Abstract
We herein report the synthesis of a 7-chloro-aminoquinoline triazine conjugate. The s-triazine library was generated by stepwise nucleophilic substitution of cyanuric chloride with butylamine. The structure of the compound was comprehensively determined using various analytical techniques, including proton nuclear magnetic resonance (1H NMR), carbon-13 nuclear magnetic resonance (13C NMR), heteronuclear single quantum coherence (HSQC), and Distortionless Enhancement by Polarization Transfer (DEPT-135) experiments. Additionally, ultraviolet (UV) spectroscopy, Fourier-transform infrared (FTIR) spectroscopy, and high-resolution mass spectrometry (HRMS) were employed for full characterization. Preliminary studies explored the potential interaction of the molecule with dihydrofolate reductase (DHFR) using molecular modeling. Furthermore, its drug-likeness was assessed by predicting relevant pharmacokinetic properties.
1. Introduction
Malaria is a potentially life-threatening disease caused by parasites from the Plasmodium genus, which are transmitted to humans through the bites of Anopheles mosquitoes. It affects nearly half of the global population, leading to 249 million clinical cases and 608,000 deaths, according to the World Malaria Report 2023 [1].
Five species of Plasmodium are known to infect humans, with P. vivax, most common in South America and Asia, and P. falciparum, responsible for most malaria-related deaths globally. A growing concern is the increasing resistance of malaria parasites, particularly P. falciparum, to nearly all existing treatments. Due to the persistent parasites that emerged from western Cambodia over the past century, nearly all first-line antimalarial medications, including chloroquine (CQ), primaquine, artemisinin (ART), sulfadoxine, and pyrimethamine (Figure 1a–f), have become less effective [2]. As a result, artemisinin combination therapy (ACT), a combination of medications, is now frequently used to treat malaria [3].
The discovery and development of new, effective, and affordable antimalarial drugs are crucial for controlling and ultimately eradicating the disease [4].
Developing hybrid antimalarial agents has been explored as a strategy to enhance potency and combat the rise of drug resistance [5,6]. The hybrid molecule potentially possesses a dual mode of action, enabling it to target multiple pathways simultaneously, resulting in enhanced effectiveness and reduced susceptibility to resistance. In addition, hybrid compounds have the ability to target various stages of the life cycle of the parasite, a crucial feature for achieving the ultimate goal of malaria eradication.
Dihydrofolate reductase (DHFR) is a crucial enzyme involved in the de novo synthesis of nucleotides in P. falciparum, playing an essential role in cell proliferation.
DHFR is a well-established antimalarial target for drugs such as pyrimethamine and triazine cycloguanil (Figure 1e,f), which exert their pharmacological effects by inhibiting this enzyme [7].
Various N-heterocyclic moieties have been broadly examined for their antimalarial potential. In this connection, s-Triazine (1,3,5 triazine) derivatives represent a vital class of nitrogen-containing heterocyclic molecules, known for their substantial impact as antibacterial [8], antitumor [9] and antimalarial [10] agents. Moreover, quinoline hybrids were previously developed (Figure 1g,h) [6,11,12,13].
Structure–activity relationship (SAR) studies on 4-aminoquinolines indicate that the presence of a 7-chloro group and a 4-amino group in the quinoline nucleus is crucial for antimalarial efficacy. These moieties work together to inhibit β-hematin formation and facilitate the accumulation of the drug within the acidic food vacuole of the parasite (Figure 1ii) [14].
The current work is based on the preparation of compound 5, which was designed, synthesized, and fully characterized by NMR, MS, IR, and UV. Then, the interaction of the molecule with DHFR and its drug-likeness were assayed by means of computational tools.
