Abstract
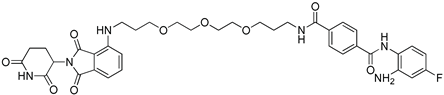
The design of proteolysis targeting chimeras (PROTACs) has become a promising technology for modifying a protein of interest (POI) through protein degradation. Herein, we describe the synthetic pathway to develop N4-(2-amino-4-fluorophenyl)-N1-(3-{2-[2-(3-{[2-(2,6-dioxo-3-piperidyl)-1,3-dioxoisoindolin-4-yl]amino}propoxy)ethoxy]ethoxy}propyl)terephthalamide, which was designed to work as a selective degrader of histone deacetylase-3 (HDAC3). The newly synthesized compounds were characterized by 1H-NMR, 13C-NMR, IR and HRMS. The title compound was tested in vitro against human class-I HDACs isoforms and showed IC50 = 3.4 µM against HDAC3; however, it did not show degradation for the targeted HDACs.
1. Introduction
Class-I histone deacetylases (including HDAC1, 2, 3, and 8 isoforms) are among eleven zinc-dependent histone deacetylases that catalyze the hydrolysis of acetyl groups from histone lysine residues [1]. They play an important role in the regulation of gene expression and cell proliferation [2]. Dysregulation of their epigenetic activity has been involved in a wide range of diseases [3,4,5], including cancer [2]. Several HDAC inhibitors (HDACis) have been developed and identified as potential anticancer therapeutics [6]. Most HDACis share a common pharmacophoric scaffold consisting of three different parts as follows: a zinc binding group (ZBG) that is responsible for chelating of zinc ion in the active site of HDACs, a capping group that usually induce hydrophobic interactions at the rim of the HDAC enzyme, in addition to a linker connecting both groups [7]. HDACis can be classified based on their zinc binding groups (ZBG) into different groups, mainly hydroxamates, 2-aminobenzamides, thiols, cyclic peptides, and others [8]. To date, four HDACis have been approved by US Food and Drug Administration (FDA) for treatment of different hematological malignancies including the hydroxamate-based vorinostat, belinostat and panobinostat, in addition to the cyclic peptide romidepsin [9]. Although the hydroxamic acids exhibit potent ZBG activity, it was observed that hydroxamate-based HDACis lack isoform selectivity, and this might contribute to the off-target side effect associated with such drugs [10,11]. It has been reported that 2-aminobenzamides can enhance class-I HDAC selectivity and strongly inhibit HDAC subtypes HDACs1, -2, and -3 [12]. Tacedinaline (Figure 1) is the first reported 2-aminobenzamide-based HDAC inhibitor, which is currently in a clinical trial (phase II) for treatment of patients with multiple myeloma [13]. BRD3308 is an analog of tacedinaline with higher selectivity against HDAC3 (IC50 HDAC3 = 64 nM) (Figure 1) [14].
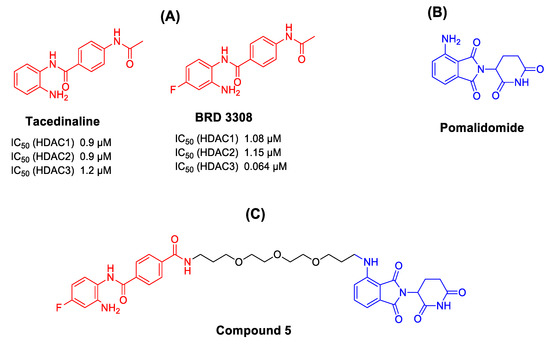
Figure 1.
Design of HDACs-PROTACs. (A) Examples of 2-aminobenzamide-based HDAC inhibitors. (B) Example of CRBN-based E3 ligase ligand. (C) Our designed HDAC-PROTACs.
