Abstract
We describe a straightforward synthesis of the title compound, incorporating a relatively rare 2-methyl-2H-1,2,3-triazole heterocylic motif as a potential kinase inhibitor motif. The small molecule crystal structure has been resolved, revealing an interesting packing arrangement and overall conformation. We also performed routine characterization with 1H/13C-NMR and liquid chromatography (LC) and high-resolution mass spectra (HRMS).
1. Introduction
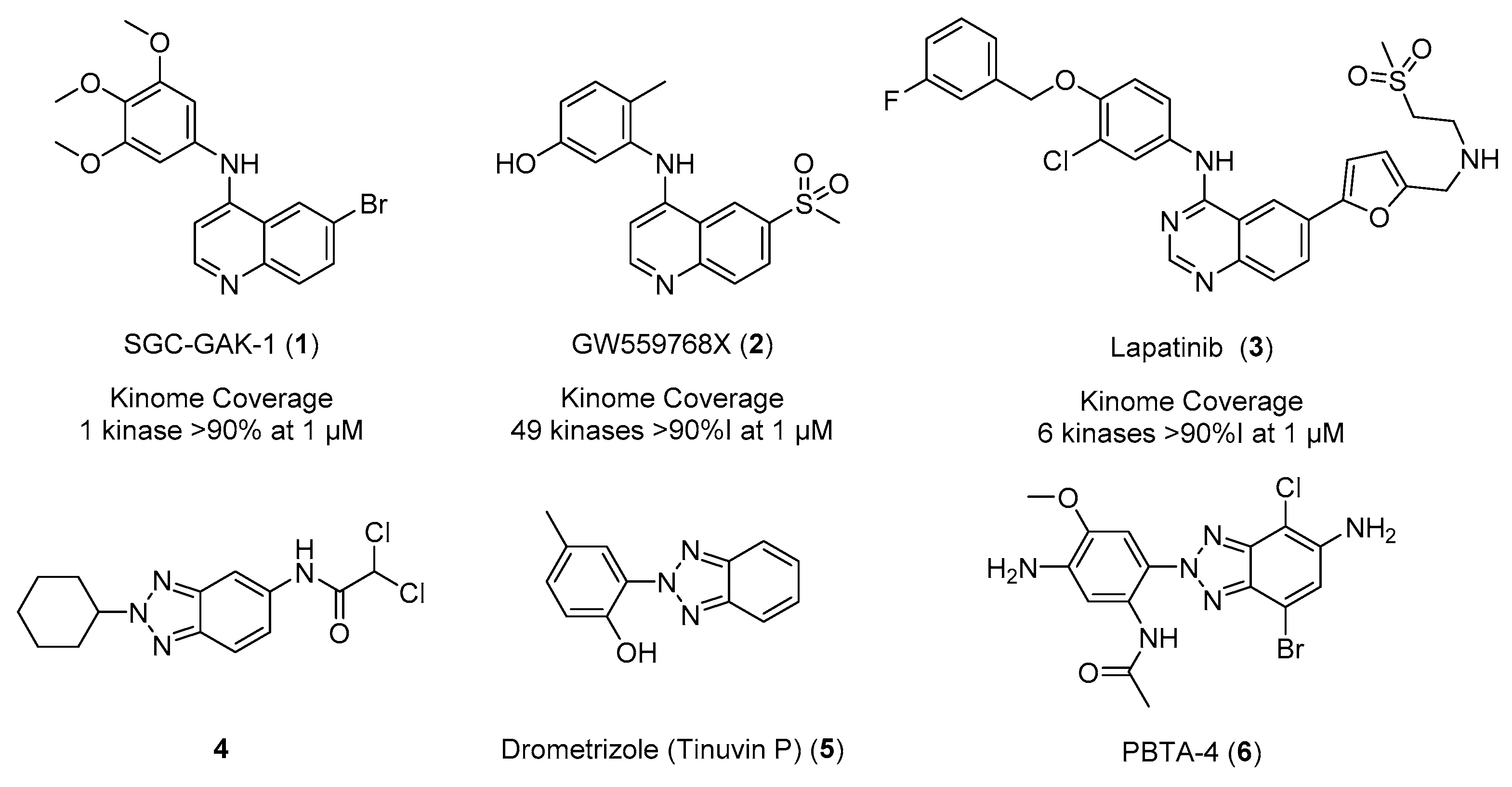
Kinases have been successfully utilized as drug targets for the past 30 years, with >40 kinase inhibitors approved by the FDA to date [1]. These drugs are predominantly multi-targeted tyrosine kinase inhibitors for the treatment of cancer [1]. The need for highly selective compounds is of paramount importance in the development of the field beyond oncology [2]. The 4-anilino-quin(az)oline scaffold has shown an ability to modulate kinome promiscuity to be both selective, such as SGC-GAK-1 (1), and broad spectrum, such as GW559768X (2). There are also examples of fine-tuned inhibitors including clinical epidermal growth factor receptor (EGFR) inhibitor lapatinib (3), which demonstrates a relatively narrow spectrum kinome profile (Figure 1) [3,4,5,6,7]. This kinome profile is mainly dependent on the substitution patterns and electronics of the pendant arms of the hinge binding scaffold [6,7].