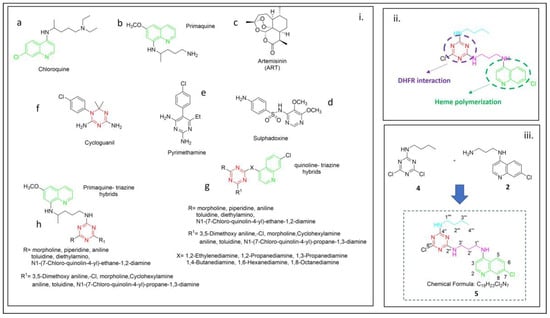
Figure 1.
(i) Structures of classical antimalarial drugs (a–f) and example of triazine quinoline derivatives reported in the literature: (g) [11] and (h) [13]. (ii) Functional groups that are important for the antimalarial activity [7,14]. (iii) Chemical structure of 5 is reported in the inset of the figure.
2. Results and Discussion
The synthesis of the triazine hybrid compound, 2-(Butylamino)-6-chloro-4-[3-(7-chloro-4-quinolylamino)propylamino]-1,3,5-triazine 5, involved the initial preparation of the precursors N1-(7-chloroquinolin-4-yl)propane-1,3-diamine 2 and N-butyl-4,6-dichloro-1,3,5-triazin-2-amine 4.
Quinoline 2 was obtained applying the modified method reported by Biot et al. [15] and N’Da et al. [16], where 4,7-dichloroquinoline 1 was reacted with 10 equivalents of 1,3 propandiamine in ethanol at 80 °C for 3 h (Scheme 1a) to afford 2 in an 80% yield.
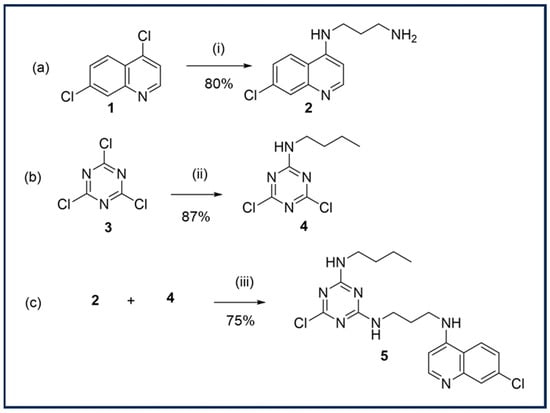
Scheme 1.
(a) Synthesis of N1-(7-chloroquinolin-4-yl)propane-1,3-diamine. (i) 1,3-propanediamine, EtOH, reflux 3 h; (b) synthesis of N-butyl-4,6-dichloro-1,3,5-triazin-2-amine. (ii) Butylamine, acetone, DIPEA, 0 °C; (c) synthesis of 2-(Butylamino)-6-chloro-4-[3-(7-chloro-4-quinolylamino)propylamino]-1,3,5-triazine; (iii) CHCl3, DIPEA and RT to reflux.
Then, 4 was prepared by nucleophilic substitution of cyanuric chloride with butylamine at 0 °C to yield monosubstitued triazine in an 87% yield (Scheme 1b). The hybrid compound 5 was finally obtained by nucleophilic substitution of 4 applying the modified method reported by Rodrigues et al. [13]. Quinoline 2 and three equivalent of 4 were dissolved in CHCl3 and were treated with N,N diisopropylethylamine (DIPEA) (Scheme 1c). The reaction mixture was then stirred vigorously at room temperature (RT) and then refluxed for 20 h. Compound 5 was isolated by column chromatography in a high yield (75%).
The structure of 5 was verified by 1H and 13C NMR analyses (Supplementary Materials, Figures S8 and S10). The 1H-NMR spectrum of 5 showed peaks above 7 ppm corresponding to aromatic protons of the quinoline ring and methylene groups (1‴–3‴ and 1′–3′) corresponding to the six chain groups at 3.41–3.44, 3.06, 1.92, 1.55 and 1.38 ppm, respectively.