PROTACs are hybrid bifunctional molecules connecting a protein of interest (POI) ligand to an E3 ubiquitin ligase (E3) recruiting ligand using a certain linker [15]. The von Hippel–Lindau (VHL) and cereblon (CRBN) ligands are the most frequently used E3 ligands for PROTACs design [16]. Binding of a PROTACs molecule to both a protein of interest and E3 ligase induce the formation of ternary complex. Formation of such a complex hijacks subsequent ubiquitination of the target protein and degradation via the ubiquitin-proteasome system (UPS) [16]. Due to this characteristic mechanism of action, the PROTACs approach has several advantages compared to conventional inhibitors. Firstly, PROTACs technology has been demonstrated to overcome the problem of resistance encountered in most current therapeutics through elimination of the entire target, which results in deletion of both enzymatic and non-enzymatic activity of the target protein [15]. In addition, the degradation of a POI through PROTACs is a catalytic process which means that only low doses of PROTACs are required; therefore, PROTACs are less prone to target overexpression and mutations [15]. In the past few years, targeting different HDAC isoforms using the PROTAC approach has attracted great interest. In 2018, we reported the development of the first Sirtuin-2 deacetylase degrader [17]. Very recently we have also reported the design of novel HDAC8 degraders [18].
In the present study, we planned to design a selective degrader for class-I HDACs isoforms. For the PROTAC design, the 2-aminobenzamide selective HDAC-3 inhibitor (BRD3308) was chosen and connected to the (CRBN) E3 ligase ligand (pomalidomide) through a polyethylene glycol linker (Figure 1).
2. Results and Discussion
2.1. Chemistry
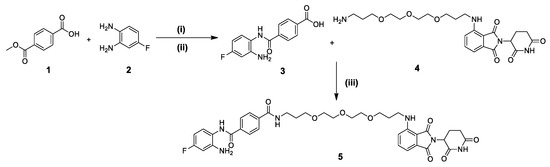
To obtain the desired compound, a synthetic strategy was used that involved three main steps, as shown in Scheme 1. The first step involved synthesis of the HDAC3 inhibitor part functionalized with carboxylic acid group 3. This intermediate was synthesized via the amide coupling reaction between 4-methoxycarbonylbenzoic acid (1) and 4-fluorobenzene-1,2-diamine (2) using hexafluorophosphate azabenzotriazole tetramethyl uronium (HATU) as a carboxylic acid activating agent and DIPEA, followed by the alkaline hydrolysis of the methyl ester using lithium hydroxide to afford the desired carboxylic acid 3. The second step included the preparation of the pomalidomide connected to the polyethylene glycol linker functionalized with terminal primary amine intermediate 4. This intermediate was synthesized according to the previously reported method [19]. Finally, amide coupling between the obtained carboxylic acid (3) and the pomalidomide-linker-amine 4 was carried out using the method mentioned above to afford compound 5.
Scheme 1.
The synthetic pathway toward the synthesis of compound 5 involved the synthesis of intermediates 3 and 4 followed by amide coupling to obtain the final compound. Reagents and conditions: (i) HATU, DIPEA, DMF, RT, 3 h; (ii) LiOH.H2O, THF, H2O, RT, 5 h; (iii); HATU, DIPEA, DMF, RT, 3 h.
2.2. Biological Evaluation
2.2.1. In Vitro HDAC Inhibition Assay
Compound 5 was subjected to in vitro HDAC inhibition activity against human class-I HDACs (HDAC1, 2, and 3) using a fluorogenic peptide derived from p53 (Ac-RHKK(Acetyl)-AMC) HDAC1-3 isoforms as demonstrated in Table 1 [20]. Compound 5 showed moderate inhibitory activity against HDAC1 and 2, while it showed an IC50 = 3.4 µM for HDAC3.

Table 1.
Inhibitory activity of compound (5) against HDAC 1, 2, and 3.
2.2.2. Cellular Testing
In addition, compound 5 was tested against a pancreatic cancer cell line (PSN1). In order to evaluate the degradation capability of synthesized PROTAC for HDAC1-3, the cellular levels of HDAC1-3 in PSN1 cell line were analyzed by Western blot. When tested in human HCT116 cells, compound 5 unfortunately showed no degradation of HDAC1-3 (data not shown). Further structural modifications might be tested for compound 5 (e.g., using different linkers with different lengths, further ubiquitin E3 ligase ligands) to obtain the desired degradation activity. In summary, we established a synthetic route for class I HDAC degraders with good yields that can be used for the development of further analogs.
