5-[(5-Chloro-1H-pyrrolo[2,3-b]pyridin-3-yl)methyl]-N-[[6-(trifluoromethyl)pyridin-3-yl]methyl]pyridin-2-amine Dihydrochloride Dihydrate
Abstract
1. Introduction
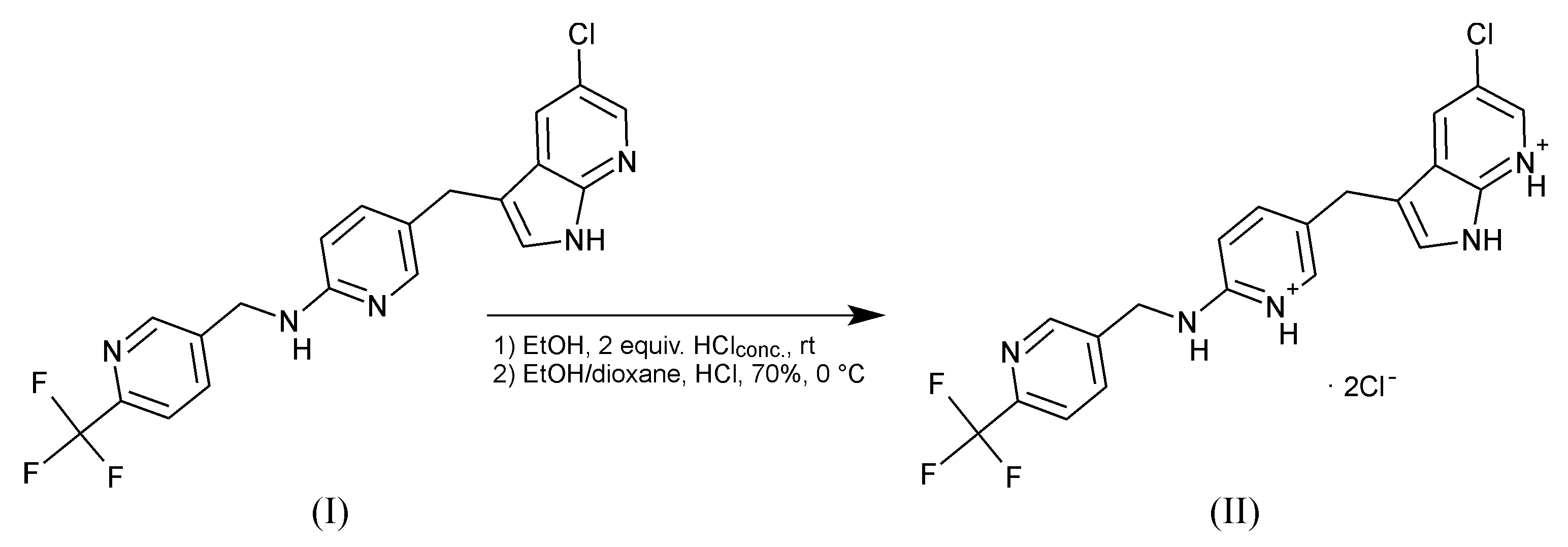
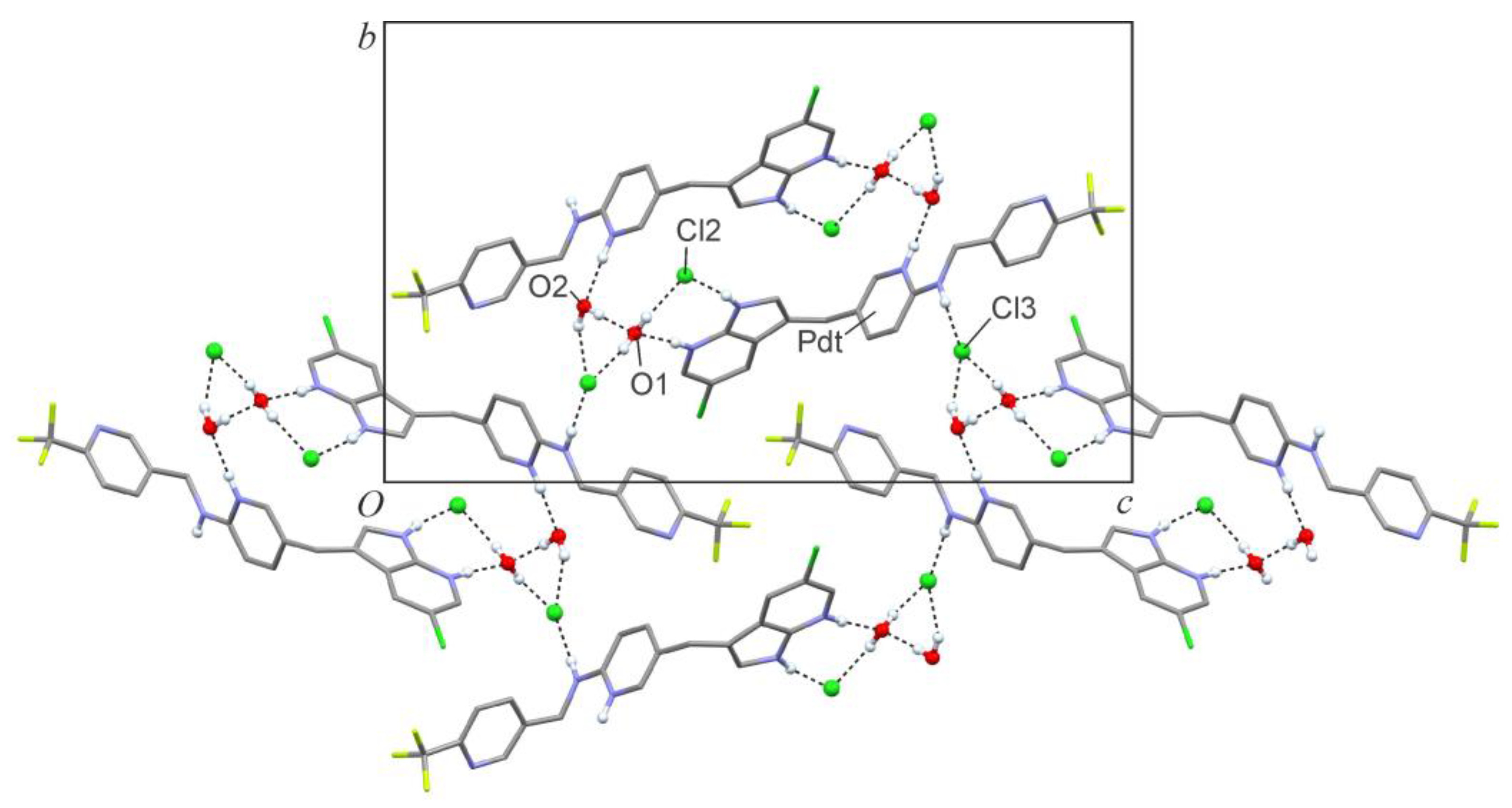
2. Results
3. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mok, S.; Koya, R.C.; Tsui, C.; Xu, J.; Robert, L.; Wu, L.; Graeber, T.G.; West, B.L.; Bollag, G.; Ribas, A. Inhibition of CSF-1 Receptor Improves the Antitumor Efficacy of Adoptive Cell Transfer Immunotherapy. Cancer Res. 2014, 74, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Tap, W.D.; Wainberg, Z.A.; Anthony, S.P.; Ibrahim, P.N.; Zhang, C.; Healey, J.H.; Chmielowski, B.; Staddon, A.P.; Cohn, A.L.; Shapiro, G.I.; et al. Structure-guided blockade of CSF1R kinase in tenosynovial giant-cell tumor. N. Engl. J. Med. 2015, 373, 428–437. [Google Scholar] [CrossRef] [PubMed]
- Lamb, Y.N. Pexidartinib: First Approval. Drugs 2019, 79, 1805–1812. [Google Scholar] [CrossRef] [PubMed]