Regarding the 13C NMR signals, the appearance of C-13 aromatic signals at 98–152 ppm, and the C-1‴–3‴ and C-1′–3′ methylene signal at 40.5–43.7, 40.8, 31.4, 29.9 and 19.9 ppm were considered as relevant to assess product formation. A methyl group was present, as testified by the peak at a low field at 13.8 ppm. Moreover, heteronuclear single quantum coherence spectroscopy (HSQC) was used to assign 13C signals of compound 5 as shown in Table S1 (see Supplementary Materials for 2D spectra, Figures S11 and S12). The 13C-NMR spectrum of 5 exhibited nineteen carbon signals, classified by DEPT experiments as one methyl group, six methylenes, five methines and five aromatic, and seven quaternary carbons.
The IR spectrum of compound 5 showed peculiar signals, i.e., N–H stretching at 3410 cm−1 and 3246 cm−1, C-H stretching at 2939 cm−1 and C-Cl stretching at 802 cm−1 (Supplementary Materials, Figure S17). Other vibrational peaks at 1534 and 1481 cm−1 were identified as corresponding to the N–H bending in the amine.
The UV and HRMS spectra of 5 were also recorded for further characterization (Supplementary Materials, Figures S18 and S19).
Then, in order to inspect whether compound 5 could be a good candidate as a DHFR inhibitor, we performed computational studies to assess its binding mode with the target protein (Figure 2).
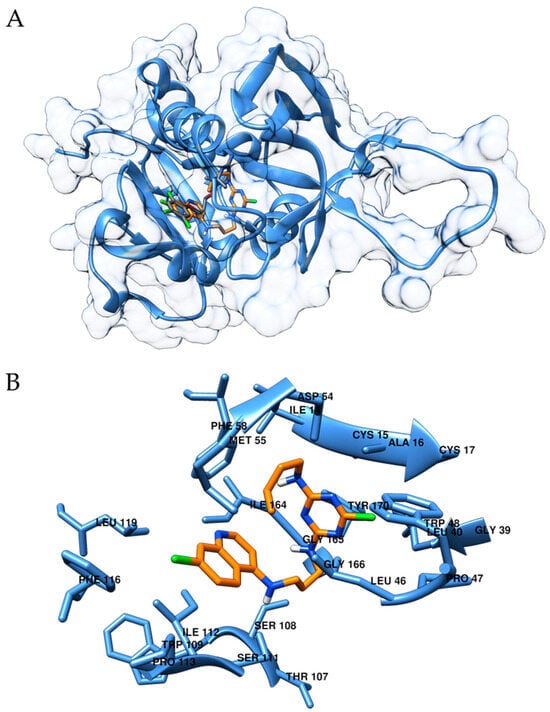
Figure 2.
(A): Predicted binding mode of compound 5 (orange) in comparison with the cognate ligand (grey). (B): Detailed view of the binding pocket of compound 5, in which the residues at a distance <5 Å have been labelled.
Furthermore, the site-specific molecular docking technique was adopted to investigate, from a theoretical point of view, the interaction motif of compound 5 with DHFR. Thus, the X-ray crystallographic structure of DHFR in complex with the ligand 6,6-dimethyl-1-[3-(2,4,5-trichlorophenoxy)propoxy]-1,6-dihydro-1,3,5-triazine-2,4-diamine was considered (PDB ID 1J3I) [17]. The robustness of the docking protocol was assessed by re-docking this cognate ligand, as detailed in Section 3. For the studied compound 5, a calculated binding energy value of −8.6 kcal/mol was computed (docking score for cognate ligand: −9.0 kcal/mol). As can be observed in Figure 2A, compound 5 (orange) and the cognate ligand (grey) interact with the same region of the protein. On the other hand, even if the halogenated rings of the cognate ligand and of compound 5 partially co-localize, the two molecules do not perfectly superimpose. Indeed, this could have been expected given the chemical diversity of the derivatives. In more detail, compound 5 interacts with several hydrophobic residues such as Leu46 Ile112, Phe116 and Leu119, Ile 164, Gly165 and Gly166. Trp48, Ser108, Ser111 and Tyr170 are also within interaction distance in the binding pocket (Figure 2B). As mentioned in the Introduction, the design of hybrid compounds has been previously explored in the literature. To assess the relevance of the scaffold of the hybrid antimalarial, we designed a putative primaquine-based analog of compound 5 (inspired to structure h in Figure 1) and docked it to DHFR. As depicted in Figure S21 in Supplementary Materials, this compound also interacts with the same binding pocket, and a comparable docking score was retrieved (−8.7 kcal/mol). The docking findings aligns with the gastrointestinal absorption results from SwissADME, where compound 5 was predicted to have high absorption according to the BOILED-Egg model [18] (Figure S20). Additionally, data from admetSAR 2 [19] predicted that the compound is orally bioavailable and absorbed in the human intestine. Furthermore, compound 5 was evaluated by predicting its physicochemical properties and oral bioavailability. Based on the calculated physicochemical properties (Supplementary Materials Table S2), compound 5 adhered to all of Lipinski’s rules [20], suggesting its drug-like characteristics and a strong potential for oral administration.
