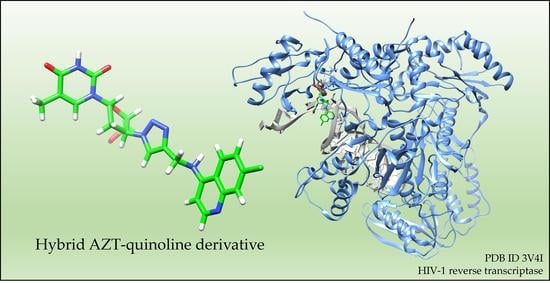
(2R, 4S, 5S) 1-(4-(4-(((7-Chloroquinolin-4-yl)amino)methyl)-1H-1,2,3-triazol-1-yl)-5-(hydroxymethyl)tetrahydrofuran-2-yl)-5-methylpyrimidine-2,4(1H,3H)-dione
Abstract
1. Introduction
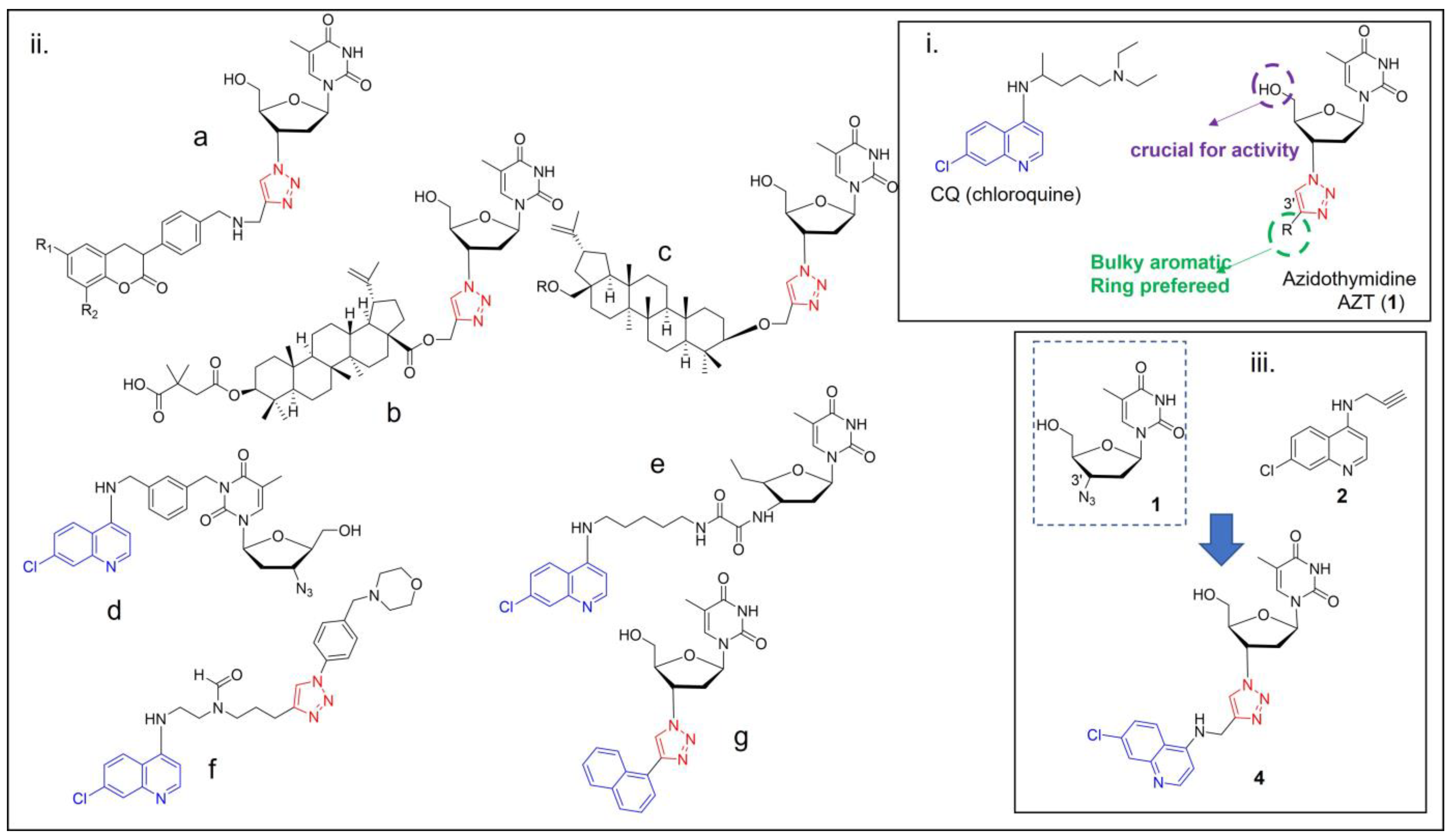
2. Results and Discussion
3. Materials and Methods
3.1. Chemistry
3.1.1. Synthesis of N-(Prop-2-yn-1-yl)-7-chloroquinolin-4-amine (3)
3.1.2. Synthesis of 1-(4-(4-(((7-Chloroquinolin-4-yl)amino)methyl)-1H-1,2,3-triazol-1-yl)-5-(hydroxymethyl)tetrahydrofuran-2-yl)-5-methylpyrimidine-2,4(1H,3H)-dione (4)
3.2. Computational Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- World Health Organization. Available online: https://www.who.int/news-room/fact-sheets/details/hiv-aids (accessed on 30 May 2023).
