Abstract
On the basis of the knowledge from traditional herbal and folk medicine, flavonoids are among the most studied chemical classes of natural compounds for their potential activity as phosphodiesterase 5 (PDE5) inhibitors. We here describe the preparation of a semi-synthetic hydrazone derivative of quercetin, 2-(3,4-dihydroxyphenyl)-4-(2-(4-nitrophenyl)hydrazono)-4H-chromene-3,5,7-triol, that was obtained via a single-step modification of the natural compound. The product was characterized by NMR, mass spectrometry and HPLC. Preliminary molecular modeling studies suggest that this compound could efficiently interact with PDE5.
1. Introduction
Phosphodiesterase (PDE) inhibitors contrast the degradation of 3′,5′-cyclic adenosine monophosphate (cAMP) and/or 3′,5′-cyclic guanosine monophosphate (cGMP), thus promoting several downstream effects. The inhibitors of PDE5 isoform, in particular, interfere with cGMP hydrolysis and induce smooth-muscle relaxation in specific tissues [1,2]. These compounds find clinical applications in the treatment of erectile dysfunction and pulmonary hypertension, and they are being studied as potential treatments against other diseases [3,4,5]. Repurposing of approved drugs is becoming an attractive strategy for identifying new applications for compounds with proved safety [6], and in this context PDE5 inhibitors are currently under investigation to contrast neurodegeneration [7,8], depression [9], diabetes [10] and rare pathologies such as Duchenne muscular dystrophy [11].
The development of synthetic PDE5 inhibitors, such as sildenafil (Figure 1A) and its analogues, has been paralleled by the exploration of the potential activity of natural compounds from traditional and folk medicine [12,13,14]. Flavonoids, in particular, have been known as PDE inhibitors for decades [15]. Natural glycosylated flavonoids and aglycones [16,17,18,19], as well as semi-synthetic flavones [20] and isoflavones [21,22,23], have been studied in silico, in vitro and in vivo for their inhibitory activity towards PDE5 and other isoforms.
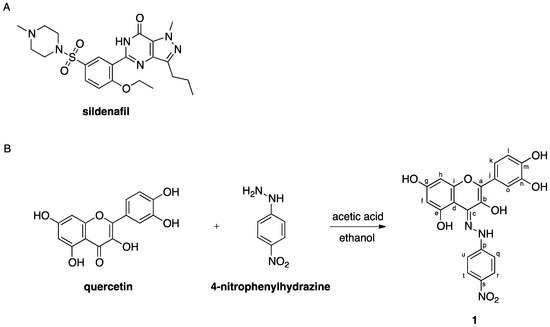
Figure 1.
Chemical structure of sildenafil (A) and synthetic scheme for the preparation of compound 1 (B).
The single-step derivatization procedure to obtain 2-(3,4-dihydroxyphenyl)-4-(2-(4-nitrophenyl)hydrazono)-4H-chromene-3,5,7-triol (1) from quercetin is here reported. This semi-synthetic compound was characterized by NMR, mass spectrometry and HPLC. Its interaction pattern with PDE5 was investigated in silico and compared to that of quercetin and sildenafil.
2. Results and Discussion
2.1. Chemistry
Quercetin was previously reported to possess inhibitory activity on several PDE isoforms [24], and the vasorelaxant effect of this natural flavonoid and of the corresponding metabolites was demonstrated to be due to the interference with the cGMP pathway [25]. Chan et al. investigated the effects of quercetin derivatives on PDE isoforms [26], and we previously explored the semi-synthetic derivatization of flavonoids to enhance their interaction with PDE5 in silico and in vitro [21,22].