3. Materials and Methods
3.1. Chemistry
Silica gel (FCP 230–400 mesh) was used for column chromatography. Thin-layer chromatography was carried out on Merck precoated silica gel 60 F254 plates (Merck, Darmstadt, Germany), and visualized with phosphomolybdic acid, iodine, or a UV–visible lamp.
All chemicals were purchased from Bide Pharmatech., Ltd. (Shanghai, China) and J & K scientific (Hong Kong, China). 1H-NMR and 13C-NMR spectra were collected for CDCl3 at 25 °C on a Bruker Ascend®-600 (Magnet System 600’54 Ascend LH, San Jose, CA, USA) NMR spectrometer (600 MHz for 1H and 150 MHz for 13C). All chemical shifts were reported in the standard δ notation of parts per million using the peak of residual proton signals of CDCl3 as an internal reference (CDCl3, δC 77.2 ppm, δH 7.26 ppm). High-resolution mass spectra (HRMS) were measured using an Agilent 6230 electrospray ionization (ESI) time-of-flight (TOF) mass spectrometer (Santa Clara, CA, USA) with an Agilent C18 column (4.6 mm × 150 mm, 3.5 μm). The measurements were carried out in a negative ion mode (interface capillary voltage 4500 V); the mass ratio was from m/z 50 to 3000 Da; external/internal calibration was performed with Electrospray Calibration Solution.
The mobile phase was isocratic (water +0.01% TFA; CH3CN) at a flow rate of 0.35 mL/min. The peaks were determined at 254 nm under UV.
UV analysis was performed by the Shimadzu UV—2600 (Osaka, Japan) with a 1 cm quartz cell and a slit width of 2.0 nm. The analysis was carried out using a wavelength in the range of 200–700 nm.
3.1.1. Synthesis of N1-(7-Chloroquinolin-4-yl)Propane-1,3-Diamine (2)
4,7-dichloroquinoline (15.1 mmol, 3.0 g, 1 equiv.) and 1,3-diaminopropane (0.15 mol, 11.1g, 10 equiv.) were placed in a round-bottom flask and dissolved in ethanol. The mixture was stirred and reacted to reflux under N2 atmosphere for 3 h. The reaction mixture was allowed to cool and neutralized with an aqueous solution of 1 M NaOH. The product was extracted with hot ethyl acetate, (3 × 50 mL). These extracts were then combined, washed with water (2 × 50 mL), dried over anhydrous Na2SO4 and evaporated in vacuo to yield the product residue 2 as a white solid in an 80% yield δH (600 MHz, CDCl3) δH (600 MHz, CDCl3) 1.91 (2H, m, CH2), 3.06 (2H, m, CH2), 3.43 (2H, m, CH2), 6.33 (1H, d, H-3), 7.32 (1H, dd, H-6), 7.43 (1H, br, NH), 7.72 (1H, d, H-8), 7.93 (1H, d, H-5), 8.51 (1H, d, H-2); δC (150 MHz, CDCl3) 29.8, 41.3 43.9, 98.1, 117.4, 122.0, 124.9, 128.5, 134.6, 149.1, 150.4, 152.1.