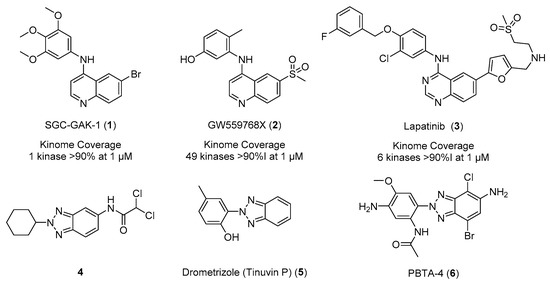
Figure 1.
Previously reported 4-anilino-quin(az)olines (1–3) and 2H-1,2,3-triazoles (4–6).
A plethora of 1- and 3-position substitution benzotriazoles have been reported in the literature, but the 2-position substitution has been largely overlooked [8,9]. This is partly due to the fact that the formation of such a heterocycle is electronically unfavorable, as it generally requires breaking of the aromaticity of the benzene ring. However, several recently developed synthetic techniques exist to access the N-2-aryl-1,2,3-triazole [10,11,12,13]. There are a number of examples of 2-position-substituted benzotriazoles, including potent anti-microbials (4), anti-infectives, and anti-ultraviolet light agents including Drometrizole (Tinuvin P) (5) (Figure 1) [14,15,16]. There have also been reports of more complex 2-position benzotriazoles as industrial pollutants and mutagens from diazo dye production, including PBTA-4 (6), isolated in a Japanese river (Figure 1) [17,18,19]. We now describe incorporation of a 2-methyl-2H-1,2,3-triazole onto a known kinase-active scaffold based on SGC-GAK-1 (1) [4,5,20].
2. Results
2.1. Synthesis of 9
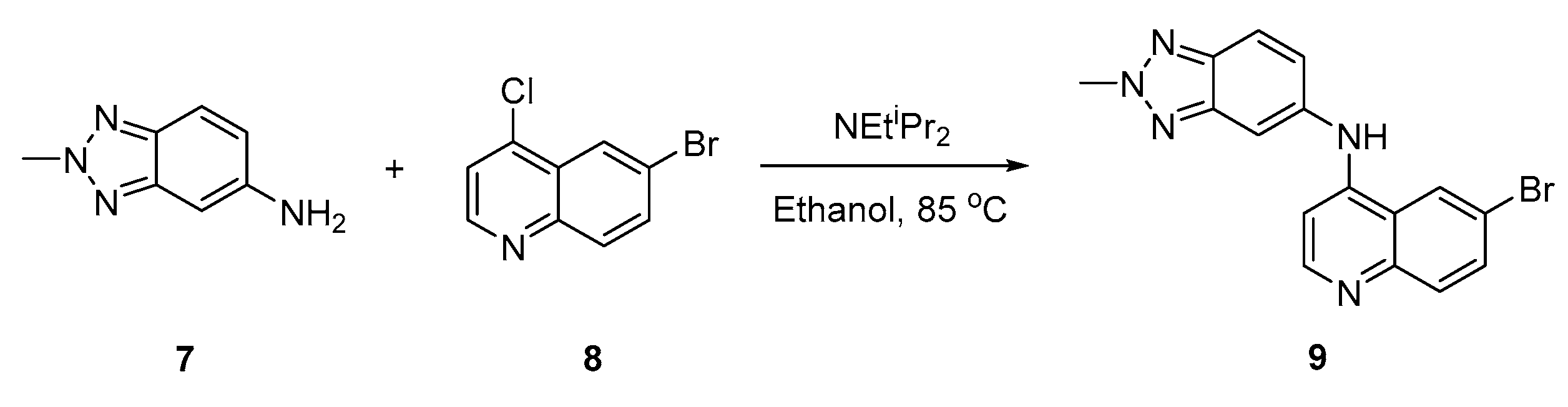
The title compound was synthesized by a one-step protocol (Scheme 1) [3,4,5,20,21,22,23]. The corresponding 2-methyl-2H-benzo[d][1,2,3]triazol-5-amine (7) and 6-bromo-4-chloroquinoline (8) were mixed with the addition of Hünig’s base (2.2 equiv.); this was then refluxed to purified to afford the title compound (9) with a good overall yield (68%) (see Supporting Information).
