Abstract
An amino acid-derived 2-thiohydantoin, 5-(2-methylsulfanylethyl)-3-prop-2-enyl-2-sulfanylideneimidazolidin-4-one, obtained from l-methionine, was synthesized in a two-step reaction protocol with allyl isothiocyanate. The compound was obtained in an 82% yield and was fully structurally characterized by NMR and IR spectroscopy. The crystal structure, molecular packing, and intermolecular interactions were characterized by X-ray diffraction analysis.
1. Introduction
2-Thiohydantoins, as part of the broader hydantoin family, are considered to be valuable and privileged scaffolds in the fields of medicinal chemistry and pharmacology, as well as in material sciences [1]. Hydantoins are known for their applications as clinically approved drugs as anticonvulsants, antiandrogens, muscle relaxants, and antibiotics [2], as well as for their use in various branches of industry as pesticides, preservatives, corrosion inhibitors, and more [3].
The hydantoin scaffold is synthetically available through various possible synthetic strategies and starting substrates, but one of the more convenient routes is synthesis from amino acids [4]. The advantages of this route include mild reaction conditions, high yields, simple reaction workup, and the availability of a large number of substituted and functionalized structures. Hydantoins and amino acids are, in this sense, interchangeable, because not only can hydantoins be synthesized from amino acids, but amino acids can be synthesized from hydantoins. In fact, highly functionalized and structurally complex amino acids can be obtained from hydantoins via hydrolysis, which are often very difficult or even impossible to obtain otherwise [5].
Amino acid-derived 2-thiohydantoins exhibit a variety of biological activities, such as antimicrobial [6], antiviral [7], anticancer [8], antimutagenic [9], and neurotropic [10], and are the subject of numerous preclinical investigations. The body of work conducted so far highlights the significance of these compounds in the effort to obtain novel bioactive compounds and therapeutics. Since hydantoins exhibit a plethora of biological activities and preclinical biological testing consumes a lot of time and resources, it is difficult to screen for the full spectrum of their bioactive potential. It is, therefore, highly beneficial to have published libraries of small molecules that are readily available for in vitro, in vivo, or even in silico screening. For this reason, in this study, we report the synthesis and structural characterization of an l-methionine-derived 2-thiohydantoin derivative, 5-(2-methylsulfanylethyl)-3-prop-2-enyl-2-sulfanylideneimidazolidin-4-one. The structure of the compound is fully characterized by NMR and IR spectroscopy, as well as X-ray diffraction analysis.
2. Results and Discussion
2.1. Synthesis and Spectral Characterization
Synthesis of the amino acid-derived 2-thiohydantoin, 5-(2-methylsulfanylethyl)-3-prop-2-enyl-2-sulfanylideneimidazolidin-4-one is depicted in Scheme 1. The first synthetic step is a conversion of l-methionine 1 to its methyl ester 2 using a methanolic HCl protocol. The reaction of acetyl chloride with methanol generates HCl, and in this acidic methanol environment, amino acids are easily converted to their corresponding methyl ester hydrochlorides. l-Methionine was converted to its methyl ester because methyl esters undergo cyclization to 2-thiohydantoins more easily than amino acids due to the methoxy group being a better leaving group than hydroxyl in the applied reaction conditions.
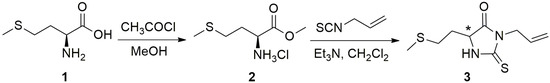
Scheme 1.
Synthesis of 5-(2-methylsulfanylethyl)-3-prop-2-enyl-2-sulfanylideneimidazolidin-4-one.
The second step is the reaction of the methyl ester with allyl isothiocyanate. l-Methionine methyl ester hydrochloride 2 is first relieved of HCl with the addition of Et3N, because the amino group needs to be deprotonated and free for the nucleophilic addition reaction. l-Methionine methyl ester then undergoes an addition and cyclization reaction to form the 2-thiohydantoin product, 5-(2-methylsulfanylethyl)-3-prop-2-enyl-2-sulfanylideneimidazolidin-4-one 3. It is obtained in a high 82% yield.