The spectral characteristics are consistent with those of 2 in the literature [21].
3.1.2. Synthesis of N-Butyl-4,6-Dichloro-1,3,5-Triazin-2-Amine Diamine (4)
To a stirred solution of cyanuric chloride (1 g, 5.4 mmol) in acetone (50 mL) held at 0 °C under a nitrogen atmosphere, we added n-butylamine (1.0 mL, 10.9 mmol) and DIPEA (10.9 mmol) over 5 min. The reaction mixture was stirred for 1 h before being washed with water (2 × 50 mL), the organic phase was dried (MgSO4) and evaporated to dryness, giving the title compound as a viscous colourless oil (1.10 g, 87%). 0.94 (3H, t), 1.39 (2H, m, CH2), 1.57 (2H, m, CH2), 3.41 (2H, m, CH2) ppm; δC (150 MHz, CDCl3) 13.7, 20.0, 31.4, 40.6, 165., 168.3 pm.
The spectral characteristics are consistent with those of 4 in the literature [22].
3.1.3. Synthesis of 2-(Butylamino)-6-Chloro-4-[3-(7-Chloro-4-Quinolylamino)Propylamino]-1,3,5-Triazine (5)
To a stirred solution of 4 (0.1 g, 0.45 mmol) in chloroform (50 mL) held at RT under a nitrogen atmosphere, we added 2 (0.31 g, 1.35 mmol) over 5 min and DIPEA (235 mL, 1.35 mmol). After 45 min of stirring, the solution was warmed up to RT, stirred for another 1.25 h, and then heated at reflux for 20 h reaction. After cooling, the mixture was washed with water (2 × 50 mL) and the organic phase was dried (MgSO4) and evaporated to dryness. The resulting crude material was purified by preparative TLC (EtOAc/hexane 1:1); the fraction at Rf = 0.5 gave the desired product. Yield 75%, δH (600 MHz, CDCl3) 0.94 (3H, t, H–4‴), 1.38 (2H, m, H–3‴), 1.55 (2H, m, H–2‴), 1.92 (2H, m, H–2′), 3.06 (H, t, H–1‴), 3.20 (1H, NH), 3.40 (4H, m, H–3′ and H–1′), 6.34 (1H, d, J = 5.3 Hz, Haromat), 7.08 (1H, NH), 7.34 (1H, dd, J = 8.9 and 2.2 Hz, Haromat), 7.43 (1H, NH), 7.72 (1H, d, J = 8.9 Hz, Haromat), 7.94 (1H, d, J = 2.1 Hz, Haromat), 8.50 (1H, d, J = 5.37 Hz, Haromat); δC (150 MHz, CDCl3) 13.8 (C–4‴), 19.9 (C–3‴), 29.9 (C–2‴), 40.8 (C–1‴), 43.7, 98.3 (C-3), 121.4 (C-6), 124.9 (C-6), 128.5 (C-5), 134.5 (C-7), 149.1 (C-8), 150.3 (C-4), 152.0 (C-2), 165.8 (C-4), 166.2 (C-2), 168.4 (C–4″) ppm; HRMS-ESI m/z 418.1356 [M–H]− (calcd. for C19H23N7Cl2, m/z 418.1319), UV (CH2Cl2) peaks 229, 258 and 331 nm, IR (KBr) 3410, 3246, 2939, 2569, 1534, 1481, 1388, 802 cm−1.