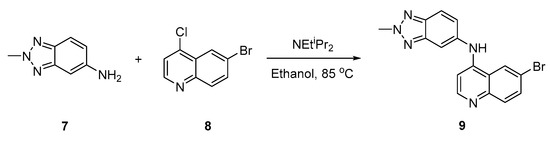
Scheme 1.
Synthesis route to access 4-anilinoquinoline (9).
2.2. Crystal Structure of 9
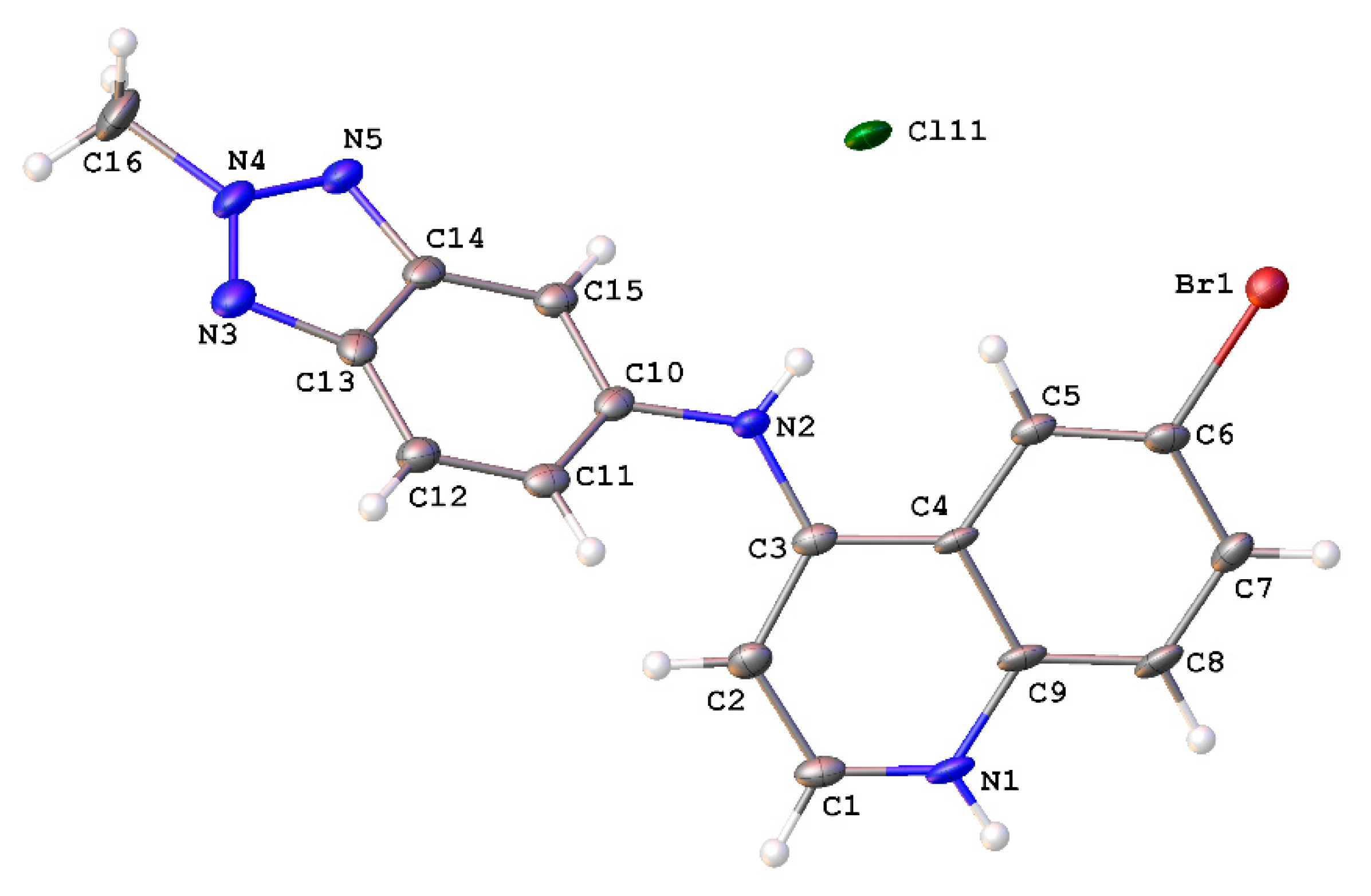
A crystallographic analysis revealed 9 crystallized as a chloride salt (Figure 2). The molecule comprises two planar moieties. The quinoline moiety, i.e., N1 and C1–C9, exhibited a root mean square deviation (r.m.s.) deviation of 0.047 Å, with the maximum deviation from the plane being 0.085 Å for the N-atom bonded C3. The r.m.s. deviation of the benzotriazole moiety, i.e., C10–C15 and N3–N5 was 0.007 Å, with the aniline N-atom bonded to C10 displaying the maximum deviation of 0.012 Å. The dihedral angle between the two aforementioned planes was 55.9(1)°. The C3–N2 bond distance of 1.354(6) Å was indicative of a double bond character in this bond, consistent with conjugation in the quinoline moiety. All other bond distances and angles were within expected limits.
