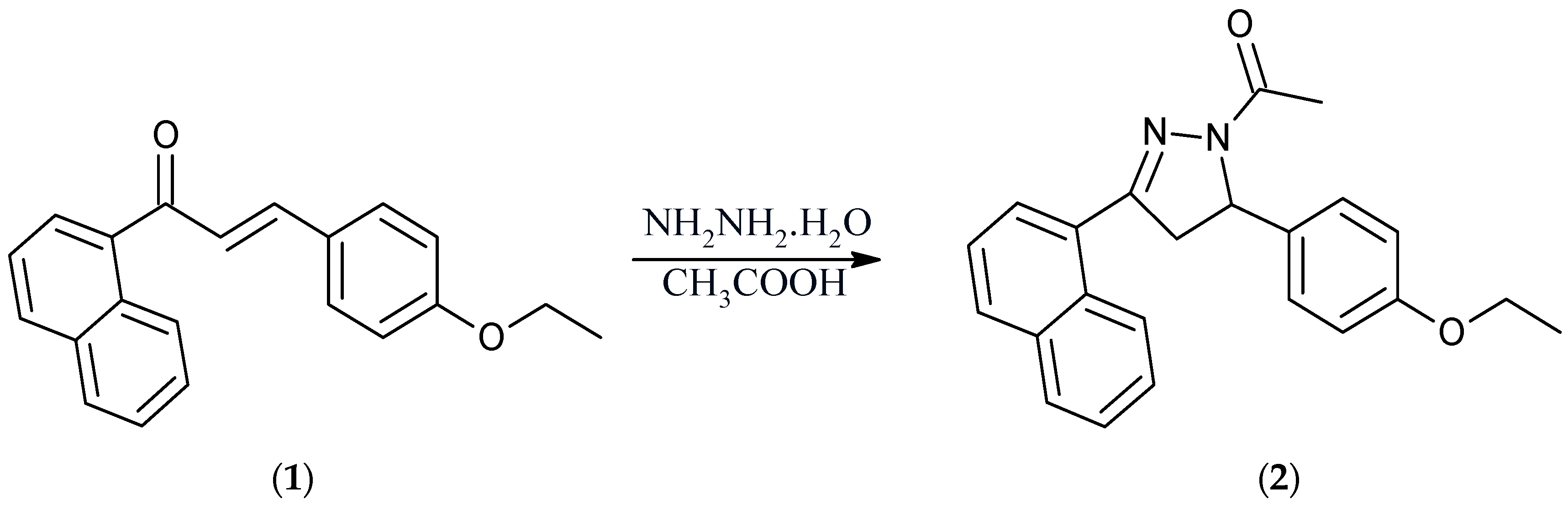
1-[5-(4-Ethoxyphenyl)-3-(naphthalen-1-yl)-4,5-dihydro-1H-pyrazol-1-yl]ethan-1-one
Abstract
:1. Introduction
2. Results
Molecular Docking
3. Discussion
4. Materials and Methods
Synthesis of 1-[5-(4-ethoxyphenyl)-3-(naphthalen-1-yl)-4,5-dihydro-1H-pyrazol-1-yl)ethan-1-one
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ahn, J.H.; Kim, H.M.; Jung, S.H.; Kang, S.K.; Kim, K.R.; Rhee, S.D.; Yang, S.D.; Cheon, H.G.; Kim, S.S. Synthesis and DP-IV inhibition of cyano-pyrazoline derivatives as potent anti-diabetic agents. Bioorg. Med. Chem. Lett. 2004, 14, 4461–4465. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.A.; Shaharyar, M.; Siddiqui, A.A. Synthesis, structural activity relationship and anti-tubercular activity of novel pyrazoline derivatives. Eur. J. Med. Chem. 2007, 42, 268–275. [Google Scholar] [CrossRef] [PubMed]
- Prasad, Y.R.; Rao, A.L.; Prasoona, L.; Murali, K.; Kumar, P.R. Synthesis and antidepressant activity of some 1,3,5-triphenyl-2-pyrazolines and 3-(2”-hydroxy naphthalene-1”-yl)-1,5-diphenyl-2-pyrazolines. Bioorg. Med. Chem. Lett. 2005, 22, 5030–5034. [Google Scholar] [CrossRef] [PubMed]