- Broder, S. The development of antiretroviral therapy and its impact on the HIV-1/AIDS pandemic. Antivir. Res. 2010, 85, 1–18. [Google Scholar] [CrossRef]
- Wright, K. AIDS therapy. First tentative signs of therapeutic promise. Nature 1986, 323, 283. [Google Scholar] [CrossRef]
- Feng, L.S.; Zheng, M.J.; Zhao, F.; Liu, D. 1,2,3-Triazole hybrids with anti-HIV-1 activity. Arch. der Pharm. 2021, 354, e2000163. [Google Scholar] [CrossRef]
- Demaray, J.A.; Thuener, J.E.; Dawson, M.N.; Sucheck, S.J. Synthesis of triazole-oxazolidinones via a one-pot reaction and evaluation of their antimicrobial activity. Bioorganic Med. Chem. Lett. 2008, 18, 4868–4871. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, R.P.; Yadav, A.K.; Ajay, A.; Bisht, S.S.; Chaturvedi, V.; Sinha, S.K. Application of Huisgen (3+2) cycloaddition reaction: Synthesis of 1-(2,3-dihydrobenzofuran-2-yl-methyl [1,2,3]-triazoles and their antitubercular evaluations. Eur. J. Med. Chem. 2010, 45, 142–148. [Google Scholar] [CrossRef]
- Herrmann, L.; Hahn, F.; Wangen, C.; Marschall, M.; Tsogoeva, S.B. Anti-SARS-CoV-2 Inhibitory Profile of New Quinoline Compounds in Cell Culture-Based Infection Models. Chem. Eur. J. 2022, 28, e202103861. [Google Scholar] [CrossRef] [PubMed]
- Fai, L.K.; Anyanwu, M.; Ai, J.; Xie, Y.; Gianoncelli, A.; Ribaudo, G.; Coghi, P. 4-(4-(((1H-Benzo[d][1,2,3]triazol-1-yl)oxy)methyl)-1H-1,2,3-triazol-1-yl)-7-chloroquinoline. Molbank 2022, 2022, M1404. [Google Scholar] [CrossRef]
- Coghi, P.; Ng, J.P.L.; Nasim, A.A.; Wong, V.K.W. N-[7-Chloro-4-[4-(phenoxymethyl)-1H-1,2,3-triazol-1-yl]quinoline]-acetamide. Molbank 2021, 2021, M1213. [Google Scholar] [CrossRef]
- Yun, X.; Xie, Y.; Ng, J.P.L.; Law, B.Y.K.; Wong, V.K.W.; Coghi, P. 2-Bromo-3-((1-(7-chloroquinolin-4-yl)-1H-1,2,3-triazol-4-yl)-methoxy)-benzaldehyde. Molbank 2022, 2022, M1351. [Google Scholar] [CrossRef]
- Boelaert, J.R.; Sperber, K.; Piette, J. The additive in vitro anti-HIV-1 effect of chloroquine, when combined with zidovudine and hydroxyurea. Biochem. Pharmacol. 2001, 61, 1531–1535. [Google Scholar] [CrossRef]
- Fehintola, F.A.; Akinyinka, O.O.; Adewole, I.F.; Maponga, C.C.; Ma, Q.; Morse, G.D. Drug interactions in the treatment and chemoprophylaxis of malaria in HIV infected individuals in sub Saharan Africa. Curr. Drug Metab. 2011, 12, 51–56. [Google Scholar] [CrossRef]
- Bori, I.D.; Hung, H.Y.; Qian, K.; Chen, C.H.; Morris-Natschke, S.L.; Lee, K.H. Anti-AIDS agents 88. Anti-HIV conjugates of betulin and betulinic acid with AZT prepared via click chemistry. Tetrahedron Lett. 2012, 53, 1987–1989. [Google Scholar] [CrossRef] [PubMed]
- Sirivolu, V.R.; Vernekar, S.K.; Ilina, T.; Myshakina, N.S.; Parniak, M.A.; Wang, Z. Clicking 3’-azidothymidine into novel potent inhibitors of human immunodeficiency virus. J. Med. Chem. 2013, 56, 8765–8780. [Google Scholar] [CrossRef] [PubMed]
- Ivasiv, V.; Albertini, C.; Gonçalves, A.E.; Rossi, M.; Bolognesi, M.L. Molecular Hybridization as a Tool for Designing Multitarget Drug Candidates for Complex Diseases. Curr. Top. Med. Chem. 2019, 19, 1694–1711. [Google Scholar] [CrossRef]
