Inductive QSAR Descriptors. Distinguishing Compounds with Antibacterial Activity by Artificial Neural Networks
Abstract
:Introduction.
Results
Dataset
Descriptors
| Descriptor | Characterization | Parental formula(s) |
|---|---|---|
| χ (electronegativity) – based | ||
| EO_Equalizeda | Iteratively equalized electronegativity of a molecule | Calculated iteratively by (7) where charges get updated according to (6); an atomic hardness in (7) is expressed through (8) |
| Average_EO_Posa | Arithmetic mean of electronegativities of atoms with positive partial charge | where is the number of atoms i in a molecule with positive partial charge |
| Average_EO_Nega | Arithmetic mean of electronegativities of atoms with negative partial charge | where is the number of atoms i in a molecule with negative partial charge |
| η (hardness) – based | ||
| Global_Hardnessa | Molecular hardness - reversed softness of a molecule | (10) |
| Sum_Hardnessa | Sum of hardnesses of atoms of a molecule | Calculated as a sum of inversed atomic softnesses in turn computed within (9) |
| Sum_Pos_Hardnessa | Sum of hardnesses of atoms with positive partial charge | Obtained by summing up the contributions from atoms with positive charge computed by (8) |
| Sum_Neg_Hardnessa | Sum of hardnesses of atoms with negative partial charge | Obtained by summing up the contributions from atoms with negative charge computed by (8) |
| Average_Hardnessa | Arithmetic mean of hardnesses of all atoms of a molecule | Estimated by dividing quantity (10) by the number of atoms in a molecule |
| Average_Pos_Hardness | Arithmetic mean of hardnesses of atoms with positive partial charge | where is the number of atoms i with positive partial charge. |
| Average_Neg_Hardnessa | Arithmetic mean of hardnesses of atoms with negative partial charge | where is the number of atoms i with negative partial charge. |
| Smallest_Pos_Hardnessa | Smallest atomic hardness among values for positively charged atoms | (8) |
| Smallest_Neg_Hardnessa | Smallest atomic hardness among values for negatively charged atoms. | (8) |
| Largest_Pos_Hardness | Largest atomic hardness among values for positively charged atoms | (8) |
| Largest_Neg_Hardness | Largest atomic hardness among values for negatively charged atoms | (8) |
| Hardness_of_Most_Pos | Atomic hardness of an atom with the most positive charge | (8) |
| Hardness_of_Most_Nega | Atomic hardness of an atom with the most negative charge | (8) |
| s (softness) - based | ||
| Global_Softness | Molecular softness – sum of constituent atomic softnesses | (11) |
| Total_Pos_Softnessa | Sum of softnesses of atoms with positive partial charge | Obtained by summing up the contributions from atoms with positive charge computed by (9) |
| Total_Neg_Softnessa | Sum of softnesses of atoms with negative partial charge | Obtained by summing up the contributions from atoms with negative charge computed by (9) |
| Average_Softness | Arithmetic mean of softnesses of all atoms of a molecule | (11) divided by the number of atoms in molecule |
| Average_Pos_Softness | Arithmetic mean of softnesses of atoms with positive partial charge | where is the number of atoms i with positive partial charge. |