The 2-thiohydantoin product 3 is expected to be obtained as a perfect racemate, as racemization is known to occur in these types of reaction systems under basic conditions. This is supported by the crystallographic data. A recemization of this type is reported in a paper by Haslak et al., where they examined a reaction of l-alanine methyl ester and aryl isothiocyanates in the presence of triethylamine [11]. They proposed that racemization occurs with triethylamine, after cyclization, as cyclic ureides are known to undergo a ring-opening racemization process in basic conditions.
5-(2-Methylsulfanylethyl)-3-prop-2-enyl-2-sulfanylideneimidazolidin-4-one was structurally characterized by NMR and IR spectroscopy. The spectra can be found in the Supplementary Material. In the 1H NMR spectrum, all expected signals can be observed. The broad singlet of the NH proton of the 2-thiohydantoin ring can be observed at 7.81 ppm. For the sake of clarity, this discussion follows the numbering scheme depicted in Figure 1. At 5.86 ppm, a complex doublet of doublet of triplets from the CH proton of the allyl group double bond (C8) can be observed. The complexity of this signal originates from the coupling of the proton with three chemically distinct neighboring protons in its environment. The triplets in this signal result from coupling with two chemically identical protons from the allyl methylene group C7 (the value of the coupling constant is J = 5.8 Hz and is characteristic for vicinal aliphatic coupling), while further splitting of the signal results from coupling with two chemically inequivalent protons from the terminal allyl double bond CH2 group C9. The coupling constant for the proton that is in the cis position is 10.2 Hz, while the coupling constant for the proton in the trans position is expectedly higher and is 17.2 Hz. A multiplet of the terminal allyl double bond CH2 group C9 can be observed at 5.18–5.32 ppm. The signal is very complex and cannot be fully characterized, but can, for the sake of discussion, be considered as two doublets of doublets for each of the protons. The signal of the C9 proton in the trans position to the proton at C8 is expectedly shifted further downfield at 5.24–5.32 ppm and is more split, while the signal of the cis proton is slightly less shifted at 5.18–5.23 ppm and is less split. Signal of the allyl methylene protons C7 is at 4.43 ppm, seemingly in the form of a doublet, with a coupling constant of J = 6.0 Hz. Upon closer look, it can be observed that the signal is actually a doublet of triplets, due to allylic coupling. The signal of the 2-thiohydantoin ring proton C3 can be observed at 4.28 ppm in the form of a doublet of doublet of doublets, with coupling constants J = 1.2, 4.2, and 7.4 Hz. The proton from the rigid ring system interacts differently with vicinal protons from the C4 methylene group, and thus, the coupling constants are different (4.2 and 7.4 Hz), while the third coupling constant of 1.2 Hz originates from long-range coupling with C5 methylene protons. Other signals in the spectrum originate from the protons from the side chain attached to the C3 carbon of the 2-thiohydantoin ring. The triplet at 2.68 ppm is from the C5 methylene protons, the multiplets at 2.18–2.34 ppm and 1.89–2.10 ppm are from the C4 methylene protons, and the singlet at 2.12 ppm is from the C6 methyl group attached to the sulfur atom.
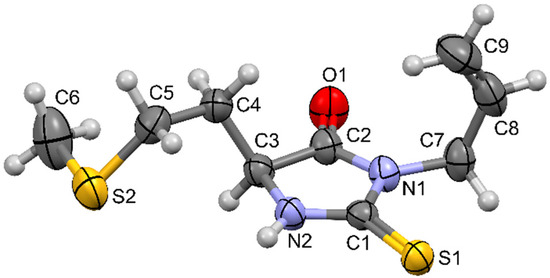
Figure 1.
Molecular structure of 5-(2-methylsulfanylethyl)-3-prop-2-enyl-2-sulfanylideneimidazolidin-4-one and atom numbering scheme.
In the 13C NMR spectrum, all expected signals can be observed. The signals of thiocarbonyl and carbonyl carbons from the 2-thiohydantoin ring are at 183.54 and 173.42 ppm. The allyl double bond carbons C8 and C9 are at 130.50 and 118.58 ppm. 2-Thiohydantoin ring carbon C3 and allyl methylene carbon C7 are at 55.52 and 43.34 ppm, side branch methylene carbons C4 and C5 are at 30.42 and 30.30 ppm, and the C6 methyl carbon is at 15.30 ppm.