- Aminake, M.N.; Mahajan, A.; Kumar, V.; Hans, R.; Wiesner, L.; Taylor, D.; de Kock, C.; Grobler, A.; Smith, P.J.; Kirschner, M.; et al. Synthesis and evaluation of hybrid drugs for a potential HIV/AIDS-malaria combination therapy. Bioorganic Med. Chem. 2012, 20, 5277–5289. [Google Scholar] [CrossRef] [PubMed]
- Fisher, G.M.; Tanpure, R.P.; Douchez, A.; Andrews, K.T.; Poulsen, S.A. Synthesis and evaluation of antimalarial properties of novel 4-aminoquinoline hybrid compounds. Chem. Biol. Drug Des. 2014, 84, 462–472. [Google Scholar] [CrossRef]
- Feldman, A.K.; Colasson, B.; Fokin, V.V. One-pot synthesis of 1,4-disubstituted 1,2,3-triazoles from in situ generated azides. Org. Lett. 2004, 6, 3897–3899. [Google Scholar] [CrossRef]
- Santos, L.H.; Ferreira, R.S.; Caffarena, E.R. Computational drug design strategies applied to the modelling of human immunodeficiency virus-1 reverse transcriptase inhibitors. Mem. do Inst. Oswaldo Cruz 2015, 110, 847–864. [Google Scholar] [CrossRef]
- Furman, P.A.; Fyfe, J.A.; St Clair, M.H.; Weinhold, K.; Rideout, J.L.; Freeman, G.A.; Lehrman, S.N.; Bolognesi, D.P.; Broder, S.; Mitsuya, H. Phosphorylation of 3’-azido-3’-deoxythymidine and selective interaction of the 5’-triphosphate with human immunodeficiency virus reverse transcriptase. Proc. Natl. Acad. Sci. USA 1986, 83, 8333–8337. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminformatics 2012, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Adasme, M.F.; Linnemann, K.L.; Bolz, S.N.; Kaiser, F.; Salentin, S.; Haupt, V.J.; Schroeder, M. PLIP 2021: Expanding the Scope of the Protein–Ligand Interaction Profiler to DNA and RNA. Nucleic Acids Research 2021, 49, W530–W534. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Zoete, V. A BOILED-Egg To Predict Gastrointestinal Absorption and Brain Penetration of Small Molecules. ChemMedChem 2016, 11, 1117–1121. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuan, H.; Xie, Y.; Guo, Y.; Gianoncelli, A.; Ribaudo, G.; Coghi, P. (2R, 4S, 5S) 1-(4-(4-(((7-Chloroquinolin-4-yl)amino)methyl)-1H-1,2,3-triazol-1-yl)-5-(hydroxymethyl)tetrahydrofuran-2-yl)-5-methylpyrimidine-2,4(1H,3H)-dione. Molbank 2023, 2023, M1681. https://doi.org/10.3390/M1681
Kuan H, Xie Y, Guo Y, Gianoncelli A, Ribaudo G, Coghi P. (2R, 4S, 5S) 1-(4-(4-(((7-Chloroquinolin-4-yl)amino)methyl)-1H-1,2,3-triazol-1-yl)-5-(hydroxymethyl)tetrahydrofuran-2-yl)-5-methylpyrimidine-2,4(1H,3H)-dione. Molbank. 2023; 2023(3):M1681. https://doi.org/10.3390/M1681
Chicago/Turabian StyleKuan, Houin, Yuhan Xie, Yuzhu Guo, Alessandra Gianoncelli, Giovanni Ribaudo, and Paolo Coghi. 2023. "(2R, 4S, 5S) 1-(4-(4-(((7-Chloroquinolin-4-yl)amino)methyl)-1H-1,2,3-triazol-1-yl)-5-(hydroxymethyl)tetrahydrofuran-2-yl)-5-methylpyrimidine-2,4(1H,3H)-dione" Molbank 2023, no. 3: M1681. https://doi.org/10.3390/M1681
APA StyleKuan, H., Xie, Y., Guo, Y., Gianoncelli, A., Ribaudo, G., & Coghi, P. (2023). (2R, 4S, 5S) 1-(4-(4-(((7-Chloroquinolin-4-yl)amino)methyl)-1H-1,2,3-triazol-1-yl)-5-(hydroxymethyl)tetrahydrofuran-2-yl)-5-methylpyrimidine-2,4(1H,3H)-dione. Molbank, 2023(3), M1681. https://doi.org/10.3390/M1681