| Average_Neg_Softness | Arithmetic mean of softnesses of atoms with negative partial charge | where is the number of atoms i with negative partial charge. |
| Smallest_Pos_Softnessa | Smallest atomic softness among values for positively charged atoms | (9) |
| Smallest_Neg_Softnessa | Smallest atomic softness among values for negatively charged atoms | (9) |
| Largest_Pos_Softness | Largest atomic softness among values for positively charged atoms | (9) |
| Largest_Neg_Softness | Largest atomic softness among values for positively charged atoms | (9) |
| Softness_of_Most_Posa | Atomic softness of an atom with the most positive charge | (9) |
| Softness_of_Most_Nega | Atomic softness of an atom with the most negative charge | (9) |
| q (charge)- based | ||
| Total_Charge | Sum of absolute values of partial charges on all atoms of a molecule | where all the contributions derived within (6) |
| Total_Charge_Formala | Sum of charges on all atoms of a molecule (formal charge of a molecule) | Sum of all contributions (6) |
| Average_Pos_Chargea | Arithmetic mean of positive partial charges on atoms of a molecule | where is the number of atoms i with positive partial charge |
| Average_Neg_Chargea | Arithmetic mean of negative partial charges on atoms of a molecule | where is the number of atoms i with negative partial charge |
| Most_Pos_Chargea | Largest partial charge among values for positively charged atoms | (6) |
| Most_Neg_Charge | Largest partial charge among values for negatively charged atoms | (6) |
| σ* (inductive parameter) – based | ||
| Total_Sigma_mol_ia | Sum of inductive parameters σ*(molecule→atom) for all atoms within a molecule | where contributions are computed by equation (2) with n=N-1 – i.e. each atom j is considered against the rest of the molecule G |
| Total_Abs_Sigma_mol_i | Sum of absolute values of group inductive parameters σ*(molecule→atom) for all atoms within a molecule | |
| Most_Pos_Sigma_mol_ia | Largest positive group inductive parameter σ*(molecule→atom) for atoms in a molecule | (2) |
| Most_Neg_Sigma_mol_ia | Largest (by absolute value) negative group inductive parameter σ*(molecule→atom) for atoms in a molecule | (2) |
| Most_Pos_Sigma_i_mola | Largest positive atomic inductive parameter σ*(atom→molecule) for atoms in a molecule | (5) |
| Most_Neg_Sigma_i_mola | Largest negative atomic inductive parameter σ*(atom→molecule) for atoms in a molecule | (5) |
| Sum_Pos_Sigma_mol_i | Sum of all positive group inductive parameters σ*( molecule →atom) within a molecule | where >0 and is the number of N-1 atomic substituents in a molecule with positive inductive effect (electron acceptors) |
| Sum_Neg_Sigma_mol_ia | Sum of all negative group inductive parameters σ*( molecule →atom) within a molecule | where <0 and is the number of N-1 atomic substituents in a molecule with negative inductive effect (electron donors) |
| Rs (steric parameter) – based | ||
| Largest_Rs_mol_ia | Largest value of steric influence Rs(molecule→atom) in a molecule | (1) where n=N-1 - each atom j is considered against the rest of the molecule G |