2.2. Molecular and Crystal Structure Description
Molecular structure of 5-(2-methylsulfanylethyl)-3-prop-2-enyl-2-sulfanylideneimidazolidin-4-one is depicted in Figure 1. All bond lengths, valence, and torsion angles are within the expected range, as found by comparison with structures available in the Cambridge structural database that have similar structural fragments, using methodology implemented in Mogul [12]. The five-membered 2-thiohydantoin ring is planar, with allyl substituent oriented perpendicular to the ring plane (torsion angle C1–N1–C7–C8 equals −88.7(2)°).
Hirshfeld surface analysis (Figure 2) reveals that most intermolecular contacts are found between hydrogen atoms (51%), followed by S∙∙∙H (26%), O∙∙∙H (12%), and C∙∙∙H (7%). Calculation of enrichment ratios [13] shows that, except H∙∙∙H contacts (EHH = 0.9), all other mentioned are favored in the crystal structure (EOH = 5.8, ESH = 1.2, ECH = 1.2). For the largest proportion of the Hirshfeld surface, it is found that dnorm ≥ 0 holds, meaning that crystal packing does not involve many short intermolecular contacts. The exception is N2–H2∙∙∙S1i hydrogen bond (symmetry operation (i) −x + 2, −y + 1, −z + 1), which shows prominent red patches (dnorm < 0) on the Hirshfeld surface in the proximity of donor and acceptor atoms (Figure 3). In the fingerprint plot, this interaction is visualized as a very sharp pair of spikes. Geometrical parameters of this interaction are N2–H2 = 0.81(2) Å, H2···S1i = 2.56(2) Å, N2···S1i = 3.3696(18) Å, N2–H2···S1i = 174(2)°. Less prominent are red patches corresponding to short contacts C3–H3∙∙∙S1ii (symmetry operation (ii) −x + 1, −y + 1, −z + 1).
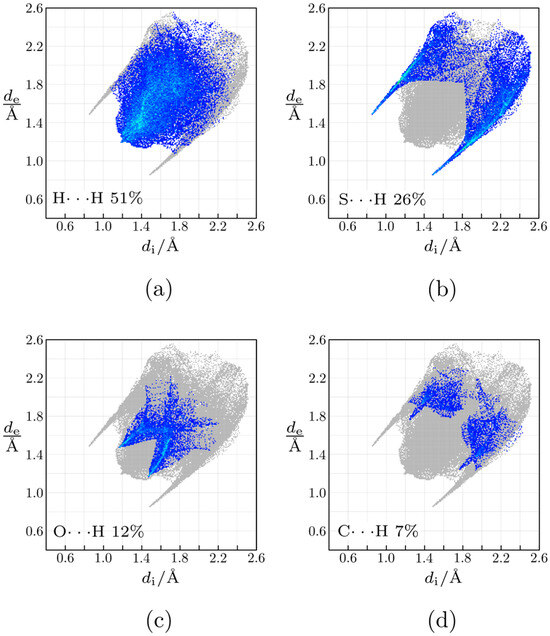
Figure 2.
Fingerprint plots of 5-(2-methylsulfanylethyl)-3-prop-2-enyl-2-sulfanylideneimidazolidin-4-one: (a) H···H contacts; (b) S···H contacts; (c) O···H contacts; (d) C···H contacts.