3.2. Computational Studies
The 3D X-ray crystal structure of wild-type Plasmodium falciparum dihydrofolate reductase-thymidylate synthase was retrieved from the RCSB Protein Data Bank (www.rcsb.org; accessed on 30 August 2024; PDB ID 1J3I, resolution 2.33 Å) and used in agreement with previous studies [23,24]. Prior to site-specific docking studies, the protein was prepared, and chain B was isolated and considered. Further, the obtained structure was processed through the DockPrep tool of UCSF Chimera (Chimera: 1.17.1, Avogadro: 1.1.1, AutoDock Vina: 1.2.5)to fix eventually missing residues in the receptor [25]. Ligands were built and optimized using Avogadro [26]. The site-specific molecular docking study was performed using AutoDock Vina [27,28]. The search grid was set according to the following parameters: x = 33.1137, y = −28.0501, z = 5.69427; size: 22.000 × 22.000 × 22.000 Å. Docking simulation was carried out with default Vina parameters, in which the number of generated docking poses was set to 8 and the docking energy conformation value was expressed in −kcal/mol. To validate the docking protocol, the cognate ligand was re-docked and a calculated binding energy value of −9.0 kcal/mol was obtained. Moreover, root mean square deviation (RMSD) was calculated with respect to the co-crystallized pose and a value of 2.203 Å was observed. The results and figures were processed with UCSF Chimera [25].
4. Conclusions
The synthesis of a triazine-based quinoline analogue with a hybrid structure and a potential application as an antimalarial was presented. The chemical structure of the synthesized compound was confirmed using NMR, HRMS, IR, and UV spectroscopies. Additionally, drug-likeness was assessed through computational analysis and molecular docking studies showed that the compound may efficiently interact with the binding site of DHFR.
Supplementary Materials
The following are available online: Figure S1: 1-H NMR compound 2 (CDCl3, 600 MHz), Figure S2: 13-C NMR compound 2 (CDCl3, 150 MHz), Figure S3:1-H NMR compound 2 (CDCl3, 600 MHz), Figure S4: An expanded view of 1-H NMR compound 2 (CDCl3, 600 MHz), Figure S5: 13-C NMR compound 2 (CDCl3, 150 MHz), Figure S6: DEPT-135 spectrum (CDCl3, 600 MHz) of compound 2, Figure S7: HSQC of compound 2 (CDCl3, 600 MHz), Figure S8: 1-H NMR compound 5 (CDCl3, 600 MHz), Figure S9: An expanded view of 1-H NMR compound 5 (CDCl3, 600 MHz), Figure S10: 13-C NMR compound 5 (CDCl3, 150 MHz), Figure S11: An expanded view of 13-C NMR compound 5 (CDCl3, 150 MHz), Figure S12: DEPT-135 spectrum (CDCl3, 600 MHz) of compound 5), Figure S13: HSQC of compound 5 (CDCl3, 600 MHz), Figure S14: An expanded view of HSQC compound 5 (CDCl3, 600 MHz), Figure S15: 1H-1H COSY compound 5 (CDCl3, 600 MHz), Figure S16: An expanded view of 1H-1H COSY compound 5 (CDCl3, 600 MHz), Figure S17: IR spectrum (KBr) of compound 5, Figure S18: UV spectrum of compound 5 (range 200–400 nm in CH2Cl2), Figure S19: HRMS of compound 5, Figure S20: BOILED-egg graph of compound 5, Figure S21: molecular modeling for analog h; Table S1 1H and 13C-nuclear magnetic spectroscopy (NMR) chemical shifts and the structure of 5, Table S2: Physicochemical properties of 5 calculated by SwissADME.
Author Contributions
Conceptualization, P.C. and G.R; methodology, Z.R., Y.G. and Y.X.; investigation, Z.R., Y.G., A.G. and Y.X.; data curation, Z.R.; writing—original draft preparation, Z.R.; writing—review and editing, P.C. and G.R.; supervision, P.C. and G.R.; project administration, P.C. and G.R.; funding acquisition, P.C. and G.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by FDCT grants from Macau Science and Technology University to P.C. (Project Code: 0005-2023-RIA1) and by the University of Brescia.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- World Malaria Report 2023. Available online: https://www.who.int/teams/global-malaria-programme/reports/world-malaria-report-2023 (accessed on 27 August 2023).