We here report the preparation of a hydrazone derivative, 2-(3,4-dihydroxyphenyl)-4-(2-(4-nitrophenyl)hydrazono)-4H-chromene-3,5,7-triol (1), that was obtained via a single-step modification procedure from quercetin (Figure 1B). Rollas et al. recently discussed the biochemical relevance, in terms of reactivity and bioactivity aspects, of hydrazones [27]. Hydrazones are generally prepared from carbonyl compounds by reaction with an opportune hydrazine in acidic conditions [28,29]. Hydrochloric, acetic or 3-chloroperbenzoic acid are usually adopted for the synthesis of hydrazones, and the use of catalysts has been reported [30,31]. Compound 1 was synthesized by reacting quercetin with an excess of 4-nitrophenylhydrazine in a 1:1 mixture of acetic acid and ethanol. Following this procedure, the compound was isolated by filtration in a good yield (56%).
Compound 1 was characterized by NMR, mass spectrometry and HPLC (see Figures S1–S4 in the Supplementary material).
2.2. Molecular Modeling
The interaction of compound 1 with PDE5 was investigated in silico following a protocol reported previously [22]. For comparison, sildenafil and quercetin were also docked to the same 3D model and the predicted interaction patterns demonstrated a good co-localization of the ligands within the protein. The calculated binding energy value was particularly encouraging for compound 1 (−10.3 kcal/mol), exceeding that predicted for sildenafil (−9.7 kcal/mol) and quercetin (−9.5 kcal/mol) (Figure 2A). More in detail, docking experiments showed that the three compounds bind to the same region of the protein, consisting in the catalytic site (Figure 2B–D and Figures S5–S10 in the Supplementary materials). Most importantly, according to the predicted models, compound 1, sildenafil and quercetin interact with the same group of residues in such a PDE5 domain. In particular, Ile778, Val782, Ala783, Leu804, Ile813, Met816, Gln817 and Phe820, which were previously reported to be relevant interacting residues for known PDE5 inhibitors [32], were highlighted within the < 5 Å region from the docked ligands (Figure 2).
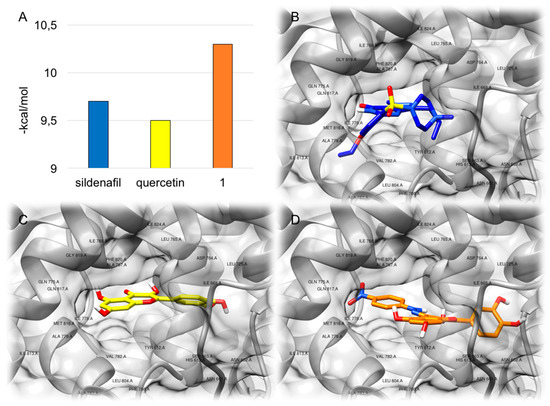
Figure 2.
Results of the docking study for compound 1 to PDE5, in comparison with sildenafil and quercetin. (A): calculated binding energy values obtained from docking experiments. (B): docking pose of sildenafil. (C): docking pose of quercetin. (D): docking pose of compound 1.
Furthermore, the stability of the complex predicted for compound 1 and PDE5 was assessed using molecular dynamics simulations [33]. The results show that the complex reached stability after 8 ns, and it was retained during the remaining simulation time (see Figures S11–S12 in the Supplementary materials)
3. Materials and Methods
3.1. Chemistry
3.1.1. General
Commercially available chemicals were purchased from Sigma–Aldrich (Saint Louis, MO, USA) and used as received, unless otherwise stated. 1H and 13C{1H} NMR spectra were recorded on an Avance III 400 MHz spectrometer (Bruker, Billerica, MA, USA). All spectra were recorded at room temperature; the solvent for each spectrum is given in parentheses. Chemical shifts are reported in ppm and are relative to tetramethylsilane (TMS) internally referenced to the residual solvent peak. Datasets were edited with iNMR (Nucleomatica, Molfetta, Italy). The multiplicity of signals is reported as a singlet (s), doublet (d), triplet (t), quartet (q), multiplet (m), broad (b) or a combination of any of these. Mass spectra were recorded by direct infusion ESI on a Xevo G2-XS (Waters, Milford, MA, USA). The purity profile (96%) was assayed by HPLC using a Pro-Star system (Varian, Palo Alto, CA, USA) equipped with a 1706 UV–VIS detector (254 nm, Bio-rad, Hercules, CA, USA) and an C-18 column (5 μm, 4.6 × 250 mm, Agilent Technologies, Santa Clara, CA, USA). An appropriate ratio of water (A) and acetonitrile (B) was used as mobile phase with an overall flow rate of 1 mL/min; the general methods for the analyses are reported here: 0 min (95% A–5% B), 5 min (95% A–5% B), 25 min (5% A–95% B), 35 min (5% A–95% B) and 40 min (95% A–5% B).