| Smallest_Rs_mol_ia | Smallest value of group steric influence Rs(molecule→atom) in a molecule | (1) where n=N-1 - each atom j is considered against the rest of the molecule G |
| Largest_Rs_i_mol | Largest value of atomic steric influence Rs(atom→molecule) in a molecule | (4) |
| Smallest_Rs_i_mola | Smallest value of atomic steric influence Rs(atom→molecule) in a molecule | (4) |
| Most_Pos_Rs_mol_ia | Steric influence Rs(molecule→atom) ON the most positively charged atom in a molecule | (1) |
| Most_Neg_Rs_mol_ia | Steric influence Rs(molecule→atom) ON the most negatively charged atom in a molecule | (1) |
| Most_Pos_Rs_i_mol | Steric influence Rs(atom→molecule) OF the most positively charged atom to the rest of a molecule | (4) |
| Most_Neg_Rs_i_mola | Steric influence Rs(atom→molecule) OF the most negatively charged atom to the rest of a molecule | (4) |
Selection of variables
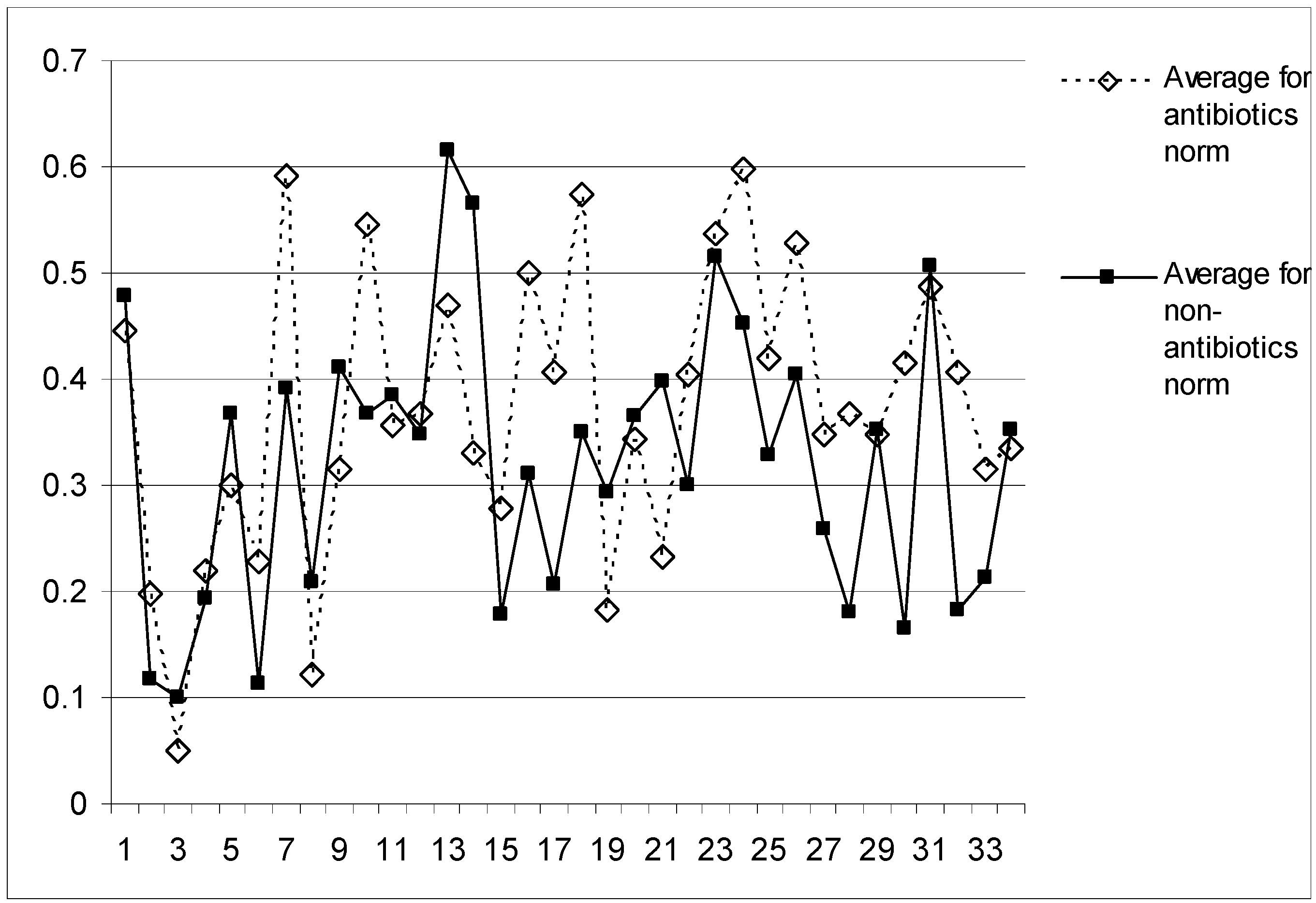
QSAR model
| Hidden nodes | Specificity | Sensitivity | Accuracy | PPV |
|---|---|---|---|---|
| 2 | 0.8 | 0.92 | 0.846 | 0.751 |
| 3 | 0.926 | 0.928 | 0.923 | 0.884 |
| 4 | 0.925 | 0.92 | 0.923 | 0.884 |
| 6 | 0.9 | 1 | 0.938 | 0.862 |
| 8 | 0.9 | 0.92 | 0.907 | 0.851 |
| 10 | 0.9 | 0.92 | 0.907 | 0.851 |
| 12 | 0.9 | 0.92 | 0.907 | 0.851 |
| 14 | 0.815 | 1 | 0.923 | 0.833 |
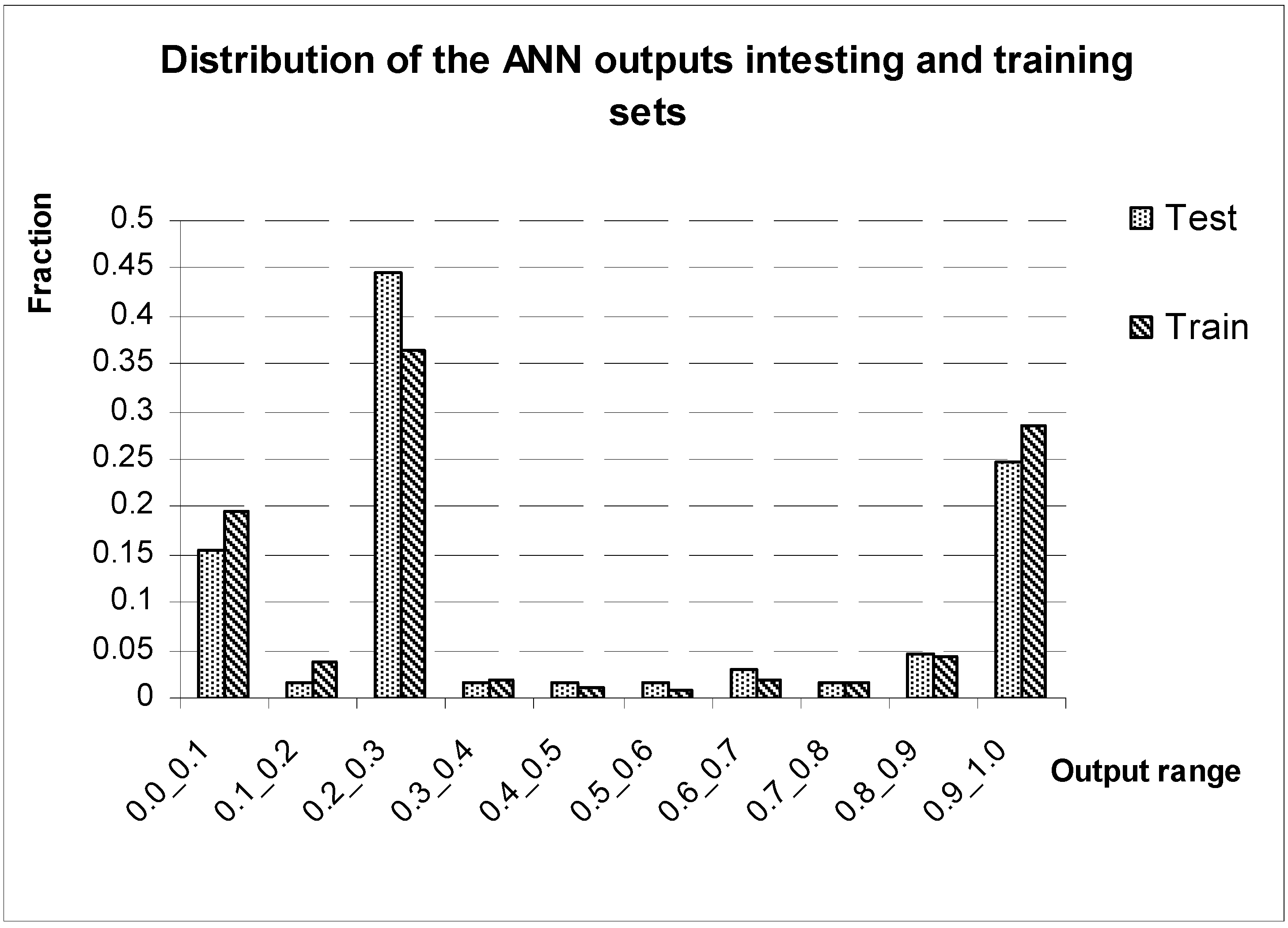
Discussion
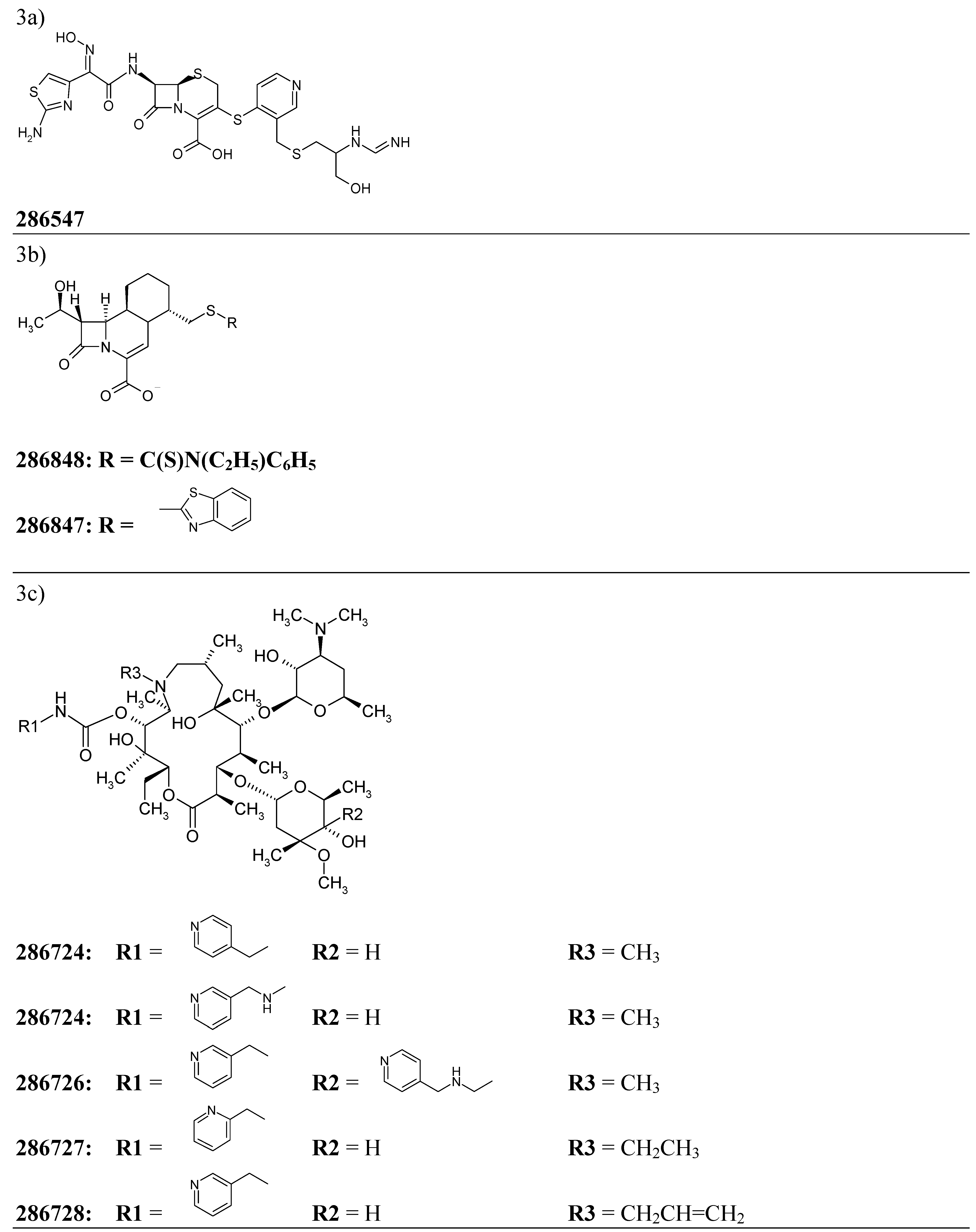
An application of the model
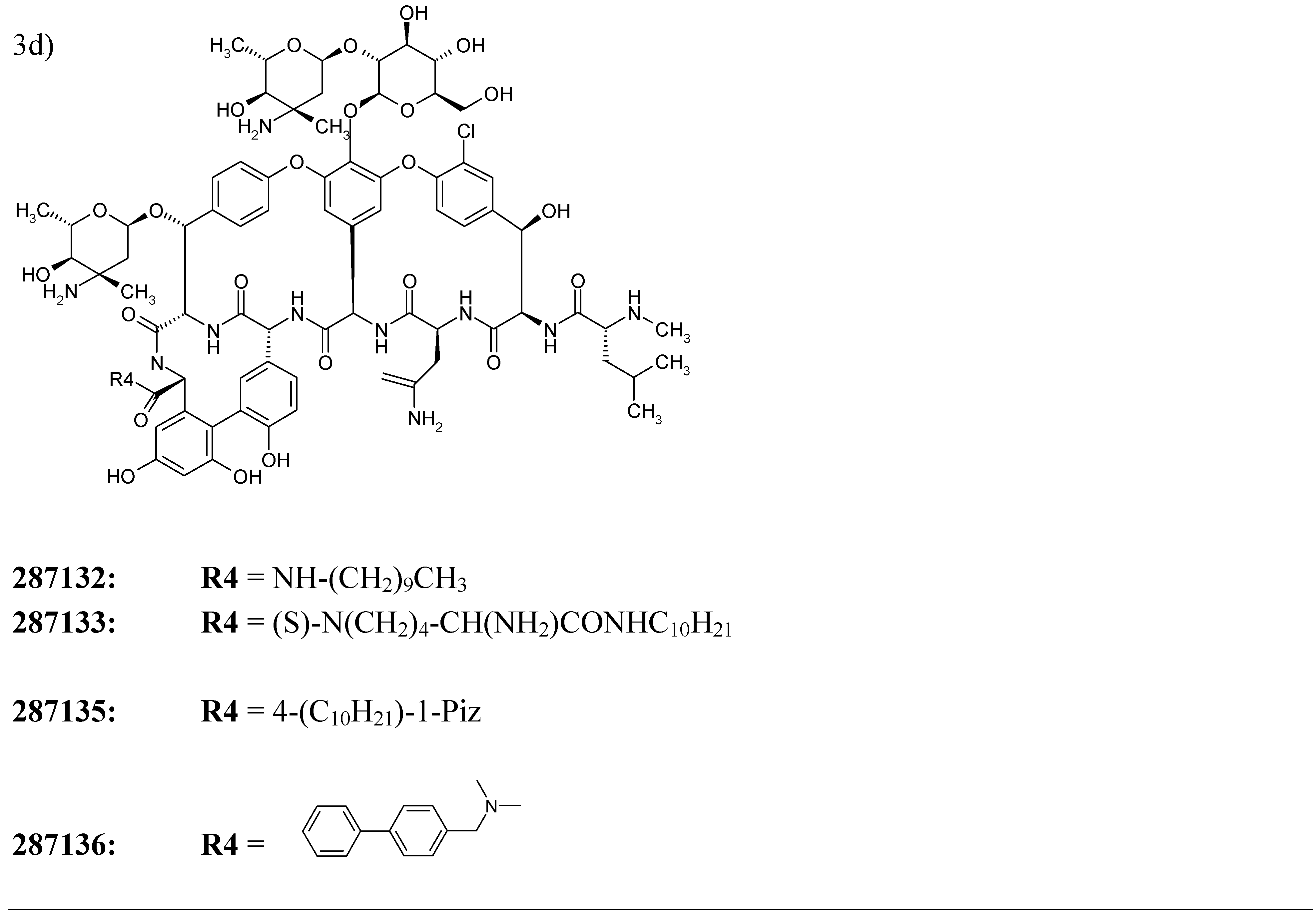
| Name | Output | Name | Output | |
|---|---|---|---|---|
| antibiotics | apicycline | 0.975 | ||
| 4'-(methylsulfamoyl)sulfanilanilide | 0.973 | apramycin | 0.980 | |
| 4'-formylsuccinanilic acid thiosemicarbazone | azidocillin | 0.979 | ||
| 0.259 | arbekacin | 0.980 | ||
| 4-sulfanilamidosalicylic acid | 0.938 | aspoxicillin | 0.975 | |
| acediasulfone | 0.828 | azidamfenicol | 0.966 | |
| acetyl sulfamethoxypyrazine | 0.855 | azlocillin | 0.850 | |
| acetyl sulfisoxazole | 0.964 | aztreonam | 0.981 | |
| amidinocillin | 0.702 | bacampicillin | 0.982 | |
| amidinocillin pivoxil | 0.938 | benzylpenicillinic acid | 0.924 | |
| amifloxacin | 0.881 | benzylsulfamide | 0.733 | |
| amikacin | 0.984 | biapenem | 0.830 | |
| apalcillin | 0.981 | brodimoprim | 0.585 | |
| butirosin | 0.984 | cephalothin | 0.977 | |
| carbenicillin | 0.974 | cephapirin sodium | 0.984 | |
| carfecillin sodium | 0.970 | cephradine | 0.897 | |
| carindacillin(a,e,f,i) | 0.938 | chloramphenicol | 0.606 | |
| carumonam | 0.985 | chloramphenicol palmitate | 0.604 | |
| cefaclor | 0.860 | chloramphenicol pantothenate | 0.983 | |
| cefadroxil | 0.915 | chlortetracycline | 0.984 | |
| cefamandole | 0.964 | cinoxacin | 0.770 | |
| cefatrizine | 0.973 | clinafloxacin | 0.920 | |
| cefazedone | 0.984 | clindamycin | 0.926 | |
| cefazolin | 0.979 | clometocillin | 0.953 | |
| cefbuperazone | 0.984 | clomocycline | 0.982 | |
| cefcapene pivoxil | 0.983 | cloxacillin | 0.935 | |
| cefclidin(a,i,j) | 0.985 | cyclacillin | 0.960 | |
| cefdinir(e,i) | 0.984 | dibekacin | 0.952 | |
| cefditoren | 0.984 | dichloramine | 0.253 | |
| cefepime | 0.982 | dicloxacillin | 0.983 | |
| cefetamet | 0.983 | difloxacin | 0.835 | |
| cefixime | 0.984 | diphenicillin sodium | 0.767 | |
| cefmenoxime | 0.984 | doxycycline | 0.981 | |