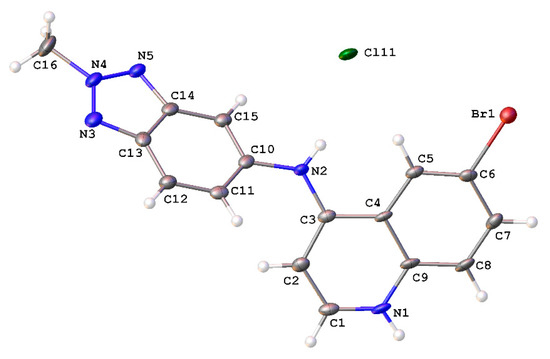
Figure 2.
Molecular structure of 9H+Cl− showing atomic labeling and displacement ellipsoids at a 50% probability level.
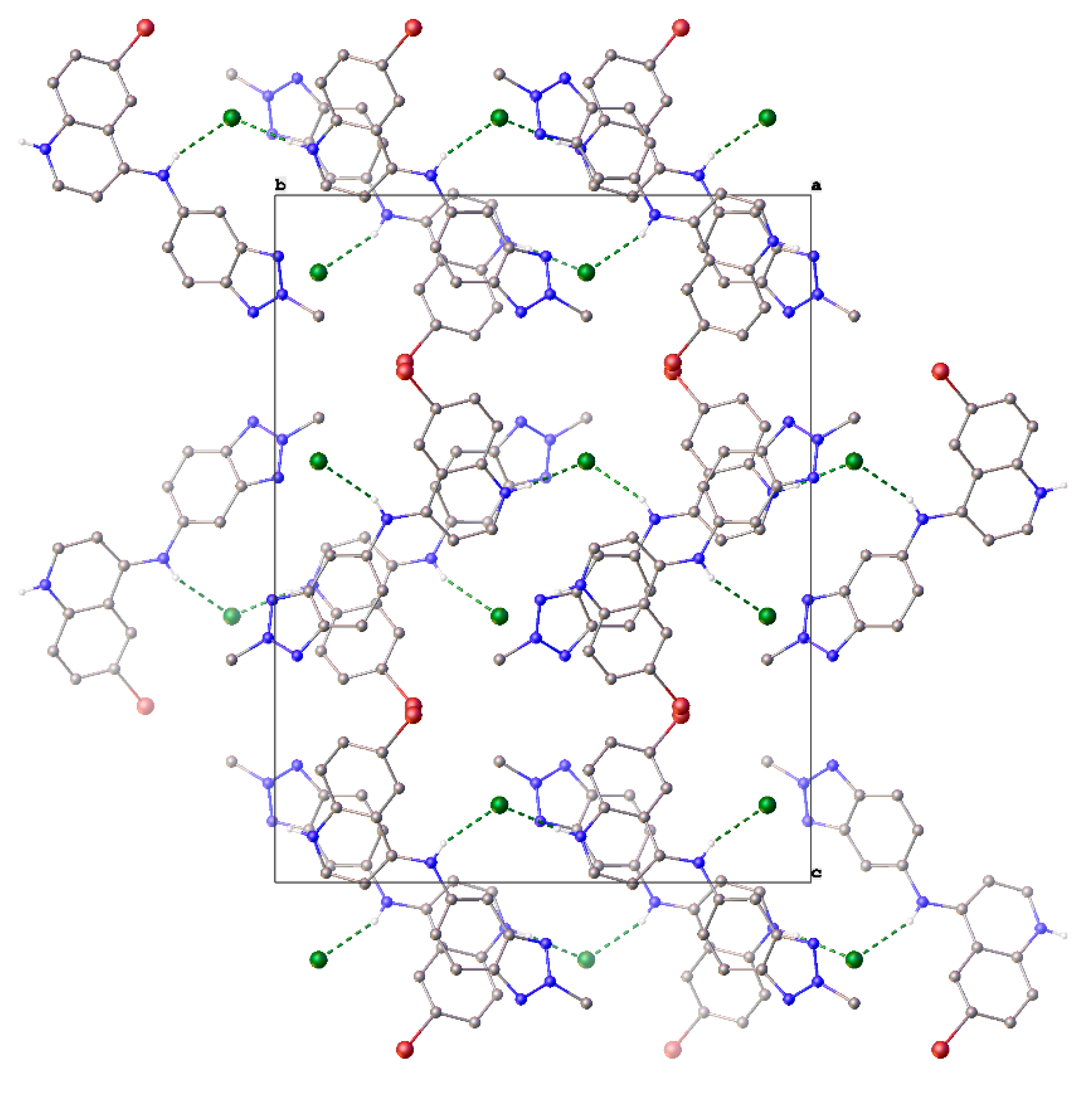
The chloride anion is integral to the solid-state structure. The structure comprises corrugated “tapes” of 9H+ hydrogen bonded to Cl− via the quinoline N–H+ and aniline N–H, respectively (N–H+…Cl−…H–N (3.047(4) Å, 3.173(4) Å)), parallel to the b-axis. These tapes then close-pack to form the structure (Figure 3).
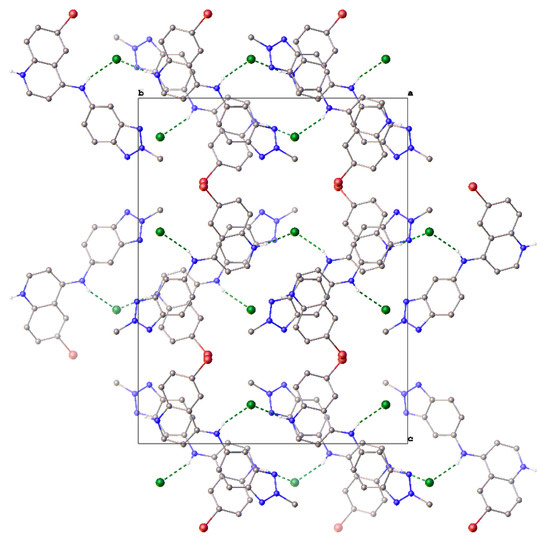
Figure 3.
Unit cell contents of 9H+Cl−, shown in projection down the a-axis. Hydrogen atoms except those involved in H-bonding have been omitted for clarity. Hydrogen bonds are shown as dashed green lines.
3. Discussion
We demonstrated a robust way to access the title compound (9) with a good yield. This methodology can be used for rapid library development as previously described [3,4,5,20,21,22,23]. This diversity of heterocycles and substitution patterns, while not endless, provides for a wealth of opportunities to tune the 4-anilino-quin(az)oline scaffold in the discovery of new chemical probes.
4. Experimental Section
4.1. Chemistry
All reactions were performed using flame-dried round-bottomed flasks or reaction vessels. Where appropriate, reactions were carried out under an inert atmosphere of nitrogen with dry solvents, unless otherwise stated. Yields refer to chromatographically and spectroscopically pure isolated yields. Reagents were purchased at the highest commercial quality and used without further purification. Reactions were monitored by thin-layer chromatography carried out on 0.25 mm E. Merck silica gel plates (60F-254) using ultraviolet light as the visualizing agent. NMR spectra were recorded on a Varian Inova 400 spectrometer (Varian, Palo Alto, CA, USA) and were calibrated using residual undeuterated solvent as an internal reference (CDCl3: 1H-NMR = 7.26, 13C-NMR = 77.16). The following abbreviations or combinations thereof have been used to explain the multiplicities observed: s = singlet, d = doublet, t = triplet, q = quartet, m = multiplet, br = broad. Liquid chromatography (LC) and high-resolution mass spectra (HRMS) were recorded on a ThermoFisher hybrid LTQ FT (ICR 7T) (ThermoFisher, Waltham, MA, USA). The University of Southampton (Southampton, UK) small molecule X-ray facility collected and analyzed all X-ray diffraction data.
