2-Bromo-3-((1-(7-chloroquinolin-4-yl)-1H-1,2,3-triazol-4-yl)-methoxy)-benzaldehyde
Abstract
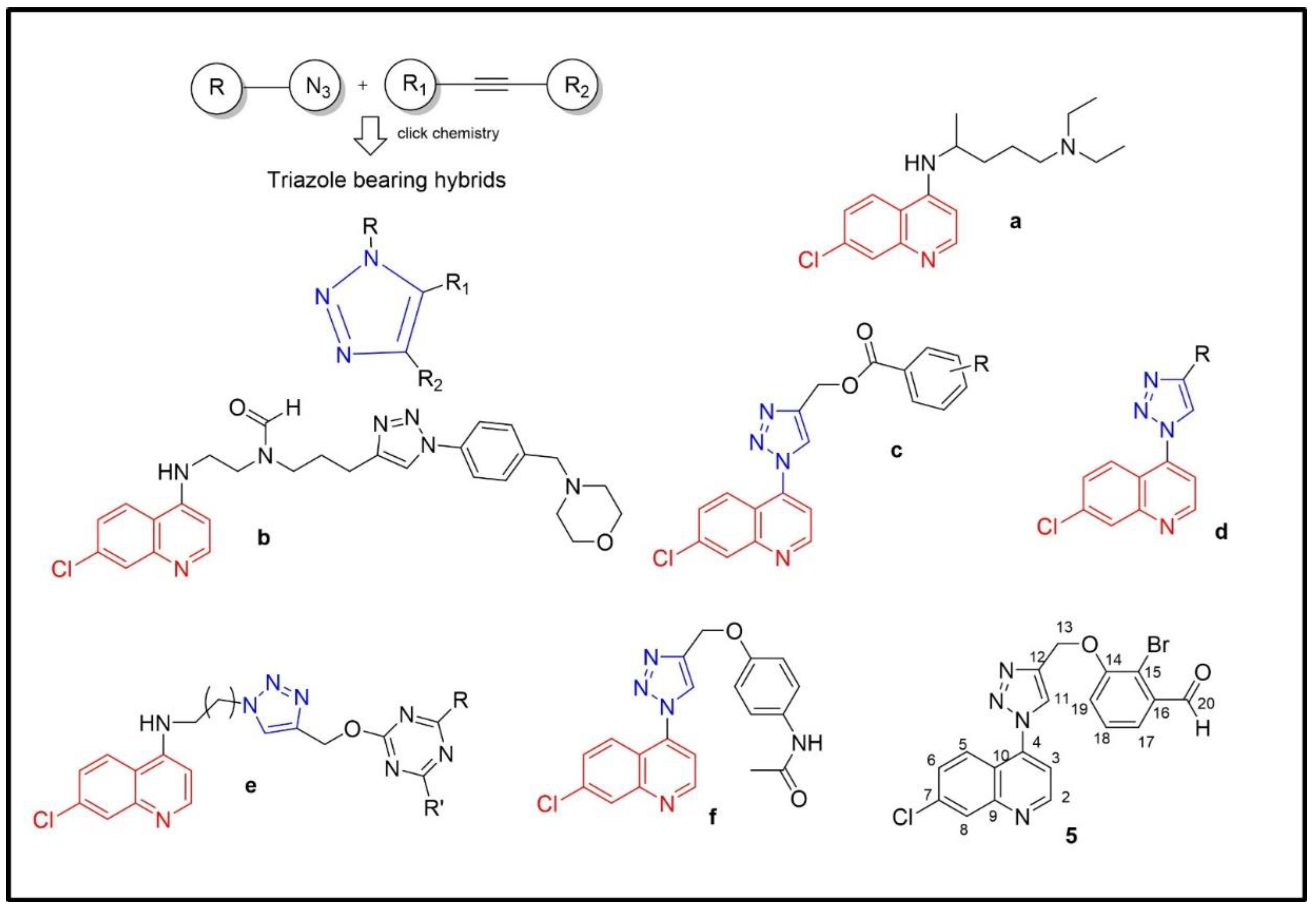
:1. Introduction
2. Results and Discussion
3. Materials and Methods
3.1. Chemistry
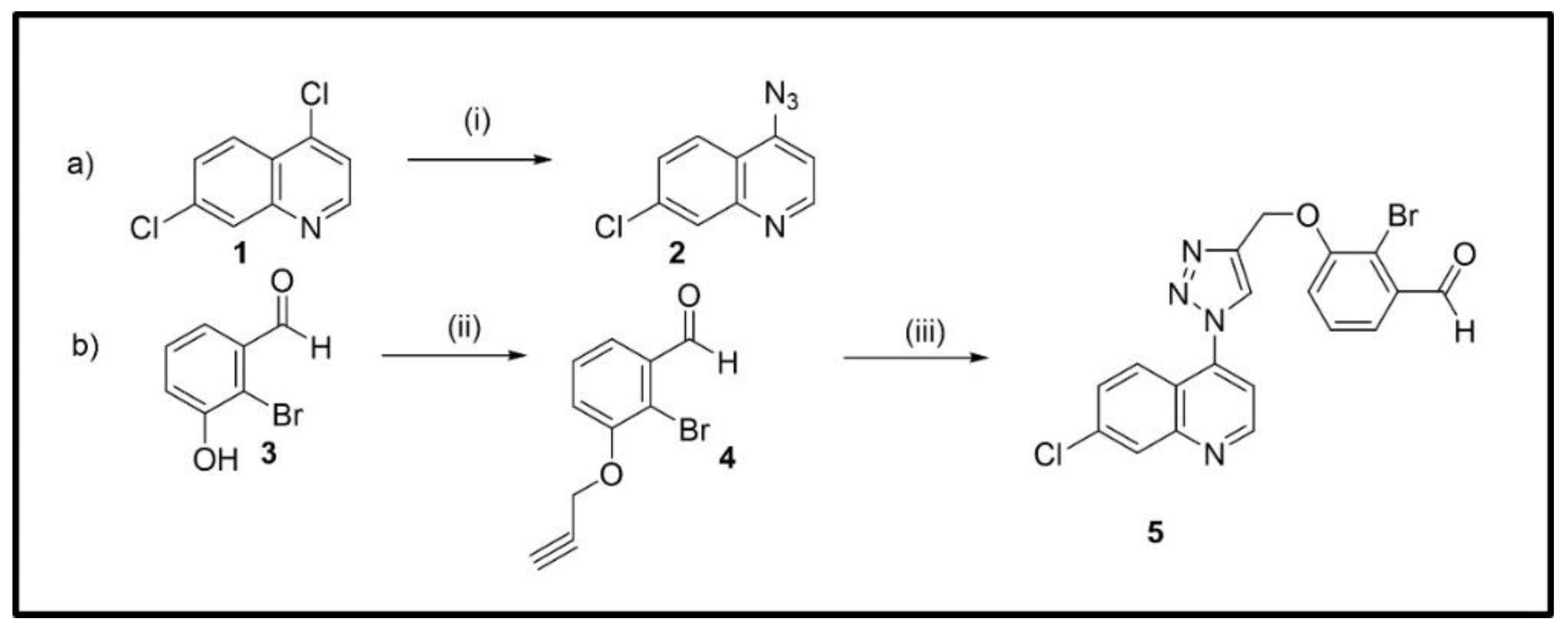
3.1.1. Synthesis of 4-Azido-7-chloroquinoline (2)
3.1.2. Synthesis of 2-Bromo-3-(prop-2-yn-1-yloxy) benzaldehyde (4)
3.1.3. Synthesis of 2-Bromo-3-((1-(7-chloroquinolin-4-yl)-1H-1,2,3-triazol-4-yl)-methoxy)-benzaldehyde (5)
3.2. Biological Studies
3.2.1. Cytotoxicity Drug Assay
3.2.2. Biolayer Interferometry Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Colson, P.; Rolain, J.M.; Raoult, D. Chloroquine for the 2019 novel coronavirus SARS-CoV-2. Int. J. Antimicrob. Agents 2020, 55, 105923. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Cao, R.; Zhang, L.; Yang, X.; Liu, J.; Xu, M.; Shi, Z.; Hu, Z.; Zhong, W.; Xiao, G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020, 30, 269–271. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, K.; Tsuno, N.H.; Sunami, E.; Tsurita, G.; Kawai, K.; Okaji, Y.; Nishikawa, T.; Shuno, Y.; Hongo, K.; Hiyoshi, M.; et al. Chloroquine potentiates the anti-cancer effect of 5-fluorouracil on colon cancer cells. BMC Cancer 2010, 10, 370. [Google Scholar] [CrossRef] [Green Version]
- Herrmann, L.; Hahn, F.; Wangen, C.; Marschall, M.; Tsogoeva, S.B. Anti-SARS-CoV-2 Inhibitory Profile of New Quinoline Compounds in Cell Culture-Based Infection Models. Chem. Eur. J. 2022, 28, e202103861. [Google Scholar] [CrossRef] [PubMed]
- Harrison, J.R.; Brand, S.; Smith, V.; Robinson, D.A.; Thompson, S.; Smith, A.; Davies, K.; Mok, N.; Torrie, L.S.; Collie, I.; et al. A Molecular Hybridization Approach for the Design of Potent, Highly Selective, and Brain-Penetrant N-Myristoyltransferase Inhibitors. J. Med. Chem. 2018, 61, 8374–8389. [Google Scholar] [CrossRef]