| cefmetazole | 0.984 | enoxacin | 0.915 | |
| cefminox | 0.985 | enrofloxacin | 0.630 | |
| cefodizime | 0.985 | epicillin | 0.963 | |
| cefonicid | 0.984 | fenbenicillin | 0.967 | |
| ceforanide | 0.974 | fleroxacin | 0.980 | |
| cefotiam | 0.985 | flomoxef | 0.985 | |
| cefoxitin | 0.984 | florfenicol | 0.955 | |
| cefozopran | 0.982 | floxacillin | 0.983 | |
| cefpimizole | 0.985 | fortimicin a | 0.978 | |
| cefpiramide | 0.985 | fortimicin b | 0.700 | |
| cefpirome | 0.984 | furaltadone | 0.901 | |
| cefpodoxime proxetil | 0.985 | gentamicin c1 | 0.850 | |
| cefprozil | 0.902 | gentamicin c2 | 0.940 | |
| cefroxadine | 0.970 | gentamicin c3 | 0.956 | |
| cefsulodin | 0.982 | grepafloxacin | 0.862 | |
| ceftazidime | 0.984 | guamecycline | 0.977 | |
| cefteram | 0.979 | imipenem | 0.577 | |
| ceftezole | 0.984 | isepamicin | 0.985 | |
| ceftizoxime | 0.984 | kanamycin a | 0.962 | |
| cefuroxime | 0.980 | kanamycin b | 0.976 | |
| cefuzonam | 0.985 | kanamycin c | 0.971 | |
| cephacetrile sodium | 0.982 | lenampicillin | 0.985 | |
| cephalexin | 0.847 | lincomycin | 0.907 | |
| cephaloglycin | 0.951 | lomefloxacin | 0.946 | |
| cephaloridine | 0.960 | loracarbef | 0.862 | |
| cephalosporin c | 0.976 | lymecycline | 0.978 | |
| meclocycline | 0.984 | propicillin | 0.814 | |
| meropenem | 0.977 | quinacillin | 0.984 | |
| methacycline | 0.983 | ribostamycin | 0.965 | |
| methicillin sodium | 0.951 | rifamide | 0.979 | |
| mezlocillin | 0.976 | rifamycin sv | 0.984 | |
| micronomicin | 0.966 | rifaximin | 0.984 | |
| miloxacin | 0.786 | ritipenem | 0.977 | |
| moxalactam | 0.984 | rolitetracycline | 0.979 | |
| n2-formylsulfisomidine | 0.919 | rosoxacin | 0.265 | |
| n4-sulfanilylsulfanilamide | 0.980 | rufloxacin | 0.975 | |
| nadifloxacin | 0.658 | salazosulfadimidine | 0.970 | |
| nafcillin sodium | 0.919 | sancycline | 0.980 | |
| nalidixic acid | 0.268 | sisomicin | 0.909 | |
| neomycin a(c,i,j) | 0.983 | sparfloxacin | 0.975 | |
| neomycin b(a,d,h,i) | 0.981 | spectinomycin | 0.628 | |
| netilmicin | 0.938 | succinylsulfathiazole | 0.977 | |
| nifuradene | 0.600 | sulbenicillin | 0.884 | |
| nifuratel | 0.980 | sulfabenzamide | 0.895 | |
| nifurfoline | 0.963 | sulfacetamide | 0.955 | |
| nifurprazine | 0.267 | sulfachlorpyridazine | 0.915 | |
| nifurtoinol | 0.694 | sulfachrysoidine | 0.975 | |
| nitrofurantoin | 0.291 | sulfacytine | 0.971 | |
| norfloxacin | 0.523 | sulfadiazine | 0.937 | |
| N-sulfanilyl-3,4-xylamide | 0.956 | sulfadicramide | 0.933 | |
| ofloxacin | 0.972 | sulfadimethoxine | 0.958 | |
| oxytetracycline | 0.984 | sulfadoxine | 0.965 | |
| panipenem | 0.939 | sulfaethidole | 0.918 | |
| paromomycin | 0.984 | sulfaguanidine | 0.904 | |
| pasiniazide | 0.236 | sulfaguanol | 0.943 | |
| pazufloxacin | 0.926 | sulfalene | 0.938 | |
| pefloxacin | 0.563 | sulfaloxic acid | 0.857 | |
| penamecillin | 0.636 | sulfamethazine | 0.912 | |
| penethamate hydriodide | 0.704 | sulfamethizole | 0.759 | |
| penicillin G potassium | 0.848 | sulfamethomidine | 0.940 | |
| penicillin N | 0.901 | sulfamethoxazole | 0.908 | |
| penicillin O | 0.978 | sulfamethoxypyridazine | 0.912 | |
| penicillin V | 0.912 | sulfamidochrysoidine | 0.952 | |