6-Bromo-N-(2-methyl-2H-benzo[d][1,2,3]triazol-5-yl)quinolin-4-amine (9): 6-bromo-4-chloroquinoline (150 mg, 0.619 mmol, 1 equiv.), 2-methyl-2H-1,2,3-benzotriazol-5-amine (101 mg, 0.680 mmol, 1.1 equiv.) and iPr2NEt (0.225 mL, 1.361 mmol, 2.2 equiv.) were suspended in ethanol (10 mL) and refluxed for 18 h. The crude mixture was reduced and extracted with ethyl acetate/saturated ammonium chloride and purified by flash chromatography using EtOAc:hexane followed by 1–5% methanol in EtOAc. After solvent removal under reduced pressure, the product was obtained as a beige solid (149 mg, 0.421 mmol, 68%). MP > 300 °C; 1H-NMR (400 MHz, DMSO-d6) δ 11.29 (s, 1H), 9.21 (d, J = 2.0 Hz, 1H), 8.54 (d, J = 6.9 Hz, 1H), 8.18 (dd, J = 9.0, 2.0 Hz, 1H), 8.16–8.05 (m, 2H), 8.06–7.89 (m, 1H), 7.53 (dd, J = 9.0, 1.9 Hz, 1H), 6.89 (d, J = 6.9 Hz, 1H), 4.54 (s, 3H). 13C-NMR (101 MHz, DMSO-d6) δ 154.2, 144.0, 143.1, 142.7, 137.3, 136.6, 135.1, 126.3, 124.9, 122.5, 120.0, 119.5, 118.7, 113.7, 100.8, 43.5. HRMS m/z [M + H]+ calculated for C16H13N5Br: 354.0354, found 354.0341, LC tR = 4.30 min, >99% Purity.
4.2. Crystallography
Single colourless plate-shaped crystals of 9H+Cl− were crystallised from EtOH/water and several drops of dioxane/HCL (4 M). A suitable crystal 0.09 × 0.01 × 0.01 mm3 was selected and mounted on a MITIGEN holder (MiTeGen, Ithaca, NY, USA) in perfluoroether oil on a Rigaku FRE+ diffractometer equipped with VHF Varimax confocal mirrors and an AFC12 goniometer and HyPix 6000 detector. The crystal was kept at a steady T = 100(2) K during data collection. The structure was resolved by the ShelXT [24] structure solution program using the using dual methods solution method, and using Olex2 [25] as the graphical interface. The model was refined with version 2018/3 of ShelXL [26], using full matrix least squares minimisation on F2 minimisation. All non-hydrogen atoms were refined anisotropically. Hydrogen atom positions were calculated geometrically and refined using the riding model, except for those bonded to N-atoms which were located in the difference map and refined with a riding model.
Crystal data for C16H13BrClN5 (9H+Cl−): Mr = 390.67; orthorhombic; Pbca (No. 61); a = 7.6840(2) Å; b = 17.9973(5) Å; c = 23.0912(6) Å; α = β = γ = 90°; V = 3193.31(15) Å3; T = 100(2) K; Z = 8, Z’ = 1; μ(Mo Kα) = 2.748 mm−1; 39,663 reflections measured; 3642 unique (Rint = 0.1010), which were used in all calculations. The final wR2 was 0.1487 (all data) and R1 was 0.0675 (I > 2(I)).
Supplementary Materials
The following are available online, crystallographic data for Compound 9H+Cl− in crystallographic information file (CIF) format. CCDC 1957384 also contains the supplementary crystallographic data for this paper. These data can be obtained free of charge via http://www.ccdc.cam.ac.uk/conts/retrieving.html.
Author Contributions
Conceptualization and Writing the Original Draft Preparation C.R.M.A.; Validation, Resources, Data Curation and Editing C.R.M.A. and G.J.T.
Funding
The SGC is a registered charity (number 1097737) that receives funds from AbbVie, Bayer Pharma AG, Boehringer Ingelheim, Canada Foundation for Innovation, Eshelman Institute for Innovation, Genome Canada, Innovative Medicines Initiative (EU/EFPIA) [ULTRA-DD grant no. 115766], Janssen, Merck KGaA Darmstadt Germany, MSD, Novartis Pharma AG, Ontario Ministry of Economic Development and Innovation, Pfizer, São Paulo Research Foundation-FAPESP, Takeda, and Wellcome [106169/ZZ14/Z].