3.1.2. Synthesis of 2-(3,4-dihydroxyphenyl)-4-(2-(4-nitrophenyl)hydrazono)-4H-chromene-3,5,7-triol (1)
A round-bottom flask was charged with quercetin (50.0 mg, 0.17 mmol) and ethanol (5 mL). A solution of 4-nitrophenylhydrazine (76.0 mg, 0.50 mmol) in acetic acid (5 mL) was added dropwise to this mixture and the reaction was refluxed under stirring for 6 h. After cooling to room temperature, the concentration of the solvent induced the formation of a precipitate. The solid, collected by filtration, was triturated using diethyl ether and the resulting product was isolated as a brown solid (42.0 mg). Yield: 56%, mp. 264–267 °C, HPLC r.t. 21.8 min. 1H-NMR (DMSO-d6, 400 MHz): δH, 12.48 (1H, bs, OH), 9.96 (1H, bs, OH), 8.99 (1H, bs, OH), 8.05 (2H, d, J 8.1 Hz, Hr and Ht), 7.67 (1H, d, J 1.8 Hz, Ho), 7.52 (1H, dd, J 8.0 Hz, J 1.8 Hz, Hk), 6.89 (1H, d, J 8.0 Hz, Hl), 6.73 (2H, d, J 8.1 Hz, Hq and Hu), 6.37 (1H, s, Hf of Hh), 6.15 (1H, s, Hh or Hf). 13C {1H}-NMR (DMSO-d6, 100 MHz): δC 177.2, 176.3, 169.5, 164.3, 161.1, 159.2, 156.5, 155.4, 148.1, 147.2, 145.5, 138.4, 136.2, 126.3, 122.4, 120.4, 116.0, 115.5, 110.9. ESI-MS found 438.454 (C21H16N3O8+. [M + H]+), calc. 438.366.
3.2. Molecular Modeling
The structure of PDE5 was obtained from the RCSB Protein Data Bank (www.rcsb.org, PDB ID: 2H42). The target and ligands were prepared for the blind docking experiment which was performed using Autodock Vina (Molecular Graphics Laboratory, Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, CA, USA) [34]. Output data (energies, interaction patterns) were analyzed and scored using a UCSF Chimera molecular viewer [35], which was also used to produce the artworks. Molecular dynamics simulations were carried out using PlayMolecule (Accelera, Middlesex, UK) starting from the output model of docking experiments. A ligand was prepared by running a Parametrize function based on GAFF2 force field [36]. The complex was prepared for the simulation using ProteinPrepare and SystemBuilder functions, setting pH = 7.4, AMBER force field and default experiment parameters [37]. A simulation of 25 ns was carried out using SimpleRun, with default settings [38].
4. Conclusions
In this short note, we reported the preparation of 2-(3,4-dihydroxyphenyl)-4-(2-(4-nitrophenyl)hydrazono)-4H-chromene-3,5,7-triol (1), a semi-synthetic hydrazone derivative of quercetin that was obtained via a single-step approach. Preliminary in silico studies suggest that this compound could efficiently interact with the catalytic domain of PDE5 and that the effect on enzymatic inhibition of quercetin derivatives bearing this or other hydrazone substituents should be evaluated in vitro.