3. Materials and Methods
3.1. General Experimental Information
Materials and reagents were purchased from Sigma-Aldrich Co., Ltd. (Darmstadt, Germany) and abcr GmbH (Karlsruhe, Germany). All solvents were analytically pure and dried before use. Thin layer chromatography was carried out on aluminum sheets coated with silica gel 60 F254 (Merck, Darmstadt, Germany). For medium pressure chromatography (MPLC), silica gel 60 (0.036e0.200 mm) was used. The melting points (mp) were determined on Boëtius hot stage apparatus (VEB Kombinat, NAGEMA, Dresden, GDR). Purity was measured by UV absorbance at 254 nm. The HPLC consisted of a LiChrosorb®® RP-18 (5 µm) 100-4.6 Merck column (Merck, Darmstadt, Germany), two LC-10AD pumps, a SPD-M10A VP PDA detector, and a SIL-HT autosampler, all from the manufacturer Shimadzu (Kyoto, Japan). The absorption spectra were recorded with a SPD-M10A diode array detector Shimadzu spectrophotometer (Kyoto, Japan). Mass spectrometry analyses were performed with a Finnigan MAT710C (Thermo Separation Products, SanJose, CA, USA) for the ESI MS spectra and with a LTQ (linear ion trap) Orbitrap XL hybrid mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) for the HRMS-ESI (high-resolution mass spectrometry) spectra. IR spectra were taken using an FTIR, using potassium bromide (KBr) as a supporting material (ATR method). For the HRMS analyses, the signal for the isotopes with the highest prevalence was given and calculated. 1H NMR spectra were taken on a Varian Inova 400 using deuterated DMSO as solvent. Chemical shifts were referenced to the residual solvent signals. The following abbreviations and formulas for solvents and reagents were used: ethyl acetate (EtOAc), N,N-dimethylformamide (DMF), dimethyl sulfoxide (DMSO), methanol (MeOH), tetrahydrofuran (THF), water (H2O), dichloromethane (DCM), N,N-diisopropylethylamine (DIPEA), O-(7-azabenzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium-hexafluorphosphate (HATU) and hydrochloric acid (HCl), trifluoroacetic acid (TFA).
3.2. Experimental Procedures and Characterization of Synthesized Compounds
4-[(2-Amino-4-fluorophenyl)carbamoyl]benzoic acid (3)
A mixture of the 4-methoxycarbonylbenzoic acid (1) (0.145 g, 0.8 mmol, 1 eq.), HATU (0.35 g, 0.92 mmol, 1.15 eq.) was dissolved in dry DMF (5 mL) and stirred at RT for 30 min. 4-Fluorobenzene-1,2-diamine (2) (0.11 g, 0.87 mmol, 1.1 eq.) and DIPEA (0.7 mL, 4.02 mmol, 5 eq.) were added and the reaction mixture was stirred at RT for 3 h. The reaction mixture was diluted with EtOAc (15 mL) and the reaction mixture was washed with 1 N NH4Cl and 1 N NaHCO3, respectively. The organic extracts were washed with brine, dried over anhydrous Na2SO4, filtered and concentrated under vacuum. The residue was purified using MPLC using (DCM—MeOH) to provide the corresponding amide, which was further dissolved in a mixture of 10 mLTHF: H2O (1:1); then, lithium hydroxide monohydrate (87 mg) was added to the solution and the reaction mixture was stirred at RT for 5 h. The TLC showed that there were no starting materials. Then, the mixture was neutralized using 1N HCl. The precipitated solid was filtered and dried under vacuum to obtain the corresponding carboxylic acid 3 as beige amorphous powder; mp. 275–277 °C; yield (0.11 g, 0.4 mmol, 50% over 2 steps); 1H NMR (400 MHz, DMSO-d6) δ 13.20 (s, 1H), 9.71 (s, 1H), 8.04 (q, J = 8.2 Hz, 4H), 7.15–7.05 (m, 1H), 6.52 (d, J = 11.2 Hz, 1H), 6.34 (t, J = 8.4 Hz, 1H), 5.24 (s, 2H); 13C NMR (101 MHz, DMSO-d6) δ 167.2, 165.4, 162.8, 160.4, 145.9, 138.8, 133.5, 129.6, 129.1, 128.5, 119.3, 102.3, 1020, 101.7; IR (KBr, ν, cm1): 3321, 3234, 3072, 2849, 2675, 2558, 1683, 1639, 1622, 1612; MS-ESI m/z: 273.18 [M–H]−; HPLC: rt 9.30 min using MeOH/H2O/0.05%TFA (purity 96.52%); UV-Vis spectra (MeOH/H2O/TFA 50: 50: 0.05), λmax: 212 nm (ε = 974 M−1 cm−1, log ε = 2.99), 226 nm (ε = 1003 M−1 cm−1, log ε = 3.00), 256 nm (ε = 411 M−1 cm−1, log ε = 2.61), 287 nm (ε = 300 M−1 cm−1, log ε = 2.48).