- Ivasiv, V.; Albertini, C.; Gonçalves, A.E.; Rossi, M.; Bolognesi, M.L. Molecular Hybridization as a Tool for Designing Multitarget Drug Candidates for Complex Diseases. Curr. Top. Med. Chem. 2019, 19, 1694–1711. [Google Scholar] [CrossRef]
- Bozorov, K.; Zhao, J.; Aisa, H.A. 1,2,3-Triazole-containing hybrids as leads in medicinal chemistry: A recent overview. Bioorg. Med. Chem. 2019, 27, 3511–3531. [Google Scholar] [CrossRef]
- Demaray, J.A.; Thuener, J.E.; Dawson, M.N.; Sucheck, S.J. Synthesis of triazole-oxazolidinones via a one-pot reaction and evaluation of their antimicrobial activity. Bioorg. Med. Chem. Lett. 2008, 18, 4868–4871. [Google Scholar] [CrossRef]
- Tripathi, R.P.; Yadav, A.K.; Ajay, A.; Bisht, S.S.; Chaturvedi, V.; Sinha, S.K. Application of Huisgen (3 + 2) cycloaddition reaction: Synthesis of 1-(2,3-dihydrobenzofuran-2-yl-methyl [1,2,3]-triazoles and their antitubercular evaluations. Eur. J. Med. Chem. 2010, 45, 142–148. [Google Scholar] [CrossRef]
- Guanti, E.M.; Ncokazi, K.; Egan, T.J.; Gut, J.; Rosenthal, P.J.; Smith, P.J.; Chibale, K. Design, synthesis and in vitro antimalarial evaluation of triazole-linked chalcone and dienone hybrid compounds. Bioorg. Med. Chem. 2010, 18, 8243–8256. [Google Scholar] [CrossRef] [PubMed]
- Coghi, P.; Ng, J.P.L.; Nasim, A.A.; Wong, V.K.W. N-[7-Chloro-4-[4-(phenoxymethyl)-1H-1,2,3-triazol-1-yl] quinoline]-acetamide. Molbank 2021, 2021, M1213. [Google Scholar] [CrossRef]