Supplementary Materials
The following are available online, Figures S1 and S2: NMR spectra, Figure S3: ESI-MS spectrum, Figure S4: HPLC profile, Figures S5–S10: docking studies, Figures S11 and S12: molecular dynamics simulations.
Author Contributions
Conceptualization, A.G. and G.R.; methodology, G.R.; software, A.O. and G.R.; investigation, A.O.; data curation, A.G.; writing—original draft preparation, A.G., A.O. and G.R.; writing—review and editing, M.M. and G.Z.; supervision, A.G. and G.R.; funding acquisition, A.G. and G.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research was granted by University of Brescia.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gur, S.; Kadowitz, P.J.; Serefoglu, E.C.; Hellstrom, W.J.G. PDE5 inhibitor treatment options for urologic and non-urologic indications: 2012 update. Curr. Pharm. Des. 2012, 18, 5590–5606. [Google Scholar] [CrossRef] [PubMed]
- Andersson, K.-E. PDE5 inhibitors - pharmacology and clinical applications 20 years after sildenafil discovery. Br. J. Pharmacol. 2018. [Google Scholar] [CrossRef]
- Ribaudo, G.; Pagano, M.A.; Bova, S.; Zagotto, G. New Therapeutic Applications of Phosphodiesterase 5 Inhibitors (PDE5-Is). Curr. Med. Chem. 2016, 23, 1239–1249. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.S. Tadalafil: 15 years’ journey in male erectile dysfunction and beyond. Drug Dev. Res. 2018, ddr.21493. [Google Scholar] [CrossRef] [PubMed]
- Nabavi, S.M.; Talarek, S.; Listos, J.; Nabavi, S.F.; Devi, K.P.; Roberto de Oliveira, M.; Tewari, D.; Argüelles, S.; Mehrzadi, S.; Hosseinzadeh, A.; et al. Phosphodiesterase inhibitors say NO to Alzheimer’s disease. Food Chem. Toxicol. 2019, 134, 110822. [Google Scholar] [CrossRef] [PubMed]
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; et al. Drug repurposing: Progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019, 18, 41–58. [Google Scholar] [CrossRef] [PubMed]
- Zuccarello, E.; Acquarone, E.; Calcagno, E.; Argyrousi, E.K.; Deng, S.X.; Landry, D.W.; Arancio, O.; Fiorito, J. Development of novel phosphodiesterase 5 inhibitors for the therapy of Alzheimer’s disease. Biochem. Pharmacol. 2020, 113818. [Google Scholar] [CrossRef] [PubMed]
- Ribaudo, G.; Ongaro, A.; Zagotto, G.; Memo, M.; Gianoncelli, A. Therapeutic Potential of Phosphodiesterase (PDE) Inhibitors Against Neurodegeneration: The Perspective of the Medicinal Chemist. ACS Chem. Neurosci. 2020, acschemneuro.0c00244. [Google Scholar]
- Duarte-Silva, E.; Filho, A.J.M.C.; Barichello, T.; Quevedo, J.; Macedo, D.; Peixoto, C. Phosphodiesterase-5 inhibitors: Shedding new light on the darkness of depression? J. Affect. Disord. 2020, 264, 138–149. [Google Scholar] [CrossRef]
- Hackett, G. Should All Men with Type 2 Diabetes Be Routinely Prescribed a Phosphodiesterase Type 5 Inhibitor? World J. Mens. Health 2020, 38. [Google Scholar] [CrossRef]
- Vitiello, L.; Tibaudo, L.; Pegoraro, E.; Bello, L.; Canton, M. Teaching an Old Molecule New Tricks: Drug Repositioning for Duchenne Muscular Dystrophy. Int. J. Mol. Sci. 2019, 20, 6053. [Google Scholar] [CrossRef] [PubMed]