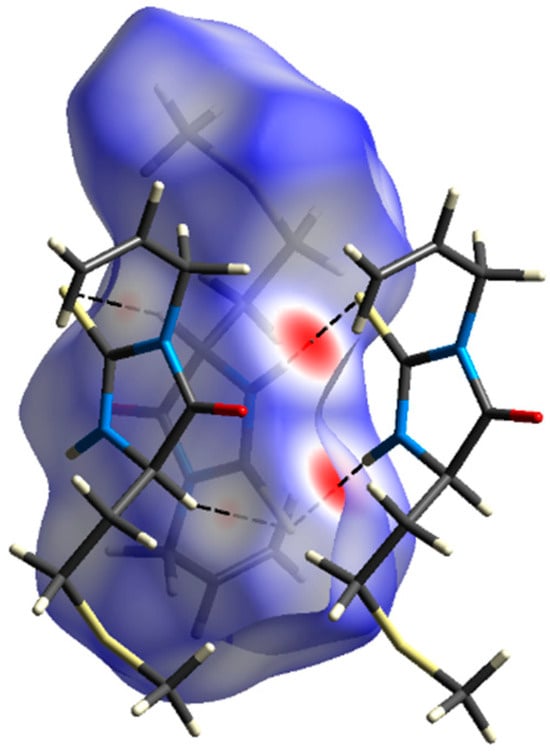
Figure 3.
Hirshfeld surface decorated with dnorm, and two neighboring molecules involved in short contacts.
There are 14 molecules in the first coordination sphere of 5-(2-methylsulfanylethyl)-3-prop-2-enyl-2-sulfanylideneimidazolidin-4-one, associated with 11 unique intermolecular interactions. Their energies are summarized in Table 1. Dissection of the molecular packing on the energetic basis reveals that this crystal structure is of a layered type. Schematically, this is visualized in the energy framework shown in Figure 4.

Table 1.
Summary of intermolecular interaction energies of the unique molecular pairs constituting the first coordination sphere for 5-(2-methylsulfanylethyl)-3-prop-2-enyl-2-sulfanylideneimidazolidin-4-one calculated using the CE-B3LYP model energy.
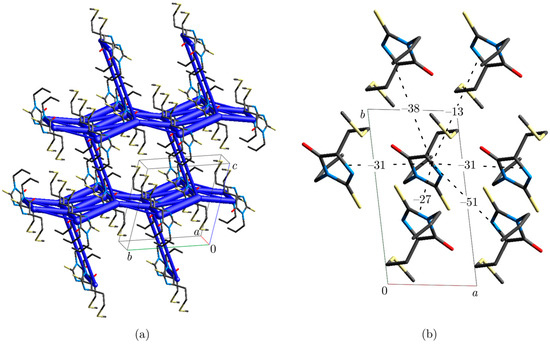
Figure 4.
(a) Energy framework. (b) Fragment of molecular packing in the crystallographic ab plane (hydrogen atoms omitted for clarity); interaction energies are expressed in kJ mol−1.
Every molecule is surrounded by six closest neighbors in the crystallographic ab plane. The strongest interaction (–51 kJ mol−1) is mediated through the mentioned N2–H2···S1i hydrogen bond formed between inversion-related molecules. The high electrostatic contribution to the stabilizing energy of this interaction is expected due to strong electrostatic complementarity between molecules, as visualized by Hirshfeld surfaces of the neighboring molecules decorated with electrostatic potential (Figure 5). Other interactions in this plane have significantly higher dispersion than the electrostatic component, but they can hardly be ascribed to some specific atom–atom interaction type. Their energies are in the range of –13 to −38 kJ mol−1. There is only one interaction between layers that has comparable energy (−20 kJ mol−1) to those found within the layer. This interaction has a similar contribution of electrostatic and dispersion terms.
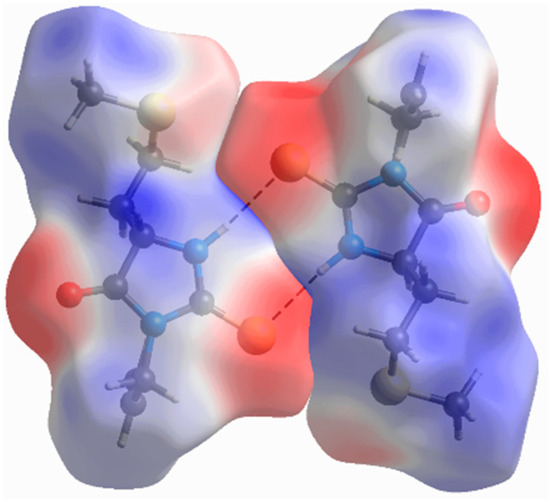
Figure 5.