- Saini, A.; Kumar, S.; Raj, R.; Chowdhary, S.; Gendrot, M.; Mosnier, J.; Fonta, I.; Pradines, B.; Kumar, V. Synthesis and antiplasmodial evaluation of 1H-1,2,3-triazole grafted 4-aminoquinoline-benzoxaborole hybrids and benzoxaborole analogues. Bioorg. Chem. 2021, 109, 104733. [Google Scholar] [CrossRef] [PubMed]
- Coghi, P.S.; Zhu, Y.; Xie, H.; Hosmane, N.S.; Zhang, Y. Organoboron Compounds: Effective Antibacterial and Antiparasitic Agents. Molecules 2021, 26, 3309. [Google Scholar] [CrossRef] [PubMed]
- De Souza, M.V.; Pais, K.C.; Kaiser, C.R.; Peralta, M.A.; de L. Ferreira, M.; Lourenço, M.C. Synthesis and in vitro antitubercular activity of a series of quinoline derivatives. Bioorg. Med. Chem. 2009, 17, 1474–1480. [Google Scholar] [CrossRef]
- Feldman, A.K.; Colasson, B.; Fokin, V.V. One-Pot Synthesis of 1,4-Disubstitued 1,2,3- Triazole from In Situ Generated Azides. Org. Lett. 2004, 6, 3897–3899. [Google Scholar] [CrossRef]
- Lipinski, C.A. Drug-like properties and the causes of poor solubility and poor permeability. J. Pharmacol. Toxicol. Methods 2000, 44, 235–249. [Google Scholar] [CrossRef]
- Daina, A.; Zoete, V.A. Boiled-egg to predict gastrointestinal absorption and brain penetration of small molecules. ChemMedChem 2016, 11, 1117–1121. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Lou, C.; Sun, L.; Li, J.; Cai, Y.; Wang, Z.; Li, W.; Liu, G.; Tang, Y. AdmetSAR 2.0: Web-service for prediction and optimization of chemical ADMET properties. Bioinformatics 2018, 35, 1067–1069. [Google Scholar] [CrossRef]
- Coghi, P.; Yang, L.J.; Ng, J.P.L.; Haynes, R.K.; Memo, M.; Gianoncelli, A.; Wong, V.K.W.; Ribaudo, G. A Drug Repurposing Approach for Antimalarials Interfering with SARS-CoV-2 Spike Protein Receptor Binding Domain (RBD) and Human Angiotensin-Converting Enzyme 2 (ACE2). Pharmaceuticals 2021, 14, 954. [Google Scholar] [CrossRef]
