. BMC Plant Biol. 2011, 11, 64. [Google Scholar] [CrossRef] [PubMed]
- Adams, K.L.; Daley, D.O.; Qiu, Y.L.; Whelan, J.; Palmer, J.D. Repeated, recent and diverse transfers of a mitochondrial gene to the nucleus in flowering plants. Nature 2000, 408, 354–357. [Google Scholar] [CrossRef] [PubMed]
- Lelandais, C.; Albert, B.; Gutierres, S.; De Paepe, R.; Godelle, B.; Vedel, F.; Chétrit, P. Organization and expression of the mitochondrial genome in the Nicotiana sylvestris CMSII mutant. Genetics 1998, 150, 873–882. [Google Scholar] [PubMed]
- Gao, L.; Yi, X.; Yang, Y.-X.; Su, Y.-J.; Wang, T. Complete chloroplast genome sequence of a tree fern Alsophila spinulosa: Insights into evolutionary changes in fern chloroplast genomes. BMC Evol. Biol. 2009, 9, 130. [Google Scholar] [CrossRef] [PubMed]
- Doolittle, W.E. You are what you eat: A gene transfer ratchet could account for bacterial genes in eukaryotic nuclear genomes. Trends Genet. 1998, 14, 307–311. [Google Scholar] [CrossRef]
- Palmer, J.D. The Molecular Biology of Plastids; Hermann, R.G., Ed.; Springer: Berlin, Germany, 1991; pp. 5–53. [Google Scholar]
- Downie, S.R.; Palmer, J.D. Molecular Systematics of Plants; Soltis, P.S., Soltis, D.E., Doyle, J.J., Eds.; Chapman and Hall: New York, NY, USA; London, UK, 1992; pp. 14–35. [Google Scholar]
- Quandt, D.; Müller, K.; Huttunen, S. Characterisation of the chloroplast DNA psbT-H region and the influence of dyad symmetrical elements on phylogenetic reconstructions. Plant Biol. 2003, 5, 400–410. [Google Scholar] [CrossRef]
- Huttunen, S.; Ignatov, M.S. Phylogeny of the Brachytheciaceae (Bryophyta) based on morphology and sequence level data. Cladistics 2004, 20, 151–183. [Google Scholar] [CrossRef]
- Hernandez-Maqueda, R.; Quandt, D.; Werner, O.; Munoz, J. Phylogeny and classification of the Grimmiaceae/Ptychomitriaceae complex (Bryophyta) inferred from cpDNA. Mol. Phylogenet. Evol. 2008, 46, 863–877. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, B.; Li, L.; Qiu, Y.-L.; Xue, J. Conservative and dynamic evolution of mitochondrial genomes in early land plants. In Genomics of Chloroplasts and Mitochondria; Springer: Dordrecht, The Netherlands, 2012; Volume 7, pp. 159–174. [Google Scholar]
- Donoghue, M.J.; Cracraft, J. Assembling the Tree of Life; Cracraft, J., Donoghue, M.J., Eds.; Oxford University Press: New York, NY, USA, 2004; pp. 1–4. [Google Scholar]
- Cronn, R.; Liston, A.; Parks, M.; Gernandt, D.S.; Shen, R.; Mockler, T. Multiplex sequencing of plant chloroplast genomes using Solexa sequencing-by synthesis technology. Nucleic Acids Res. 2008, 36, e122. [Google Scholar] [CrossRef] [PubMed]
- Wolf, P.G.; Der, J.P.; Duffy, A.M.; Davidson, J.B.; Grusz, A.L.; Pryer, K.M. The evolution of chloroplast genes and genomes in ferns. Plant Mol. Biol. 2011, 76, 251–261. [Google Scholar] [CrossRef] [PubMed]
- Henson, J.; Tischler, G.; Ning, Z. Next-generation sequencing and large genome assemblies. Pharmacogenomics 2012, 13, 901–915. [Google Scholar] [CrossRef] [PubMed]
- Lawton, E. Moss Flora of the Pacific Northwest, 1st ed.; The Hattori Botanical Laboratory: Nichican, Japan, 1971; 389p. [Google Scholar]
- Steere, W.C.; Brassard, G.R. The Mosses of Arctic Alaska, 1st ed.; Cramer Publisher: Lehre, Germany, 1978; 508p. [Google Scholar]
- Koponen, T.; Isoviita, P.; Lammes, T. The Bryophytes of Finland: An Annotated Checklist, 1st ed.; Societas pro fauna et flora Fennica: Helsinki, Finland, 1977; 77p. [Google Scholar]
- Anderson, L.E.; Crum, H.A.; Buck, W.R. List of the mosses of North America and north of Mexico. Bryologist 1990, 93, 448–499. [Google Scholar] [CrossRef]
- Derda, G.S.; Wyatt, R. Genetic variation and population structure in Polytrichum juniperinum and Polytricum strictum (Polytrichaceae). Lindbergia 2003, 28, 23–40. [Google Scholar]
- Cox, C.J.; Goffinet, B.; Wickett, N.J.; Boles, S.B.; Shaw, A.J. Moss diversity: A molecular phylogenetic analysis of genera. Phytotaxa 2010, 9, 175–195. [Google Scholar] [CrossRef]