- Roux, A.T.; Maharaj, L.; Oyegoke, O.; Akoniyon, O.P.; Adeleke, M.A.; Maharaj, R.; Okpeku, M. Chloroquine and Sulfadoxine–Pyrimethamine Resistance in Sub-Saharan Africa—A Review. Front. Genet. 2021, 12, 668574. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.D.; Gao, B.; Amaratunga, C.; Dhorda, M.; Tran, T.N.-A.; White, N.J.; Dondorp, A.M.; Boni, M.F.; Aguas, R. Preventing antimalarial drug resistance with triple artemisinin-based combination therapies. Nat. Commun. 2023, 14, 4568. [Google Scholar] [CrossRef] [PubMed]
- Siqueira-Neto, J.L.; Wicht, K.J.; Chibale, K.; Burrows, J.N.; Fidock, D.A.; Winzeler, E.A. Antimalarial drug discovery: Progress and approaches. Nat. Rev. Drug Discov. 2023, 22, 807–826. [Google Scholar] [CrossRef] [PubMed]
- Prasad Raiguru, B.; Panda, J.; Mohapatra, S.; Nayak, S. Recent developments in the synthesis of hybrid antimalarial drug discovery. Bioorg. Chem. 2023, 139, 106706. [Google Scholar] [CrossRef]
- Hu, Y.-Q.; Gao, C.; Zhang, S.; Xu, L.; Xu, Z.; Feng, L.-S.; Wu, X.; Zhao, F. Quinoline hybrids and their antiplasmodial and antimalarial activities. Eur. J. Med. Chem. 2017, 139, 22–47. [Google Scholar] [CrossRef]
- Rastelli, G.; Sirawaraporn, W.; Sompornpisut, P.; Vilaivan, T.; Kamchonwongpaisan, S.; Quarrell, R.; Lowe, G.; Thebtaranonth, Y.; Yuthavong, Y. Interaction of pyrimethamine, cycloguanil, WR99210 and their analogues with Plasmodium falciparum dihydrofolate reductase: Structural basis of antifolate resistance. Bioorg. Med. Chem. 2000, 8, 1117–1128. [Google Scholar] [CrossRef]
- Arshad, M.; Bhat, A.R.; Hoi, K.K.; Choi, I.; Athar, F. Synthesis, characterization and antibacterial screening of some novel 1,2,4-triazine derivatives. Chin. Chem. Lett. 2017, 28, 1559–1565. [Google Scholar] [CrossRef]
- Cascioferro, S.; Parrino, B.; Spanò, V.; Carbone, A.; Montalbano, A.; Barraja, P.; Diana, P.; Cirrincione, G. An overview on the recent developments of 1,2,4-triazine derivatives as anticancer compounds. Eur. J. Med. Chem. 2017, 142, 328–375. [Google Scholar] [CrossRef]
- Melato, S.; Prosperi, D.; Coghi, P.; Basilico, N.; Monti, D. A Combinatorial Approach to 2,4,6-Trisubstituted Triazines with Potent Antimalarial Activity: Combining Conventional Synthesis and Microwave-Assistance. ChemMedChem 2008, 3, 873–876. [Google Scholar] [CrossRef]
- Manohar, S.; Khan, S.I.; Rawat, D.S. Synthesis, antimalarial activity and cytotoxicity of 4-aminoquinoline-triazine conjugates. Bioorg. Med. Chem. Lett. 2010, 20, 322–325. [Google Scholar] [CrossRef]
- Feng, Y.-Y.; Dong, C.-E.; Li, R.; Zhang, X.-Q.; Wang, W.; Zhang, X.-R.; Liu, W.-W.; Shi, D.-H. Design, synthesis and biological evaluation of quinoline-1,2,4-triazine hybrids as antimalarial agents. J. Mol. Struct. 2023, 1271, 133982. [Google Scholar] [CrossRef]