- Tiwari, M.K.; Coghi, P.; Agarwal, P.; Shyamlal, R.B.K.; Yadav, L.; Sharma, R.; Yadav, D.K.; Sahal, D.; Wong, V.K.W.; Chaudhary, S. Design, Synthesis, Structure-Activity Relationship and Docking Studies of Novel Functionalized Arylvinyl-1,2,4-Trioxanes as Potent Antiplasmodial as well as Anticancer Agents. ChemMedChem 2020, 15, 1216–1228. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 3, 42717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ertl, P.; Rohde, B.; Selzer, P. Fast calculation of molecular polar surface area as a sum of fragment-based contributions and its application tothe prediction of drug transport properties. J. Med. Chem. 2000, 43, 3714–3717. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yun, X.; Xie, Y.; Ng, J.P.L.; Law, B.Y.K.; Wong, V.K.W.; Coghi, P. 2-Bromo-3-((1-(7-chloroquinolin-4-yl)-1H-1,2,3-triazol-4-yl)-methoxy)-benzaldehyde. Molbank 2022, 2022, M1351. https://doi.org/10.3390/M1351
Yun X, Xie Y, Ng JPL, Law BYK, Wong VKW, Coghi P. 2-Bromo-3-((1-(7-chloroquinolin-4-yl)-1H-1,2,3-triazol-4-yl)-methoxy)-benzaldehyde. Molbank. 2022; 2022(1):M1351. https://doi.org/10.3390/M1351
Chicago/Turabian StyleYun, Xiaoyun, Yuhan Xie, Jerome P. L. Ng, Betty Yuen Kwan Law, Vincent Kam Wai Wong, and Paolo Coghi. 2022. "2-Bromo-3-((1-(7-chloroquinolin-4-yl)-1H-1,2,3-triazol-4-yl)-methoxy)-benzaldehyde" Molbank 2022, no. 1: M1351. https://doi.org/10.3390/M1351
APA StyleYun, X., Xie, Y., Ng, J. P. L., Law, B. Y. K., Wong, V. K. W., & Coghi, P. (2022). 2-Bromo-3-((1-(7-chloroquinolin-4-yl)-1H-1,2,3-triazol-4-yl)-methoxy)-benzaldehyde. Molbank, 2022(1), M1351. https://doi.org/10.3390/M1351