Acknowledgments
We thank the EPSRC UK National Crystallography Service at the University of Southampton for the collection of the crystallographic data. We also thank Brandie Ehrmann for LC-MS/HRMS support provided by the Mass Spectrometry Core Laboratory at the University of North Carolina at Chapel Hill.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ferguson, F.M.; Gray, N.S. Kinase inhibitors: The road ahead. Nat Rev Drug Discov. 2018, 17, 353–377. [Google Scholar] [CrossRef] [PubMed]
- Cohen, P.; Alessi, D.R. Kinase drug discovery—What’s next in the field? ACS Chem. Biol. 2013, 8, 96–104. [Google Scholar] [CrossRef] [PubMed]
- Asquith, C.R.M.; Laitinen, T.; Bennett, J.M.; Godoi, P.H.; East, M.P.; Tizzard, G.J.; Graves, L.M.; Johnson, G.L.; Dornsife, R.E.; Wells, C.I.; et al. Identification and Optimization of 4-Anilinoquinolines as Inhibitors of Cyclin G Associated Kinase. ChemMedChem 2018, 13, 48–66. [Google Scholar] [CrossRef] [PubMed]
- Asquith, C.R.M.; Berger, B.T.; Wan, J.; Bennett, J.M.; Capuzzi, S.J.; Crona, D.J.; Drewry, D.H.; East, M.P.; Elkins, J.M.; Fedorov, O.; et al. SGC-GAK-1: A Chemical Probe for Cyclin G Associated Kinase (GAK). J. Med. Chem. 2019, 62, 2830–2836. [Google Scholar] [CrossRef] [PubMed]
- Asquith, C.R.M.; Treiber, D.K.; Zuercher, W.J. Utilizing comprehensive and mini-kinome panels to optimize the selectivity of quinoline inhibitors for cyclin G associated kinase (GAK). Bioorg. Med. Chem. Lett. 2019, 29, 1727–1731. [Google Scholar] [CrossRef] [PubMed]
- Drewry, D.H.; Wells, C.I.; Andrews, D.M.; Angell, R.; Al-Ali, H.; Axtman, A.D.; Capuzzi, S.J.; Elkins, J.M.; Ettmayer, P.; Frederiksen, M.; et al. Progress towards a public chemogenomic set for protein kinases and a call for contributions. PLoS ONE 2017, 12, e0181585. [Google Scholar] [CrossRef]
- Fabian, M.A.; Biggs, W.H., 3rd; Treiber, D.K.; Atteridge, C.E.; Azimioara, M.D.; Benedetti, M.G.; Carter, T.A.; Ciceri, P.; Edeen, P.T.; Floyd, M.; et al. A small molecule–kinase interaction map for clinical kinase inhibitors. Nat. Biotechnol. 2005, 23, 329–336. [Google Scholar] [CrossRef]
- Agalave, S.G.; Maujan, S.R.; Pore, V.S. Click chemistry: 1,2,3-triazoles as pharmacophores. Chem. Asian J. 2011, 6, 2696–2718. [Google Scholar] [CrossRef]
- Dheer, D.; Singh, V.; Shankar, R. Medicinal attributes of 1,2,3-triazoles: Current developments. Bioorg. Chem. 2017, 71, 30–54. [Google Scholar] [CrossRef]
- Liu, Y.; Yan, W.; Chen, Y.; Petersen, J.L.; Shi, X. Efficient synthesis of N-2-aryl-1,2,3-triazole fluorophores via post-triazole arylation. Org. Lett. 2008, 10, 5389–5392. [Google Scholar] [CrossRef]
- Wang, X.J.; Zhang, L.; Krishnamurthy, D.; Senanayake, C.H.; Wipf, P. General solution to the synthesis of N-2-substituted 1,2,3-triazoles. Org. Lett. 2010, 12, 4632–4635. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.J.; Sidhu, K.; Zhang, L.; Campbell, S.; Haddad, N.; Reeves, D.C.; Krishnamurthy, D.; Senanayake, C.H. Bromo-directed N-2 alkylation of NH-1,2,3-triazoles: Efficient synthesis of poly-substituted 1,2,3-triazoles. Org. Lett. 2009, 11, 5490–5493. [Google Scholar] [CrossRef] [PubMed]
- Cai, R.; Yan, W.; Bologna, M.G.; de Silva, K.; Ma, Z.; Finklea, H.O.; Petersen, J.L.; Li, M.; Shi, X. Synthesis and characterization of N-2-aryl-1,2,3-triazole based iridium complexes as photocatalysts with tunable photoredox potential. Org. Chem. Front. 2015, 2, 141–144. [Google Scholar] [CrossRef]