- Rodrigues, C.A.; Frade, R.F.; Albuquerque, I.S.; Perry, M.J.; Gut, J.; Machado, M.; Rosenthal, P.J.; Prudêncio, M.; Afonso, C.A.; Moreira, R. Targeting the erythrocytic and liver stages of malaria parasites with s-triazine-based hybrids. ChemMedChem 2015, 10, 883–890. [Google Scholar] [CrossRef]
- Egan, T.J.; Marques, H.M. The role of haem in the activity of chloroquine and related antimalarial drugs. Coord. Chem. Rev. 1999, 190–192, 493–517. [Google Scholar] [CrossRef]
- Biot, C.; Daher, W.; Ndiaye, C.M.; Melnyk, P.; Pradines, B.; Chavain, N.; Pellet, A.; Fraisse, L.; Pelinski, L.; Jarry, C.; et al. Probing the role of the covalent linkage of ferrocene into a chloroquine template. J. Med. Chem. 2006, 49, 4707–4714. [Google Scholar] [CrossRef]
- Smit, F.J.; N’Da, D.D. Synthesis, in vitro antimalarial activity and cytotoxicity of novel 4-aminoquinolinyl-chalcone amides. Bioorg. Med. Chem. 2014, 22, 1128–1138. [Google Scholar] [CrossRef]
- Yuvaniyama, J.; Chitnumsub, P.; Kamchonwongpaisan, S.; Vanichtanankul, J.; Sirawaraporn, W.; Taylor, P.; Walkinshaw, M.D.; Yuthavong, Y. Insights into antifolate resistance from malarial DHFR-TS structures. Nat. Struct. Biol. 2003, 10, 357–365. [Google Scholar] [CrossRef]
- Daina, A.; Zoete, V. A BOILED-Egg To Predict Gastrointestinal Absorption and Brain Penetration of Small Molecules. ChemMedChem 2016, 11, 1117–1121. [Google Scholar] [CrossRef]
- Yang, H.; Lou, C.; Sun, L.; Li, J.; Cai, Y.; Wang, Z.; Li, W.; Liu, G.; Tang, Y. admetSAR 2.0: Web-service for prediction and optimization of chemical ADMET properties. Bioinformatics 2019, 35, 1067–1069. [Google Scholar] [CrossRef]
- Lipinski, C.A. Drug-like properties and the causes of poor solubility and poor permeability. J. Pharmacol. Toxicol. Methods 2000, 44, 235–249. [Google Scholar] [CrossRef]
- Rojas Ruiz, F.A.; García-Sánchez, R.N.; Estupiñan, S.V.; Gómez-Barrio, A.; Torres Amado, D.F.; Pérez-Solórzano, B.M.; Nogal-Ruiz, J.J.; Martínez-Fernández, A.R.; Kouznetsov, V.V. Synthesis and antimalarial activity of new heterocyclic hybrids based on chloroquine and thiazolidinone scaffolds. Bioorg. Med. Chem. 2011, 19, 4562–4573. [Google Scholar] [CrossRef]
- Aldilla, V.R.; Bhadbhade, M.; Bhattacharyya, S.; Kumar, N.; Rich, A.M.; Marjo, C.E. Controlling the distance between hydrogen-bonded chloro-s-triazine tapes: Crystal engineering using N-alkyl chains and the influence of temperature. CrystEngComm 2017, 19, 4749–4758. [Google Scholar] [CrossRef]
- Joshi, A.A.; Viswanathan, C.L. Docking studies and development of novel 5-heteroarylamino-2,4-diamino-8-chloropyrimido-[4,5-b]quinolines as potential antimalarials. Bioorg. Med. Chem. Lett. 2006, 16, 2613–2617. [Google Scholar] [CrossRef]
- Hadni, H.; Elhallaoui, M. Molecular docking and QSAR studies for modeling the antimalarial activity of hybrids 4-anilinoquinoline-triazines derivatives with the wild-type and mutant receptor pf-DHFR. Heliyon 2019, 5, e02357. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).