- Roskoski, R. Properties of FDA-approved small molecule protein kinase inhibitors: A 2020 update. Pharmacol. Res. 2020, 152, 104609. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, P.N.; Visor, G.C. Solid Forms of a Compound Modulating Kinases. International Patent WO2016179415, 10 November 2016. [Google Scholar]
- Chen, M.; Zhang, Y.; Zou, P.; Zhang, X. Preparation of PLX3397 Hydrochloride Crystal Forms. International Patent WO2017215521, 21 December 2017. [Google Scholar]
- Li, H.; Li, Y.; Huang, X. Green Preparation of Pexidartinib as Tyrosine Kinase Inhibitor. China Patent CN113444083, 28 September 2021. [Google Scholar]
- Sheldrick, G.M. SHELXT—Integrated space-group and crystal-structure determination. Acta Crystallogr. A 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Sheldrick, G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. C 2015, 71, 3–8. [Google Scholar] [CrossRef] [PubMed]
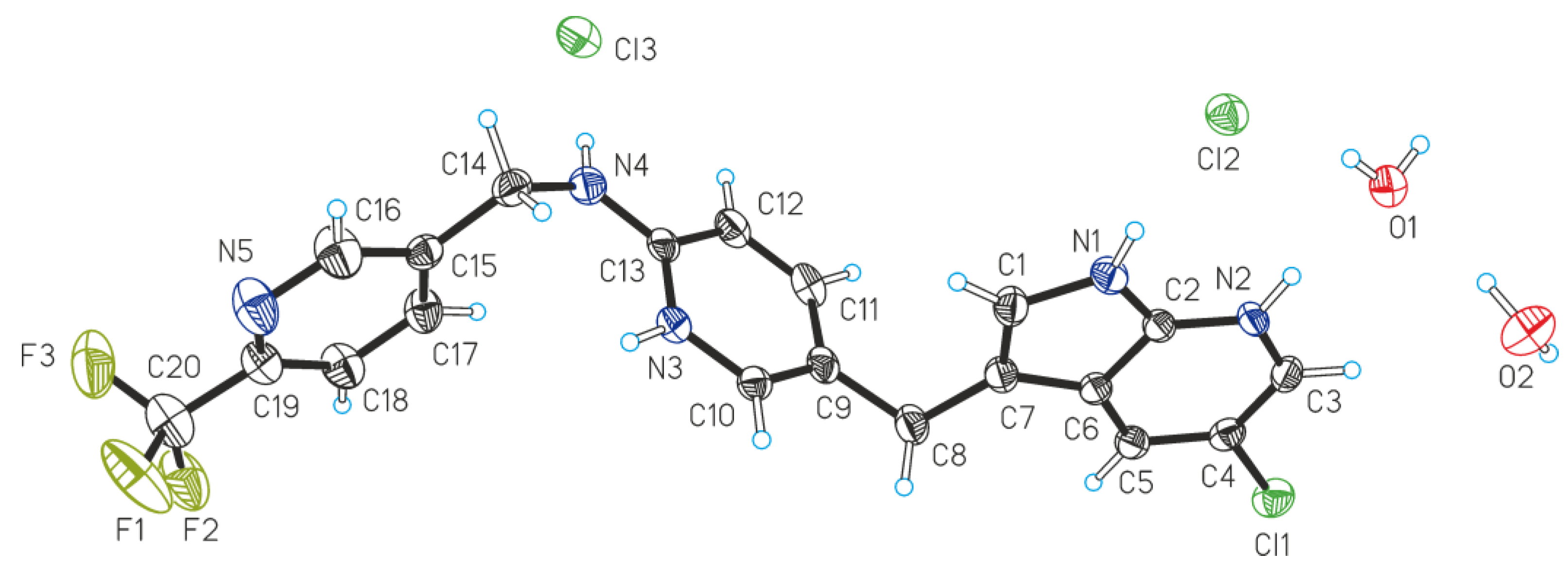
| Compound | (II)·2H2O |
|---|---|
| Moiety formula | (C20H17ClF3N5)2+ · 2Cl− · 2H2O |
| Empirical formula | C20H21Cl3F3N5O2 |
| Formula weight | 526.77 |
| Temperature (K) | 183(2) |
| Wavelength (Å) | 0.71073 |
| Crystal system | Monoclinic |