| phenethicillin potassium | 0.822 | sulfamoxole | 0.954 | |
| phthalylsulfathiazole | 0.976 | sulfanilamide | 0.653 | |
| pipacycline | 0.921 | sulfanilic acid | 0.841 | |
| pipemidic acid | 0.882 | sulfanilylurea | 0.938 | |
| piperacillin | 0.982 | sulfaphenazole | 0.929 | |
| piromidic acid | 0.696 | sulfaproxyline | 0.957 | |
| pivampicillin | 0.916 | sulfapyrazine | 0.934 | |
| pivcefalexin | 0.946 | sulfathiazole | 0.873 | |
| p-nitrosulfathiazole | 0.893 | sulfathiourea | 0.849 | |
| sulfisomidine | 0.909 | bamipine | 0.036 | |
| sulfisoxazole | 0.963 | biclofibrate | 0.247 | |
| sultamicillin | 0.983 | befunolol | 0.252 | |
| talampicillin | 0.911 | benfluorex | 0.258 | |
| temocillin | 0.985 | benorylate | 0.259 | |
| tetracycline | 0.983 | benserazide | 0.259 | |
| tetroxoprim | 0.837 | benzitramide | 0.259 | |
| thiamphenicol | 0.942 | benzotropine mesylate | 0.000 | |
| ticarcillin | 0.983 | benzpiperylon | 0.000 | |
| tigemonam | 0.985 | benzydamine | 0.000 | |
| trimethoprim | 0.739 | bermoprofen | 0.257 | |
| trospectomycin | 0.850 | betaxolol | 0.174 | |
| trovafloxacin(b) | 0.960 | bevantolol | 0.154 | |
| non-antibiotics | bevonium methyl sulfate | 0.032 | ||
| 2-amino-4-picoline | 0.258 | bezafibrate | 0.256 | |
| 5-bromosalicylic acid acetate | 0.258 | binifibrate | 0.319 | |
| 5-nitro-2propoxyacetanilide | 0.280 | bisoprolol | 0.184 | |
| acecarbromal | 0.259 | bitolterol | 0.004 | |
| aceclofenac | 0.431 | bucloxic acid | 0.258 | |
| acefylline(c,d,e,g) | 0.841 | bopindolol | 0.001 | |
| acetaminophen(b,i) | 0.258 | bromfenac | 0.258 | |
| acetanilide | 0.258 | bromisovalum | 0.258 | |
| acetazolamide | 0.023 | bromodiphenhydramine | 0.057 | |
| acetophenazine | 0.265 | brompheniramine | 0.006 | |
| acetylsalicylic acid | 0.258 | bucetin | 0.247 | |
| acrivastine | 0.260 | bucolome | 0.253 | |
| ahistan | 0.000 | bucumolol | 0.256 | |
| albuterol | 0.258 | bufetolol | 0.157 | |
| alclofenac | 0.258 | bufexamac | 0.258 | |
| alminoprofen | 0.256 | bufuralol | 0.008 | |
| alphaprodine | 0.106 | bumadizon | 0.205 | |
| alprenolol | 0.239 | bunitrolol | 0.258 | |
| aminochlorthenoxazin | 0.257 | butabarbital | 0.258 | |
| aminopyrine | 0.000 | butaclamol | 0.123 | |
| amosulalol | 0.078 | butallylonal | 0.262 | |
| amtolmetin guacil | 0.001 | butanilicaine | 0.206 | |
| anileridine | 0.262 | butibufen | 0.255 | |
| antipyrine | 0.017 | butidrine hydrochloride | 0.183 | |
| antrafenine | 0.283 | butoctamide | 0.252 | |
| apazone | 0.001 | butofilolol | 0.256 | |
| apronalide | 0.258 | caffeine | 0.159 | |
| arotinolol | 0.293 | capuride | 0.257 | |
| atenolol | 0.258 | carazolol | 0.027 | |
| atropine | 0.258 | carbamazepine | 0.015 | |
| bambuterol | 0.032 | carbidopa | 0.259 | |
| bamifylline | 0.290 | carbinoxamine | 0.066 | |
| carbiphene | 0.258 | diethylbromoacetamide | 0.257 | |
| carbocloral | 0.313 | difenamizole | 0.006 | |
| carbromal | 0.257 | difenpiramide | 0.009 | |
| carbuterol | 0.258 | diflunisal | 0.258 | |
| carfimate | 0.258 | dilevalol | 0.255 | |
| carphenazine | 0.263 | dioxadrol | 0.000 | |
| carprofen | 0.258 | dipyrocetyl | 0.315 | |
| carsalam | 0.258 | dipyrone | 0.041 | |