- Bhandari, S.; Tripathi, A.C.; Saraf, S.K. Novel 2-pyrazoline derivatives as potential anticonvulsant agents. Med. Chem. Res. 2013, 22, 5290–5296. [Google Scholar] [CrossRef]
- Salian, V.V.; Narayana, B.; Sarojini, B.K.; Kumar, M.S.; Nagananda, G.S.; Byrappa, K.; Kudva, A.K. Spectroscopic, single crystal X-ray, Hirshfeld, in vitro and in silico biological evaluation of a new series of potent thiazole nucleus integrated with pyrazoline scaffolds. Spectrochim. Acta A Mol. Biomol. 2017, 174, 254–271. [Google Scholar] [CrossRef] [PubMed]
- Chandra, T.; Garg, N.; Lata, S.; Saxena, K.K.; Kumar, A. Synthesis of substituted acridinyl pyrazoline derivatives and their evaluation for anti-inflammatory activity. Eur. J. Med. Chem. 2010, 45, 1772–1776. [Google Scholar] [CrossRef] [PubMed]
- Bansal, E.; Srivastava, V.K.; Kumar, A. Synthesis and anti-inflammatory activity of 1-acetyl-5-substituted aryl-3-(β-aminonaphthyl)-2-pyrazolines and β-(substituted aminoethyl) amidonaphthalenes. Eur. J. Med. Chem. 2001, 36, 81–92. [Google Scholar] [CrossRef]
- Havrylyuk, D.; Zimenkovsky, B.; Vasylenko, O.; Zaprutko, L.; Gzella, A.; Lesyk, R. Synthesis of novel thiazolone-based compounds containing pyrazoline moiety and evaluation of their anticancer activity. Eur. J. Med. Chem. 2009, 44, 1396–1404. [Google Scholar] [CrossRef] [PubMed]