| Space group | P21/c |
| a (Å) | 5.4641(3) |
| b (Å) | 16.2163(10) |
| c (Å) | 26.3877(17) |
| β (°) | 93.0718(18) |
| Unit cell volume (Å3) | 2334.8(2) |
| Z/Z’ | 4/1 |
| Reflections collected/Rint | 34,479/0.0479 |
| Data/restraints/parameters | 4145/8/331 |
| Goodness-of-fit on F2 | 1.044 |
| R1 [I > 2 σ(I)] | 0.0365 |
| wR2 (all data) | 0.0879 |
| Largest diff. peak and hole (e · Å−3) | 0.444 and −0.368 |
| CCDC no. | 2265163 |
| D—H···A | dD—H | dH···A | dD···A | <(DHA) |
|---|---|---|---|---|
| N1—H1N···Cl2 | 0.878(16) | 2.242(17) | 3.1109(19) | 171(2) |
| N2—H2N···O1 | 0.869(16) | 1.895(17) | 2.762(2) | 175(2) |
| N3—H3N···O2 i | 0.891(16) | 1.895(17) | 2.774(3) | 168(2) |
| N4—H4N···Cl3 | 0.871(16) | 2.267(17) | 3.138(2) | 178(2) |
| O1—H1OA···Cl2 | 0.836(17) | 2.182(19) | 3.0014(18) | 167(3) |
| O1—H1OB···Cl3 ii | 0.828(18) | 2.304(19) | 3.1217(18) | 169(3) |
| O2—H2OA···O1 | 0.854(18) | 1.902(19) | 2.754(2) | 176(3) |