| carteolol | 0.259 | disulfiram | 0.001 | |
| carvedilol | 0.000 | doxefazepam | 0.270 | |
| celiprolol | 0.211 | doxofylline | 0.629 | |
| cetamolol | 0.245 | doxylamine(b,f,g,i) | 0.000 | |
| cetirizine | 0.261 | droperidol | 0.259 | |
| chlorhexadol | 0.288 | droxicam | 0.022 | |
| chlorobutanol | 0.258 | dyphylline | 0.410 | |
| chloropyramine | 0.050 | ectylurea | 0.244 | |
| chlorothen | 0.070 | embramine | 0.122 | |
| chlorpheniramine | 0.095 | emorfazone | 0.010 | |
| chlorprothixene | 0.017 | enfenamic acid | 0.256 | |
| chlorthenoxacin | enprofylline | 0.246 | ||
| (chlorthenoxazine) | 0.258 | epanolol | 0.258 | |
| chlorcyclizine | 0.078 | ephedrine | 0.229 | |
| cinchophen | 0.251 | epirizole | 0.002 | |
| cinmetacin | 0.248 | eprozinol | 0.237 | |
| cinnarizine | 0.388 | estazolam | 0.000 | |
| cinromida | 0.197 | etafedrine | 0.179 | |
| ciprofibrate | 0.251 | etamiphyllin | 0.118 | |
| clemastine | 0.039 | etaqualone | 0.000 | |
| clenbuterol | 0.234 | eterobarb | 0.001 | |
| clidanac | 0.258 | etersalate | 0.260 | |
| clinofibrate | 0.282 | ethenzamide | 0.243 | |
| clofibric acid | 0.256 | ethinamate | 0.258 | |
| clometacin | 0.292 | ethoheptazine | 0.000 | |
| clometiazol | 0.255 | ethoxazene | 0.248 | |
| clonixin | 0.254 | etodolac | 0.259 | |
| clopirac | 0.257 | etofibrate | 0.260 | |
| cloranolol | 0.247 | etofylline | 0.266 | |
| clordesmetildiazepam | 0.257 | etomidate | 0.000 | |
| clorprenaline | 0.249 | etymemazine | 0.002 | |
| clothiapine | 0.003 | felbinac | 0.258 | |
| clozapine | 0.051 | fenadiazole | 0.230 | |
| codeine | 0.062 | fenbufen | 0.258 | |
| cropropamide | 0.002 | fenclofenac | 0.259 | |
| crotethamide | 0.035 | fenethazine | 0.000 | |
| deserpidine | 0.005 | fenofibrate | 0.254 | |
| diclofenac | 0.262 | fenoprofen | 0.258 | |
| fenoterol | 0.258 | lornoxicam | 0.031 | |
| fentanyl | 0.066 | loxapina | 0.004 | |
| fentiazac | 0.259 | loxoprofen | 0.258 | |
| floctafenine | 0.266 | mazindol(i) | 0.162 | |
| flufenamic acid | 0.259 | meclofenamic acid(f) | 0.276 | |
| fluoresone | 0.459 | mecloqualone | 0.000 | |
| fluphenazine | 0.260 | medibazine | 0.004 | |
| flupirtine | 0.260 | medrylamine | 0.001 | |
| fluproquazone | 0.258 | meparfynol | 0.258 | |
| flurazepam | 0.010 | mepindolol | 0.211 | |
| flurbiprofen | 0.258 | meprobamate | 0.259 | |
| fluspirilene | 0.259 | mequitazine | 0.001 | |
| flutropium bromide | 0.259 | methafurylene | 0.000 | |
| formoterol | 0.259 | methaphenilene | 0.000 | |
| fosazepam | 0.258 | methotrimeprazine | 0.002 | |
| fusaric acid | 0.258 | methoxyphenamine | 0.000 | |
| gemfibrozil | 0.248 | methyldopa | 0.258 | |
| gentisic acid | 0.258 | methyltyrosine | 0.256 | |
| glafenine | 0.259 | methyprylon | 0.232 | |
| glucametacin | 0.335 | metiapine | 0.002 | |
| glutethimide | 0.258 | metipranolol | 0.258 | |
| haloperidide | 0.259 | metofoline | 0.094 | |
| haloperidol | 0.258 | metoprolol | 0.179 | |
| hexapropymate | 0.258 | metron | 0.275 | |
| hexobarbital | 0.274 | mexiletine | 0.251 | |
| hexoprenaline | 0.258 | mofezolac | 0.340 | |
| histapyrrodine | 0.004 | molindone | 0.000 | |
| hydroxyethylpromethazine | moperone | 0.259 | ||