- Shaharyar, M.; Abdullah, M.M.; Bakht, M.A.; Majeed, J. Pyrazoline bearing benzimidazoles: Search for anticancer agent. Eur. J. Med. Chem. 2010, 45, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Havrylyuk, D.; Zimenkovsky, B.; Vasylenko, O.; Gzella, A.; Lesyk, R. Synthesis of new 4-thiazolidinone-, 4-pyrazoline-, and isatin-based conjugates with promising antitumor activity. J. Med. Chem. 2012, 55, 8630–8641. [Google Scholar] [CrossRef] [PubMed]
- Abid, M.; Azam, A. Synthesis and antiamoebic activities of 1-N-substituted cyclised pyrazoline analogues of thiosemicarbazones. Bioorg. Med. Chem. 2005, 13, 2213–2220. [Google Scholar] [CrossRef] [PubMed]
- Bano, S.; Alam, M.S.; Javed, K.; Dudeja, M.; Das, A.K.; Dhulap, A. Synthesis, biological evaluation and molecular docking of some substituted pyrazolines and isoxazolines as potential antimicrobial agents. Eur. J. Med. Chem. 2015, 95, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Jayashree, A.; Narayana, B.; Sarojini, B.K.; Kotai, L.; Anthal, S.; Kant, R. Synthesis and crystal structure of (E)-3-(4-butoxyphenyl)-1-(naphthalen-1-yl)-prop-2-en-1-one. Eur. Chem. Bull. 2016, 12, 501–504. [Google Scholar]