Molecular dimer that has the strongest binding energy (−51 kJ mol−1). Hirshfeld surfaces are decorated with molecular electrostatic potential in the range from −0.05 to 0.05 Hartree.
The compound crystallizes in the centrosymmetric space group, which implies that the solid phase is obtained as a racemate containing equimolar amounts of both enantiomers.
3. Materials and Methods
3.1. Chemicals
All the reagents used in this study were analytical grade or higher purity, obtained from Sigma–Aldrich (St. Louis, MO, USA) and Merck (Darmstadt, Germany).
3.2. Instrumentations
The 1H NMR spectra were acquired on a Varian Gemini-2000 spectrometer (Palo Alto, CA, USA) at 200 MHz. All chemical shifts are referenced to the solvent dimethylsulfoxide-d6 (DMSO-d6), and downfield shifts were recorded as positive numbers. IR data were obtained using PerkinElmer® Spectrum One FTIR spectrometer (Shelton, CT, USA). Diffraction measurements were performed on a Gemini S (Oxford Diffraction) single crystal X-ray diffractometer (Yarnton, England, UK).
3.3. Synthetic Protocols
3.3.1. General Procedure for the Preparation of l-Methionine Methyl Ester
Amino acid methyl esters were prepared according to a well-known methanolic HCl method. A total of 5 mL of methanol was added to a round-bottom flask and cooled to 0 °C. Acetyl chloride (2 mL) was added slowly to the stirred solution and then stirred for another 20 min at 0 °C to generate methanolic HCl. An amino acid (5 mmol) was added in one portion, and the reaction was stirred overnight at room temperature. The solvent was removed in vacuo, and solid amino acid methyl ester hydrochloride was used without further purification. Ester purity was confirmed by 1H NMR spectroscopy.
3.3.2. General Procedure for the Preparation of 5-(2-Methylsulfanylethyl)-3-prop-2-enyl-2-sulfanylideneimidazolidin-4-one
5-(2-Methylsulfanylethyl)-3-prop-2-enyl-2-sulfanylideneimidazolidin-4-one was synthesized by following a lightly modified published protocol [15]. A mixture of 5 mmol of l-methionine methyl ester, 5 mmol Et3N, and 15 mL of CH2Cl2 was stirred for about 20 min at room temperature until all of the ester was dissolved. Allyl isothiocyanate (5 mmol) was added dropwise, and the reaction mixture was heated under reflux for 7 h. The solution was cooled at room temperature, and the solvent was evaporated. The residue was dissolved in CH2Cl2, washed with water and brine, and dried over anhydrous Na2SO4. The solvent was once again evaporated, leaving a crude solid product that was recrystallized from CH2Cl2/hexane.
3.3.3. Compound Data of 5-(2-Methylsulfanylethyl)-3-prop-2-enyl-2-sulfanylideneimidazolidin-4-one
Light orange needle crystals; Yield 82%; m.p. = 119–120 °C; IR (KBr) νmax: 3169, 3002, 2921, 1741, 1646, 1531, 1432, 1346, 1255, 1165, 923, 650 cm−1; 1H NMR (200 MHz, CDCl3) δ 7.81 (bs, 1H), 5.86 (ddt, J = 5.8, 10.2, 17.2 Hz, 1H), 5.18–5.32 (m, 2H), 4.43 (d, J = 6.0 Hz, 2H), 4.28 (ddd, J = 1.2, 4.2 and 7.4 Hz, 1H), 2.68 (t, J = 7.4 Hz, 2H), 2.18–2.34 (m, 1H), 2.12 (s, 3H), 2.01 (septet, J = 6.8 Hz, 1H) ppm. 13C NMR (50 MHz, CDCl3) δ 183.54, 173.42, 130.50, 118.58, 58.52, 43.34, 30.42, 30.30, 15.30 ppm.