| (N-Hydroxyethylpromethazine) | 0.261 | moprolol | 0.213 | |
| hydroxyzine | 0.261 | morazone | 0.000 | |
| ibufenac | 0.258 | morphine | 0.289 | |
| ibuprofen | 0.258 | moxastine | 0.000 | |
| ibuproxam | 0.258 | nadoxolol | 0.258 | |
| indenolol | 0.179 | naproxen | 0.256 | |
| indomethacin | 0.323 | narcobarbital | 0.265 | |
| ipratropium bromide | 0.259 | nefopam | 0.000 | |
| isoetharine | 0.258 | niceritrol | 0.981 | |
| isofezolac | 0.183 | nicoclonate | 0.095 | |
| isonixin | 0.125 | nicofibrate | 0.214 | |
| isopromethazine | 0.000 | nifenalol | 0.256 | |
| isoxicam | 0.003 | nifenazone | 0.000 | |
| ketoprofen | 0.250 | niflumic acid | 0.260 | |
| ketorolac | 0.259 | nimetazepam | 0.512 | |
| labetalol | 0.252 | nipradilol | 0.611 | |
| lefetamine | 0.068 | nitrazepam | 0.337 | |
| lorazepam | 0.268 | nordiazepam | 0.254 | |
| novonal | 0.255 | propyphenazone | 0.000 | |
| octopamine | 0.258 | protokylol | 0.260 | |
| orphenadrine | 0.000 | proxibarbital | 0.262 | |
| oxaceprol | 0.259 | proxyphylline | 0.210 | |
| oxametacine | 0.284 | pyrilamine | 0.000 | |
| oxanamide | 0.254 | pyrrobutamine | 0.000 | |
| oxaprozin | 0.259 | quazepam | 0.331 | |
| oxitropium bromide | 0.265 | ramifenazone | 0.000 | |
| oxprenolol | 0.221 | reproterol | 0.296 | |
| oxypertine | 0.000 | rimiterol | 0.258 | |
| paramethadione | 0.258 | ronifibrate | 0.259 | |
| parsalmide | 0.259 | salacetamide | 0.258 | |
| p-bromoacetanilide | 0.258 | salicylamide | 0.257 | |
| pemoline | 0.258 | salicylamide O-acetic acid | 0.258 | |
| penbutolol | 0.099 | salsalate | 0.258 | |
| penfluridol | 0.259 | salverine | 0.000 | |
| perisoxal | 0.034 | scopolamine | 0.278 | |
| perphenazine | 0.284 | secobarbital | 0.257 | |
| phenacemide | 0.258 | setastine | 0.035 | |
| phenacetin | 0.247 | simetride | 0.028 | |
| phenoperidine | 0.191 | simfibrate | 0.259 | |
| phenopyrazone | 0.243 | simvastatin | 0.355 | |
| phenylbutazone | 0.000 | sotalol | 0.013 | |
| phenyltoloxamine(a,c,g) | 0.000 | soterenol | 0.099 | |
| pindolol | 0.055 | sulfinalol | 0.062 | |
| pipebuzone | 0.001 | sulpiride | 0.017 | |
| piperacetazine | 0.261 | suprofen | 0.258 | |
| piperidione | 0.253 | talastine | 0.000 | |
| piperylone | 0.000 | talinolol | 0.245 | |
| pirbuterol | 0.259 | talniflumate | 0.399 | |
| pirifibrate(g,h) | 0.258 | temazepam | 0.207 | |
| piroxicam | 0.013 | tenoxicam | 0.008 | |
| pirprofen | 0.258 | terbutaline | 0.258 | |
| p-lactophenetide | 0.257 | tertatolol | 0.129 | |
| p-methyldiphenhydramine | 0.000 | tetrabarbital | 0.257 | |
| pravastatin | 0.438 | thenaldine | 0.000 | |
| prazepam | 0.008 | thenyldiamine | 0.000 | |
| primidone | 0.133 | theobromine | 0.251 | |
| probucol | 0.000 | theofibrate(b,f,i) | 0.435 | |
| procaterol | 0.260 | theophylline(f,h,i,j) | 0.224 | |
| proglumetacin | 0.292 | thioridazine | 0.003 | |
| prolintane | 0.024 | thiothixene | 0.003 | |
| promazine | 0.000 | thonzylamine | 0.001 | |
| pronethalol | 0.237 | tiaprofenic acid | 0.258 | |
| propanolol | 0.067 | timolol | 0.030 | |
| toliprolol | 0.127 | tripelennamine | 0.000 | |
| tolmetin | 0.254 | triprolidine | 0.000 | |
| tolpropamine | 0.001 | tulobuterol | 0.169 | |
| tretoquinol | 0.418 | viminol | 0.028 | |