Sample Availability: Samples of the compounds are available from the authors. |
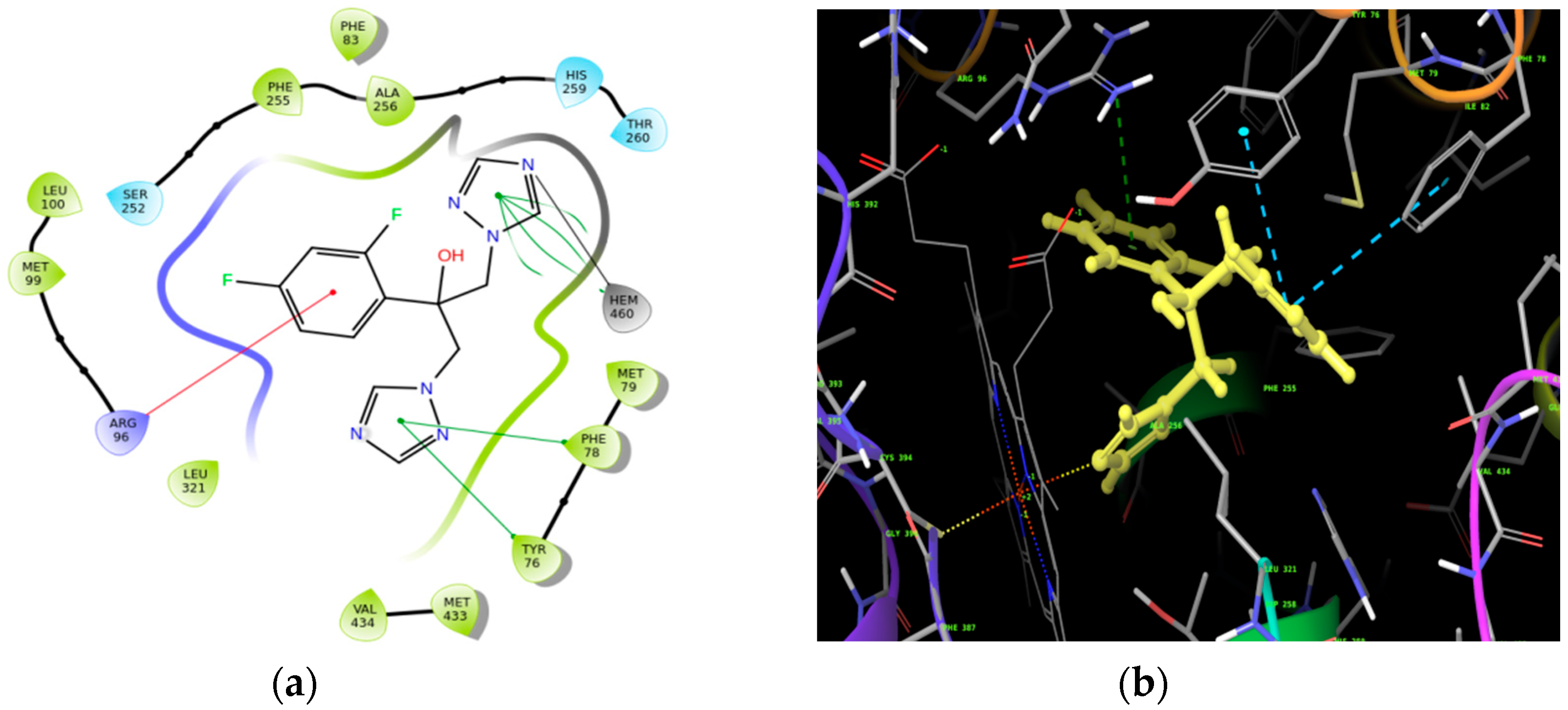
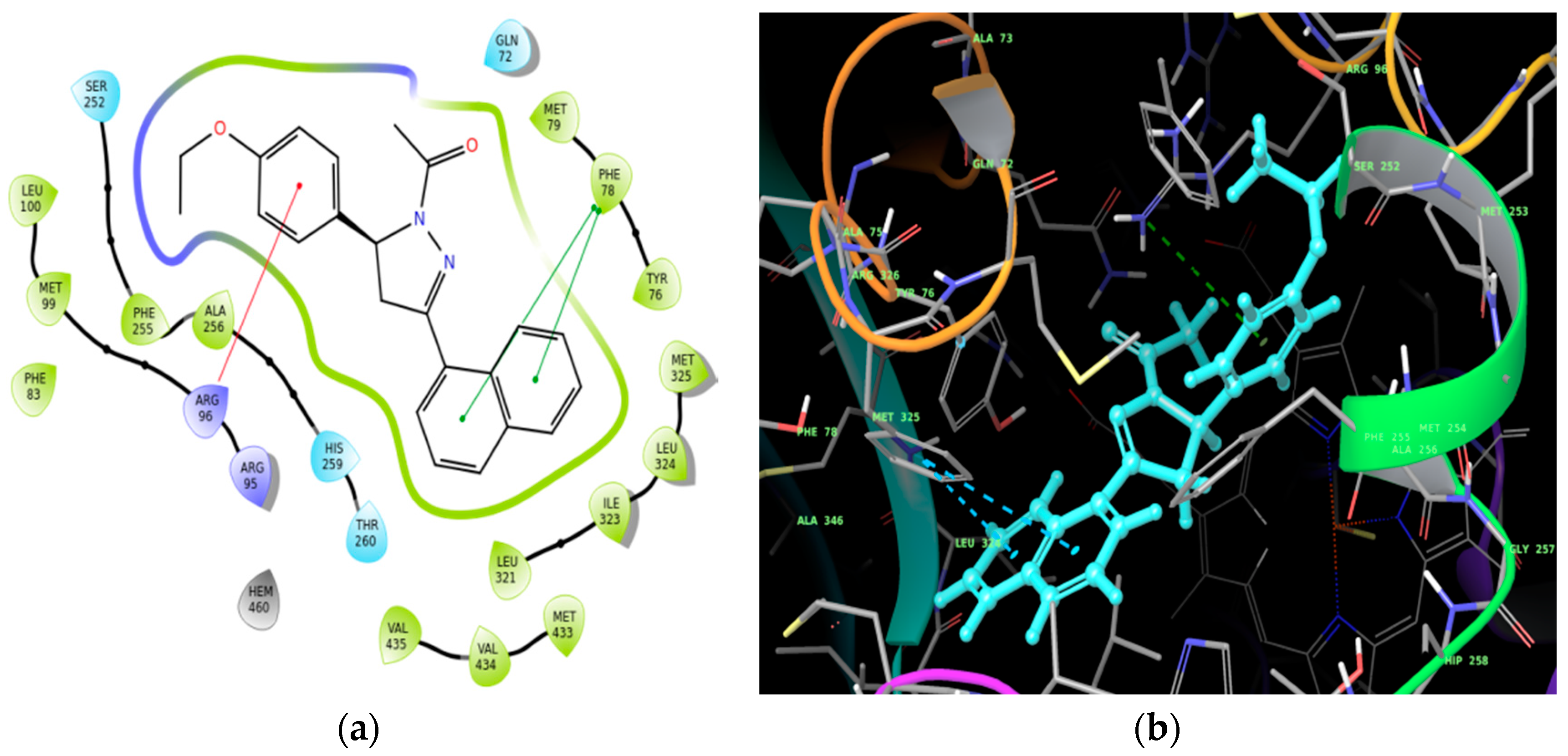
| Ligand | Docking Score (kcal/mol) |
|---|---|
| Compound (2) | −7.501 |
| Fluconazole | −5.823 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karanth, S.; Narayana, B.; Avvadukkam, J.; Sarojini, B.K. 1-[5-(4-Ethoxyphenyl)-3-(naphthalen-1-yl)-4,5-dihydro-1H-pyrazol-1-yl]ethan-1-one. Molbank 2018, 2018, M1000. https://doi.org/10.3390/M1000
Karanth S, Narayana B, Avvadukkam J, Sarojini BK. 1-[5-(4-Ethoxyphenyl)-3-(naphthalen-1-yl)-4,5-dihydro-1H-pyrazol-1-yl]ethan-1-one. Molbank. 2018; 2018(2):M1000. https://doi.org/10.3390/M1000
Chicago/Turabian StyleKaranth, Sangeetha, Badiadka Narayana, Jayashree Avvadukkam, and Balladka Kunhanna Sarojini. 2018. "1-[5-(4-Ethoxyphenyl)-3-(naphthalen-1-yl)-4,5-dihydro-1H-pyrazol-1-yl]ethan-1-one" Molbank 2018, no. 2: M1000. https://doi.org/10.3390/M1000
APA StyleKaranth, S., Narayana, B., Avvadukkam, J., & Sarojini, B. K. (2018). 1-[5-(4-Ethoxyphenyl)-3-(naphthalen-1-yl)-4,5-dihydro-1H-pyrazol-1-yl]ethan-1-one. Molbank, 2018(2), M1000. https://doi.org/10.3390/M1000