3.4. Crystal Structure Determinations
The monochromatic Mo Kα X-radiation was used as a probe, with the diffractometer operating in a ω-scan mode. Instrument control and data reduction were performed with the CrysAlisPRO [16]. Structure was solved with SHELXT [17] and refined with SHELXL [18]. A summary of crystallographic and refinement details is listed in Table 2. Crystallographic data have been deposited at the Cambridge Crystallographic Data Centre (CCDC number 2474856). Hirshfeld surface analysis and assessment of intermolecular interaction energies were performed with CrystalExplorer [19], using TONTO [20] as the backend program for quantum chemistry calculations.

Table 2.
Crystallographic and refinement details of 5-(2-methylsulfanylethyl)-3-prop-2-enyl-2-sulfanylideneimidazolidin-4-one.
Supplementary Materials
The following supporting information can be downloaded at: Figure S1. IR spectrum of 5-(2-methylsulfanylethyl)-3-prop-2-enyl-2-sulfanylideneimidazolidin-4-one; Figure S2. 1H NMR spectrum of 5-(2-methylsulfanylethyl)-3-prop-2-enyl-2-sulfanylideneimidazolidin-4-one; Figure S3. 13C NMR spectrum of 5-(2-methylsulfanylethyl)-3-prop-2-enyl-2-sulfanylideneimidazolidin-4-one.
Author Contributions
Conceptualization, P.S. and B.Š.; methodology, P.S. and M.V.R.; validation, P.S., M.V.R. and B.Š.; formal analysis, P.S., M.V.R. and B.Š.; investigation, P.S., M.V.R. and B.Š.; writing—original draft preparation, P.S. and M.V.R.; writing—review and editing, P.S., M.V.R. and B.Š.; visualization, P.S. and M.V.R.; supervision, B.Š.; funding acquisition, B.Š. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
The original contributions presented in this study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Acknowledgments
The authors are grateful to the Ministry of Science, Technological Development, and Innovation of the Republic of Serbia for support (Grant Nos. 451-03-136/2025-03/200378 and 451-03-137/2025-03/200125).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Konnert, L.; Lamaty, F.; Martinez, J.; Colacino, E. Recent Advances in the Synthesis of Hydantoins: The State of the Art of a Valuable Scaffold. Chem. Rev. 2017, 117, 13757–13809. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.H.; Kim, S.H.; Shin, D. Recent Applications of Hydantoin and Thiohydantoin in Medicinal Chemistry. Eur. J. Med. Chem. 2019, 164, 517–545. [Google Scholar] [CrossRef] [PubMed]
- Gawas, P.P.; Ramakrishna, B.; Veeraiah, N.; Nutalapati, V. Multifunctional Hydantoins: Recent Advances in Optoelectronics and Medicinal Drugs from Academia to the Chemical Industry. J. Mater. Chem. C 2021, 9, 16341–16377. [Google Scholar] [CrossRef]
- Metwally, M.A.; Abdel-Latif, E. Thiohydantoins: Synthetic Strategies and Chemical Reactions. J. Sulfur Chem. 2012, 33, 229–257. [Google Scholar] [CrossRef]
- Šmit, B.; Rodić, M.; Pavlović, R. Synthesis of Angularly Fused (Homo)Triquinane-Type Hydantoins as Precursors of Bicyclic Prolines. Synth. 2015, 48, 387–393. [Google Scholar] [CrossRef]
- de Carvalho, P.G.C.; Ribeiro, J.M.; Garbin, R.P.B.; Nakazato, G.; Yamada Ogatta, S.F.; de Fátima, Â.; de Lima Ferreira Bispo, M.; Macedo, F. Synthesis and Antimicrobial Activity of Thiohydantoins Obtained from L-Amino Acids. Lett. Drug Des. Discov. 2020, 17, 94–102. [Google Scholar] [CrossRef]