4-(3-{2-[2-(3-Aminopropoxy)ethoxy]ethoxy}propylamino)-2-(2,6-dioxo-3-piperidyl)isoindoline-1,3-dione (4) was synthesized according to the previously reported method (Supplementary Materials) [19].
N4-(2-Amino-4-fluorophenyl)-N1-(3-{2-[2-(3-{[2-(2,6-dioxo-3-piperidyl)-1,3-dioxoisoindolin-4-yl]amino}propoxy)ethoxy]ethoxy}propyl)terephthalamide (5)
A mixture of compound 3 (0.11, 0.4 mmol, 1.0 eq.) and HATU (0.167 g, 0.44 mmol, 1.1 eq.) was dissolved in dry DMF (5 mL) and stirred at RT for 30 min. Intermediate 4 (0.21 g, 0.44 mmol, 1.1 eq.) and DIPEA (0.35 mL, 2 mmol, 5.0 eq.) were added and the reaction mixture was stirred at RT for 3 h. The reaction mixture was diluted with ethyl acetate (30 mL) and the reaction mixture was washed with 1 N NH4Cl and saturated NaHCO3, respectively. The combined organic extracts were washed with brine, dried over anhydrous Na2SO4, filtered, and concentrated under vacuum. The residue was purified using MPLC (Chloroform—MeOH) to provide the targeted compound as yellow amorphous powder; mp. 96–98 °C; yield (0.12 g, 0.16 mmol, 40.9%); 1H NMR (400 MHz, DMSO-d6) δ 11.06 (s, 1H), 9.66 (s, 1H), 8.54 (t, J = 5.5 Hz, 1H), 8.02 (d, J = 8.4 Hz, 2H), 7.92 (d, J = 8.4 Hz, 2H), 7.55 (dd, J = 8.5, 7.2 Hz, 1H), 7.14–7.05 (m, 2H), 7.00 (d, J = 6.9 Hz, 1H), 6.64 (t, J = 5.9 Hz, 1H), 6.53 (dd, J = 11.2, 2.9 Hz, 1H), 6.34 (td, J = 8.5, 2.9 Hz, 1H), 5.23 (s, 2H), 5.03 (dd, J = 12.8, 5.4 Hz, 1H), 3.57–3.42 (m, 12H), 3.39–3.30 (m, 4H), 2.86 (ddd, J = 17.6, 14.1, 5.5 Hz, 1H), 2.62–2.51 (m, 2H), 2.00 (ddd, J = 10.6, 7.2, 4.8 Hz, 1H), 1.83–1.71 (m, 4H); 13C NMR (101 MHz, DMSO-d6) δ 173.2, 170.5, 169.3, 167.8, 165.9, 165.4, 146.9, 146.1, 146.0, 137.4, 137.1, 136.7, 132.6, 129.1, 128.2, 127.4, 117.5, 110.8, 109.5, 102.6, 102.3, 102.0, 101.7, 70.3, 70.2, 70.1, 70.0, 68.7, 68.7, 49.0, 37.2, 31.4, 29.8, 29.3, 22.6; IR (KBr, ν, cm−1): 3360, 2870, 1693., 1624, 1511; HRMS m/z: 733.299 [M + H]+; calculated C37H42FN6O9+: 733.299; HPLC: rt 12.26 min using MeOH/H2O/0.05%TFA (purity 96.59%); UV-Vis spectra (MeOH/H2O/TFA 50: 50: 0.05), λmax: 229 nm (ε = 2839 M−1 cm−1, log ε = 3.45), 260 nm (ε = 1109 M−1 cm−1, log ε = 3.04), 420 nm (ε = 569 M−1 cm−1, log ε = 2.76).