- Pavan, V.; Mucignat-Caretta, C.; Redaelli, M.; Ribaudo, G.; Zagotto, G. The Old Made New: Natural Compounds against Erectile Dysfunction. Arch. Pharm. (Weinheim) 2015, 348, 607–614. [Google Scholar] [CrossRef] [PubMed]
- Ribaudo, G.; Zanforlin, E.; Canton, M.; Bova, S.; Zagotto, G. Preliminary studies of berberine and its semi-synthetic derivatives as a promising class of multi-target anti-parkinson agents. Nat. Prod. Res. 2018, 32, 1395–1401. [Google Scholar] [CrossRef]
- Ribaudo, G.; Ongaro, A.; Zagotto, G. Natural Compounds Promoting Weight Loss: Mechanistic Insights from the Point of View of the Medicinal Chemist. Nat. Prod. J. 2019, 9, 78–85. [Google Scholar] [CrossRef]
- Beretz, A.; Anton, R.; Stoclet, J.C. Flavonoid compounds are potent inhibitors of cyclic AMP phosphodiesterase. Experientia 1978, 34, 1054–1055. [Google Scholar] [CrossRef] [PubMed]
- Sabphon, C.; Temkitthawon, P.; Ingkaninan, K.; Sawasdee, P. Phosphodiesterase Inhibitory Activity of the Flavonoids and Xanthones from Anaxagorea luzonensis. Nat. Prod. Commun. 2015, 10, 1934578X1501000222. [Google Scholar] [CrossRef]
- Ko, W.-C.; Shih, C.-M.; Lai, Y.-H.; Chen, J.-H.; Huang, H.-L. Inhibitory effects of flavonoids on phosphodiesterase isozymes from guinea pig and their structure–activity relationships. Biochem. Pharmacol. 2004, 68, 2087–2094. [Google Scholar] [CrossRef]
- Ribaudo, G.; Vendrame, T.; Bova, S. Isoflavones from Maclura pomifera: Structural elucidation and in silico evaluation of their interaction with PDE5. Nat. Prod. Res. 2017, 31, 1988–1994. [Google Scholar] [CrossRef] [PubMed]
- Ongaro, A.; Zagotto, G.; Memo, M.; Gianoncelli, A.; Ribaudo, G. Natural phosphodiesterase 5 (PDE5) inhibitors: A computational approach. Nat. Prod. Res. 2019, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Chau, Y.; Li, F.-S.; Levsh, O.; Weng, J.-K. Exploration of icariin analog structure space reveals key features driving potent inhibition of human phosphodiesterase-5. PLoS ONE 2019, 14, e0222803. [Google Scholar] [CrossRef]
- Ribaudo, G.; Pagano, M.A.; Pavan, V.; Redaelli, M.; Zorzan, M.; Pezzani, R.; Mucignat-Caretta, C.; Vendrame, T.; Bova, S.; Zagotto, G. Semi-synthetic derivatives of natural isoflavones from Maclura pomifera as a novel class of PDE-5A inhibitors. Fitoterapia 2015, 105, 132–138. [Google Scholar] [CrossRef]
- Ribaudo, G.; Ongaro, A.; Zagotto, G. 5-Hydroxy-3-(4-hydroxyphenyl)-8,8-dimethyl-6-(3-methylbut-2-enyl)pyrano [2,3-h]chromen-4-one. Molbank 2018, 2018, M1004. [Google Scholar] [CrossRef]
- Ribaudo, G.; Coghi, P.; Zanforlin, E.; Law, B.Y.K.; Wu, Y.Y.J.; Han, Y.; Qiu, A.C.; Qu, Y.Q.; Zagotto, G.; Wong, V.K.W. Semi-synthetic isoflavones as BACE-1 inhibitors against Alzheimer’s disease. Bioorg. Chem. 2019, 87, 474–483. [Google Scholar] [CrossRef] [PubMed]
- Adefegha, S.A.; Oboh, G.; Fakunle, B.; Oyeleye, S.I.; Olasehinde, T.A. Quercetin, rutin, and their combinations modulate penile phosphodiesterase-5′, arginase, acetylcholinesterase, and angiotensin-I-converting enzyme activities: A comparative study. Comp. Clin. Path. 2018, 27, 773–780. [Google Scholar] [CrossRef]