| O2—H2OB···Cl3 iii | 0.851(18) | 2.35(2) | 3.1561(19) | 158(3) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kozanecka-Okupnik, W.; Wurst, K.; Neuner, S.; Nerdinger, S.; Gelbrich, T. 5-[(5-Chloro-1H-pyrrolo[2,3-b]pyridin-3-yl)methyl]-N-[[6-(trifluoromethyl)pyridin-3-yl]methyl]pyridin-2-amine Dihydrochloride Dihydrate. Molbank 2023, 2023, M1673. https://doi.org/10.3390/M1673
Kozanecka-Okupnik W, Wurst K, Neuner S, Nerdinger S, Gelbrich T. 5-[(5-Chloro-1H-pyrrolo[2,3-b]pyridin-3-yl)methyl]-N-[[6-(trifluoromethyl)pyridin-3-yl]methyl]pyridin-2-amine Dihydrochloride Dihydrate. Molbank. 2023; 2023(2):M1673. https://doi.org/10.3390/M1673
Chicago/Turabian StyleKozanecka-Okupnik, Weronika, Klaus Wurst, Sandro Neuner, Sven Nerdinger, and Thomas Gelbrich. 2023. "5-[(5-Chloro-1H-pyrrolo[2,3-b]pyridin-3-yl)methyl]-N-[[6-(trifluoromethyl)pyridin-3-yl]methyl]pyridin-2-amine Dihydrochloride Dihydrate" Molbank 2023, no. 2: M1673. https://doi.org/10.3390/M1673
APA StyleKozanecka-Okupnik, W., Wurst, K., Neuner, S., Nerdinger, S., & Gelbrich, T. (2023). 5-[(5-Chloro-1H-pyrrolo[2,3-b]pyridin-3-yl)methyl]-N-[[6-(trifluoromethyl)pyridin-3-yl]methyl]pyridin-2-amine Dihydrochloride Dihydrate. Molbank, 2023(2), M1673. https://doi.org/10.3390/M1673