- La Farge, C.; Mishler, B.D.; Wheeler, J.A.; Wall, D.P.; Johannes, K.; Schaffer, S.; Shaw, A.J. Phylogenetic relationships within the haplolepideous mosses. Bryologist 2000, 103, 257–276. [Google Scholar] [CrossRef]
- De Luna, E.; Newton, A.E.; Withey, A.; Gonzalez, D.; Mishler, B.D. The transition to pleurocarpy: A phylogenetic analysis of the main Diplolepidous lineages based on rbcL sequences and morphology. Bryologist 1999, 102, 634–650. [Google Scholar] [CrossRef]
- De Luna, E.; Buck, W.R.; Akiyama, H.; Arikawa, T.; Tsubota, H.; Gonzalez, D.; Newton, A.E.; Shaw, A.J. Ordinal phylogeny within the Hypnobryalean pleurocarpous mosses inferred from cladistic analyses of three chloroplast DNA sequence data sets: trnL-F, rps4, and rbcL. Bryologist 2000, 103, 242–256. [Google Scholar] [CrossRef]
- Vaidya, G.; Lohman, D.L.; Meier, R. SequenceMatrix: Concatenation software for the fast assembly of multigene datasets with character set and codon information. Cladistics 2010, 27, 171–180. [Google Scholar] [CrossRef]
- Nishiyama, T.; Wolf, P.G.; Kugita, M.; Sinclair, R.B.; Sugita, M.; Sugiura, C.; Wakasugi, T.; Yamada, K.; Yoshinaga, K.; Yamaguchi, K.; et al. Chloroplast phylogeny indicates that bryophytes are monophyletic. Mol. Biol. Evol. 2004, 21, 1813–1819. [Google Scholar] [CrossRef] [PubMed]
- Wahrmund, U.; Rein, T.; Müller, K.F.; Groth-Malonek, M.; Knoop, V. Fifty mosses on five trees: Comparing phylonetic information in three types of non-coding mitochondrial DNA and two chloroplast loci. Plant Syst. Evol. 2009, 282, 241–255. [Google Scholar] [CrossRef]
- Shaw, A.J.; Allen, B.H. Phylogenetic relationships, morphological incongruence, and geographic speciation in the Fontinalaceae (Bryophyta). Mol. Syst. Evol. 2000, 16, 225–237. [Google Scholar] [CrossRef] [PubMed]
- Sawicki, J.; Szczecińska, M.; Bednarek-Ochyra, H.; Ochyra, R. Mitochondrial phylogenomics supports splitting the traditionally conceived genus Racomitrium (Bryophyta: Grimmiaceae). Nova Hedwig. 2015, 100, 293–317. [Google Scholar] [CrossRef]
- Molina, J.; Hazzouri, K.M.; Nickrent, D.; Geisler, M.; Meyer, R.S.; Pentony, M.M.; Flowers, J.M.; Pelser, P.; Barcelona, J.; Inovejas, S.A.; et al. Possible loss of the chloroplast genome in the parasitic flowering plant Rafflesia lagascae (Rafflesiaceae). Mol. Biol. Evol. 2014, 31, 793–803. [Google Scholar] [CrossRef] [PubMed]
- Tsujimura, M.; Mori, N.; Yamagishi, H.; Terachi, T. A possible breakage of linkage disequilibrium between mitochondrial and chloroplast genomes during Emmer and Dinkel wheat evolution. Genome 2013, 56, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Jankowiak, K.; Rybarczyk, A.; Wyatt, R.; Odrzykoski, I.; Pacak, A.; Szweykowska-Kulinska, Z. Organellar inheritance in the allopolyploid moss Rhizomnium pseudopunctatum. Taxon 2005, 54, 383–388. [Google Scholar] [CrossRef]
- Guillon, J.M.; Raquin, C. Maternal inheritance of chloroplasts in the horsetail Equisetum variegatum (Schleich). Curr. Genet. 2000, 37, 53–56. [Google Scholar] [CrossRef] [PubMed]
- Natcheva, R.; Cronberg, N. Maternal transmission of cytoplasmic DNA in interspecific hybrids of peat moss, Sphagnum (Bryophyta). J. Evol. Biol. 2007, 20, 1613–1616. [Google Scholar] [CrossRef] [PubMed]
- Mahoney, L.L.; Quimby, M.L.; Shields, M.E.; Davis, T.M. Mitochondrial DNA transmission, ancestry, and sequences in Fragaria. Acta Hortic. 2010, 859, 301–308. [Google Scholar] [CrossRef]
- Njuguna, W.; Liston, A.; Cronn, R.; Ashman, T.L.; Bassil, N. Insights into phylogeny, sex function and age of Fragaria based on whole chloroplast genome sequencing. Mol. Phylogenet. Evol. 2013, 66, 17–29. [Google Scholar] [CrossRef] [PubMed]
- Govindarajulu, R.; Parks, M.; Tennessen, J.A.; Liston, A.; Ashman, T.L. Comparison of nuclear, plastid, and mitochondrial phylogenies and the origin of wild octaploid strawberry species. Am. J. Bot. 2015, 102, 544–554. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).