- Suri, S.; Liu, X.; Rayment, S.; Hughes, D.; Kroon, P.; Needs, P.; Taylor, M.; Tribolo, S.; Wilson, V. Quercetin and its major metabolites selectively modulate cyclic GMP-dependent relaxations and associated tolerance in pig isolated coronary artery. Br. J. Pharmacol. 2010, 159, 566–575. [Google Scholar] [CrossRef] [PubMed]
- Chan, A.L.F.; Huang, H.L.; Chien, H.C.; Chen, C.M.; Lin, C.N.; Ko, W.C. Inhibitory effects of quercetin derivatives on phosphodiesterase isozymes and high-affinity [3H]-rolipram binding in guinea pig tissues. Invest. New Drugs 2008, 26, 417–424. [Google Scholar] [CrossRef] [PubMed]
- Rollas, S.; Küçükgüzel, Ş.G. Biological activities of hydrazone derivatives. Molecules 2007, 12, 1910–1939. [Google Scholar] [CrossRef]
- Newkome, G.R.; Fishel, D.L. Synthesis of Simple Hydrazones of Carbonyl Compounds by an Exchange Reaction. J. Org. Chem. 1966, 31, 677–681. [Google Scholar] [CrossRef]
- Hajipour, A.R.; Mohammadpoor-Baltork, I.; Bigdeli, M. A Convenient and Mild Procedure for the Synthesis of Hydrazones and Semicarbazones from Aldehydes or Ketones under Solvent-free Conditions. J. Chem. Res. 1999, 570–571. [Google Scholar] [CrossRef]
- Zhang, M.; Shang, Z.-R.; Li, X.-T.; Zhang, J.-N.; Wang, Y.; Li, K.; Li, Y.-Y.; Zhang, Z.-H. Simple and efficient approach for synthesis of hydrazones from carbonyl compounds and hydrazides catalyzed by meglumine. Synth. Commun. 2017, 47, 178–187. [Google Scholar] [CrossRef]
- Ribaudo, G.; Scalabrin, M.; Pavan, V.; Fabris, D.; Zagotto, G. Constrained bisantrene derivatives as G-quadruplex binders. Arkivoc 2016, 2016, 145. [Google Scholar]
- Cahill, K.B.; Quade, J.H.; Carleton, K.L.; Cote, R.H. Identification of Amino Acid Residues Responsible for the Selectivity of Tadalafil Binding to Two Closely Related Phosphodiesterases, PDE5 and PDE6. J. Biol. Chem. 2012, 287, 41406–41416. [Google Scholar] [CrossRef] [PubMed]
- Salmaso, V.; Moro, S. Bridging Molecular Docking to Molecular Dynamics in Exploring Ligand-Protein Recognition Process: An Overview. Front. Pharmacol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Galvelis, R.; Doerr, S.; Damas, J.M.; Harvey, M.J.; De Fabritiis, G. A Scalable Molecular Force Field Parameterization Method Based on Density Functional Theory and Quantum-Level Machine Learning. J. Chem. Inf. Model. 2019, 59, 3485–3493. [Google Scholar] [CrossRef]
- Martínez-Rosell, G.; Giorgino, T.; De Fabritiis, G. PlayMolecule ProteinPrepare: A Web Application for Protein Preparation for Molecular Dynamics Simulations. J. Chem. Inf. Model. 2017, 57, 1511–1516. [Google Scholar] [CrossRef]
- Doerr, S.; Harvey, M.J.; Noé, F.; De Fabritiis, G. HTMD: High-Throughput Molecular Dynamics for Molecular Discovery. J. Chem. Theory Comput. 2016, 12, 1845–1852. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds of compound 1 are available from the authors. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).