3.3. Biological Testing
In Vitro HDAC Inhibition Assay
Recombinant human HDAC1, HDAC2, and HDAC3/NCOR1 were purchased from ENZO Life Sciences AG (Lausen, CH). The in vitro testing on recombinant HDACs 1-3 was performed as described in (Supplementary Materials) [12].
Supplementary Materials
The following supporting information can be downloaded online. Synthesis of intermediate 4, in vitro HDAC inhibitory assay and copies of 1H-NMR, 13C-NMR, HPLC, IR, MS, and UV spectra.
Author Contributions
M.A. carried out the synthesis and analytical characterization and wrote the manuscript, M.Z. carried out the in vitro HDAC testing, M.S. (Matthias Schmidt) contributed in synthesis planning analytical characterization, M.S. (Mike Schutkowski) supervised the in vitro testing, W.S. carried out data analysis, supervised the project and manuscript writing. All authors have read and agreed to the published version of the manuscript.
Funding
German Academic Exchange Service: Fellowship Mohamed Abdelsalam (DAAD-GERLS 2018).
Data Availability Statement
Not applicable.
Acknowledgments
M.A. appreciates the support of DAAD and the Ministry of Higher Education and Scientific Research (Egypt) scholarship (GERLS).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Pant, K.; Peixoto, E.; Richard, S.; Gradilone, S.A. Role of histone deacetylases in carcinogenesis: Potential role in cholangiocarcinoma. Cells 2020, 9, 780. [Google Scholar] [CrossRef] [PubMed]
- Fraga, M.F.; Ballestar, E.; Villar-Garea, A.; Boix-Chornet, M.; Espada, J.; Schotta, G.; Bonaldi, T.; Haydon, C.; Ropero, S.; Petrie, K. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat. Genet. 2005, 37, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Abel, T.; Zukin, R.S. Epigenetic targets of HDAC inhibition in neurodegenerative and psychiatric disorders. Curr. Opin. Pharmacol. 2008, 8, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Miao, X.; Liu, Y.; Li, F.; Liu, Q.; Sun, J.; Cai, L. Dysregulation of Histone Acetyltransferases and Deacetylases in Cardiovascular Diseases. Oxid. Med. Cell. Longev. 2014, 2014, 641979. [Google Scholar] [CrossRef] [PubMed]
- Zeng, C.; Tsoi, L.C.; Gudjonsson, J.E. Dysregulated epigenetic modifications in psoriasis. Exp. Dermatol. 2021, 30, 1156–1166. [Google Scholar] [CrossRef] [PubMed]
- Gryder, B.E.; Sodji, Q.H.; Oyelere, A.K. Targeted cancer therapy: Giving histone deacetylase inhibitors all they need to succeed. Future Med. Chem. 2012, 4, 505–524. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, J.; Jiang, Q.; Zhang, L.; Song, W. Zinc binding groups for histone deacetylase inhibitors. J. Enzym. Inhib. Med. Chem. 2018, 33, 714–721. [Google Scholar] [CrossRef] [PubMed]
- Wagner, F.F.; Weïwer, M.; Lewis, M.C.; Holson, E.B. Small molecule inhibitors of zinc-dependent histone deacetylases. Neurotherapeutics 2013, 10, 589–604. [Google Scholar] [CrossRef] [PubMed]
- Ho, T.C.S.; Chan, A.H.Y.; Ganesan, A. Thirty Years of HDAC Inhibitors: 2020 Insight and Hindsight. J. Med. Chem. 2020, 63, 12460–12484. [Google Scholar] [CrossRef] [PubMed]