- Caliendo, G.; Novellino, E.; Sagliocco, G.; Santagada, V.; Silipo, C.; Vittoria, A. Synthesis, antimicrobial data and correlation analysis in a set of 2-alkyl-5-amidobenzotriazoles. Eur. J. Med. Chem. 1992, 27, 161–166. [Google Scholar] [CrossRef]
- Nuvole, A.; Sanna, P.; Paglietti, G.; Juliano, C.; Zanetti, S.; Cappuccinelli, P. 1,2,3-Triazolo [4,5-f]quinolines. II. Preparation and antimicrobial evaluation of 6-ethyl-6,9-dihydro-1(2)(3)-R-1(2) (3)H-triazolo[4,5-f]quinolin-9-one-8-carboxylic acids as anti-infectives of the urinary tract. Il Farmaco 1989, 44, 619–632. [Google Scholar]
- María, D.S.; Claramunt, R.M.; Bobosik, V.; Torralba, C.M.; Torres, M.R.; Alkorta, I.; Elguero, J. Synthesis and structural study of 2-arylbenzotriazoles related to Tinuvins. Tetrahedron 2013, 69, 3027–3038. [Google Scholar] [CrossRef]
- Nukaya, H.; Shiozawa, T.; Tada, A.; Terao, Y.; Ohe, T.; Watanabe, T.; Asanoma, M.; Sawanishi, H.; Katsuhara, T.; Sugimura, T.; et al. Identification of 2-[2-(acetylamino)-4-amino-5-methoxyphenyl]-5-amino-7-bromo-4-chloro-2H-benzotriazole (PBTA-4) as a potent mutagen in river water in Kyoto and Aichi prefectures, Japan. Mutat. Res. 2001, 492, 73–80. [Google Scholar] [CrossRef]
- Ohe, T.; Takeuchi, N.; Watanabe, T.; Tada, A.; Nukaya, H.; Terao, Y.; Sawanishi, H.; Hirayama, T.; Sugimura, T.; Wakabayashi, K. Quantification of two aromatic amine mutagens, PBTA-1 and PBTA-2, in the yodo river system. Environ. Health Perspect. 1999, 107, 701–704. [Google Scholar] [CrossRef]
- Shiozawa, T.; Tada, A.; Nukaya, H.; Watanabe, T.; Takahashi, Y.; Asanoma, M.; Ohe, T.; Sawanishi, H.; Katsuhara, T.; Sugimura, T.; Wakabayashi, K.; Terao, Y. Isolation and identification of a new 2-phenylbenzotriazole-type mutagen (PBTA-3) in the Nikko river in Aichi, Japan. Chem. Res. Toxicol. 2000, 13, 535–540. [Google Scholar] [CrossRef]
- Asquith, C.R.M.; Naegeli, N.; East, M.P.; Laitinen, T.; Havener, T.M.; Wells, C.I.; Johnson, G.L.; Drewry, D.H.; Zuercher, W.J.; Morris, D.C. Design of a cyclin G associated kinase (GAK)/epidermal growth factor receptor (EGFR) inhibitor set to interrogate the relationship of EGFR and GAK in chordoma. J. Med. Chem. 2019, 62, 4772–4778. [Google Scholar] [CrossRef]
- Asquith, C.R.M.; Fleck, N.; Torrice, C.D.; Crona, D.J.; Grundner, C.; Zuercher, W.J. Anti-tubercular activity of novel 4-anilinoquinolines and 4-anilinoquinazolines. Bioorg. Med. Chem. Lett. 2019, 18, 2695–2699. [Google Scholar] [CrossRef] [PubMed]
- Asquith, C.R.M.; Maffuid, K.A.; Laitinen, T.; Torrice, C.D.; Tizzard, G.J.; Crona, D.J.; Zuercher, W.J. Targeting an EGFR water network using novel 4-anilinoquin(az)olines inhibitors for chordoma. ChemMedChem 2019, 14, 1693–1700. [Google Scholar] [CrossRef] [PubMed]
- Asquith, C.R.M.; Bennett, J.M.; Su, L.; Laitinen, T.; Elkins, J.M.; Pickett, J.E.; Wells, C.I.; Li, Z.; Willson, T.M.; Zuercher, W.J. Development of SGC-GAK-1 as an orally active in vivo probe for cyclin G associated kinase through cytochrome P450 inhibition. bioRxiv 2019, 629220. [Google Scholar] [CrossRef]
- Sheldrick, G.M. ShelXT-Integrated space-group and crystal-structure determination. Acta Crystallogr. 2015, A71, 3–8. [Google Scholar] [CrossRef]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H. Olex2: A complete structure solution, refinement and analysis program. J. Appl. Cryst. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Crystal structure refinement with ShelXL. Acta Crystallogr. 2015, C27, 3–8. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).