- Rajic, Z.; Zorc, B.; Raic-Malic, S.; Ester, K.; Kralj, M.; Pavelic, K.; Balzarini, J.; De Clercq, E.; Mintas, M. Hydantoin Derivatives of L- and D-Amino Acids: Synthesis and Evaluation of Their Antiviral and Antitumoral Activity. Molecules 2006, 11, 837–848. [Google Scholar] [CrossRef] [PubMed]
- Elbadawi, M.M.; Khodair, A.I.; Awad, M.K.; Kassab, S.E.; Elsaady, M.T.; Abdellatif, K.R.A. Design, Synthesis and Biological Evaluation of Novel Thiohydantoin Derivatives as Antiproliferative Agents: A Combined Experimental and Theoretical Assessments. J. Mol. Struct. 2022, 1249, 131574. [Google Scholar] [CrossRef]
- Takahashi, A.; Matsuoka, H.; Uda, Y. Antimutagenicity of 3-Allyl-5-Substituted 2-Thiohydantoins Derived from Allyl Isothiocyanate and Amino Acids in Salmonella Assay. Environ. Mutagen Res. 2004, 26, 1–8. [Google Scholar] [CrossRef]
- Paronikyan, R.; Grigoryan, A.; Barkhudaryants, I.; Arshakyan, L.; Harutyunyan, A. Synthesis and Neurotropic Activity of New Derivatives of Some Amino Acid Hydantoins and Their Lithium Salts. Bioact. Compd. Health Dis. 2024, 7, 274–288. [Google Scholar] [CrossRef]
- Haslak, Z.P.; Agopcan Cinar, S.; Sarigul Ozbek, S.; Monard, G.; Dogan, I.; Aviyente, V. Elucidation of the Atroposelectivity in the Synthesis of Axially Chiral Thiohydantoin Derivatives. Org. Biomol. Chem. 2020, 18, 2233–2241. [Google Scholar] [CrossRef] [PubMed]
- Bruno, I.J.; Cole, J.C.; Kessler, M.; Luo, J.; Motherwell, W.D.S.; Purkis, L.H.; Smith, B.R.; Taylor, R.; Cooper, R.I.; Harris, S.E.; et al. Retrieval of Crystallographically-Derived Molecular Geometry Information. J. Chem. Inf. Comput. Sci. 2004, 44, 2133–2144. [Google Scholar] [CrossRef] [PubMed]
- Jelsch, C.; Ejsmont, K.; Huder, L. The Enrichment Ratio of Atomic Contacts in Crystals, an Indicator Derived from the Hirshfeld Surface Analysis. IUCrJ 2014, 1, 119–128. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, C.F.; Spackman, P.R.; Jayatilaka, D.; Spackman, M.A. CrystalExplorer Model Energies and Energy Frameworks: Extension to Metal Coordination Compounds, Organic Salts, Solvates and Open-Shell Systems. IUCrJ 2017, 4, 575–587. [Google Scholar] [CrossRef] [PubMed]
- Reyes, S.; Burgess, K. On Formation of Thiohydantoins from Amino Acids under Acylation Conditions. J. Org. Chem. 2006, 71, 2507–2509. [Google Scholar] [CrossRef] [PubMed]
- Rigaku Oxford Diffraction, 1.171.38.46. CrysAlisPro Software System. Rigaku Corporation: Wroclaw, Poland, 2019.
- Sheldrick, G.M. SHELXT–Integrated Space-Group and Crystal-Structure Determination. Acta Crystallogr. Sect. A Found. Adv. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Crystal Structure Refinement with SHELXL. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 3–8. [Google Scholar] [CrossRef]
- Spackman, P.R.; Turner, M.J.; McKinnon, J.J.; Wolff, S.K.; Grimwood, D.J.; Jayatilaka, D.; Spackman, M.A. CrystalExplorer: A Program for Hirshfeld Surface Analysis, Visualization and Quantitative Analysis of Molecular Crystals. J. Appl. Crystallogr. 2021, 54, 1006–1011. [Google Scholar] [CrossRef] [PubMed]
- Jayatilaka, D.; Grimwood, D.J. Tonto: A Fortran Based Object-Oriented System for Quantum Chemistry and Crystallography. In Computational Science—ICCS 2003. ICCS 2003. Lecture Notes in Computer Science; Sloot, P.M.A., Abramson, D., Bogdanov, A.V., Gorbachev, Y.E., Dongarra, J.J., Zomaya, A.Y., Eds.; Springer: Berlin/Heidelberg, Germany, 2003; Volume 2660, pp. 142–151. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).