- Perrin, J.; Werner, T.; Kurzawa, N.; Rutkowska, A.; Childs, D.D.; Kalxdorf, M.; Poeckel, D.; Stonehouse, E.; Strohmer, K.; Heller, B. Identifying drug targets in tissues and whole blood with thermal-shift profiling. Nat. Biotechnol. 2020, 38, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Becher, I.; Werner, T.; Doce, C.; Zaal, E.A.; Tögel, I.; Khan, C.A.; Rueger, A.; Muelbaier, M.; Salzer, E.; Berkers, C.R. Thermal profiling reveals phenylalanine hydroxylase as an off-target of panobinostat. Nat. Chem. Biol. 2016, 12, 908–910. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, H.S.; Abdelsalam, M.; Zeyn, Y.; Zessin, M.; Mustafa, A.-H.M.; Fischer, M.A.; Zeyen, P.; Sun, P.; Bülbül, E.F.; Vecchio, A.; et al. Synthesis, Molecular Docking and Biological Characterization of Pyrazine Linked 2-Aminobenzamides as New Class I Selective Histone Deacetylase (HDAC) Inhibitors with Anti-Leukemic Activity. Int. J. Mol. Sci. 2022, 23, 369. [Google Scholar] [CrossRef] [PubMed]
- Xie, R.; Tang, P.; Yuan, Q. Rational design and characterization of a DNA/HDAC dual-targeting inhibitor containing nitrogen mustard and 2-aminobenzamide moieties. Medchemcomm 2018, 9, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Wagner, F.F.; Lundh, M.; Kaya, T.; McCarren, P.; Zhang, Y.-L.; Chattopadhyay, S.; Gale, J.P.; Galbo, T.; Fisher, S.L.; Meier, B.C.; et al. An Isochemogenic Set of Inhibitors to Define the Therapeutic Potential of Histone Deacetylases in β-Cell Protection. ACS Chem. Biol. 2016, 11, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Gao, H.; Yang, Y.; He, M.; Wu, Y.; Song, Y.; Tong, Y.; Rao, Y. PROTACs: Great opportunities for academia and industry. Signal Transduct. Target. Ther. 2019, 4, 64. [Google Scholar] [CrossRef] [PubMed]
- Bricelj, A.; Steinebach, C.; Kuchta, R.; Gütschow, M.; Sosič, I. E3 Ligase Ligands in Successful PROTACs: An Overview of Syntheses and Linker Attachment Points. Front. Chem. 2021, 9, 707317. [Google Scholar] [CrossRef] [PubMed]
- Schiedel, M.; Herp, D.; Hammelmann, S.R.; Swyter, S.R.; Lehotzky, A.; Robaa, D.; Oláh, J.; Ovadi, J.; Sippl, W.; Jung, M. Chemically induced degradation of sirtuin 2 (Sirt2) by a proteolysis targeting chimera (PROTAC) based on sirtuin rearranging ligands (SirReals). J. Med. Chem. 2018, 61, 482–491. [Google Scholar] [CrossRef] [PubMed]
- Darwish, S.; Ghazy, E.; Heimburg, T.; Herp, D.; Zeyen, P.; Salem-Altintas, R.; Ridinger, J.; Robaa, D.; Schmidtkunz, K.; Erdmann, F.; et al. Design, Synthesis and Biological Characterization of Histone Deacetylase 8 (HDAC8) Proteolysis Targeting Chimeras (PROTACs) with Anti-Neuroblastoma Activity. Int. J. Mol. Sci. 2022, 23, 7535. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Li, Y.; Wang, X.; Dong, G.; Sheng, C. Discovery of Novel PDEδ Degraders for the Treatment of KRAS Mutant Colorectal Cancer. J. Med. Chem. 2020, 63, 7892–7905. [Google Scholar] [CrossRef] [PubMed]
- Zessin, M.; Kutil, Z.F.; Meleshin, M.; Nováková, Z.; Ghazy, E.; Kalbas, D.; Marek, M.; Romier, C.; Sippl, W.; Bařinka, C. One-atom substitution enables direct and continuous monitoring of histone deacylase activity. Biochemistry 2019, 58, 4777–4789. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).