| triazolam | 0.003 | vinylbital | 0.258 | |
| triclofos | 0.276 | xenbucin | 0.256 | |
| trifluoperazine | 0.298 | xibenolol | 0.148 | |
| trifluperidol | 0.259 | zolamine | 0.035 | |
| trimethadione | 0.258 | zomepirac | 0.263 | |
| triparanol | 0.248 | |||
| Name | Output | Name | Output | |
|---|---|---|---|---|
| antibiotics | butacetin | 0.147 | ||
| amoxicillin | 0.152 | chlorpromazine | 0.169 | |
| ampicillin | 0.728 | ciramadol | 0.150 | |
| cefoperazone | 0.997 | clocinizine | 0.125 | |
| cefotaxime | 0.999 | clofibrate | 0.142 | |
| cefotetan | 0.568 | diazepam | 0.997 | |
| cefteram | 0.999 | diphenhydramine | 0.125 | |
| ceftriaxone | 0.999 | diphenylpyraline | 0.101 | |
| ciprofloxacin | 0.999 | esmolol | 0.151 | |
| demeclocycline | 0.999 | ethclorvinol | 0.047 | |
| flumequine | 0.998 | feprazone | 0.118 | |
| hetacillin | 0.992 | flunitrazepam | 0.069 | |
| mafenide | 0.999 | fosfosal | 0.134 | |
| metampicillin | 0.978 | indoprofen | 0.287 | |
| minocycline | 0.984 | isoproterenol | 0.151 | |
| nifurpirinol | 0.998 | levobunolol | 0.150 | |
| noprylsulfamide | 0.998 | lovastatin | 0.151 | |
| oxacillin | 0.999 | mabuterol | 0.149 | |
| oxolinic acid | 0.991 | mefenamic acid | 0.097 | |
| sulfamerazine | 0.999 | mefexamide | 0.000 | |
| sulfametrole | 0.999 | meperidine | 0.146 | |
| sulfanitran | 0.998 | mephobarbital | 0.160 | |
| sulfaperine | 0.997 | methapyrilene | 0.000 | |
| temafloxacin | 0.987 | nadolol | 0.151 | |
| thiazolsulfone | 0.994 | pheniramine | 0.134 | |
| tobramycin | 0.994 | phenocoll | 0.000 | |
| tosufloxacin | 0.995 | phenyramidol | 0.000 | |
| non-antibiotics | pimozide | 0.029 | ||
| acetaminosalol | 0.110 | practolol | 0.152 | |
| acetobutolol | 0.150 | proheptazine | 0.149 | |
| aminopropylon | 0.000 | propacetamol | 0.166 | |
| benoxaprofen | 0.150 | sulindac | 0.975 | |
| brotizolam | 0.004 | talbutal | 0.063 | |
| bupranolol | 0.144 | |||
| Compound | Structural formula | Prediction |
|---|---|---|
| 286547 | 3a | 0.984 |
| 286724 | 3c | 0.985 |
| 286725 | 3c | 0.985 |
| 286726 | 3c | 0.985 |
| 286727 | 3c | 0.985 |
| 286728 | 3c | 0.985 |
| 286847 | 3b | 0.915 |
| 286848 | 3b | 0.914 |
| 287132 | 3d | 0.985 |
| 287133 | 3d | 0.985 |
| 287135 | 3d | 0.985 |
| 287136 | 3d | 0.985 |
Conclusions
Methods
References
- Kubinyi, H.; Folkers, G.; Martin, Y.C. (Eds.) 3D QSAR in Drug Design; Kluwer: Dordrecht, 2002.
- Truhlar, D.G.; Howe, W.J.; Hopfinger, A.J. (Eds.) Rational Drug Design; Springer: Berlin, 1999.
- Karelson, M. Molecular Descriptors in QSAR/QSPR; Wiley: New York, 2000; p. 448. [Google Scholar]
- Exner, O. Correlation Analysis of Chemical Data; Kluwer: Dordrecht, 1988. [Google Scholar]
- Cherkasov, A.R.; Galkin, V.I.; Cherkasov, R.A. J. Phys. Org. Chem. 1998, 11, 437. [CrossRef]
- Cherkasov, A.R.; Galkin, V.I.; Cherkasov, R.A. J. Molec. Struct. (Theochem) 1999, 489, 43. [CrossRef]
- Cherkasov, A.R.; Galkin, V.I.; Cherkasov, R.A. J. Molec. Struct. (Theochem) 2000, 497, 115. [CrossRef]
- Cherkasov, A. J. Chem. Inf. Comp. Sci. 2003, 43, 2039. [CrossRef]
- Babij, C.; Poe, A.J. J. Phys. Org. Chem. 2004, 17, 162.
- Galkin, V.I.; Sayakhov, R.D.; Cherkasov, R.A. Russ. Chem. Rev. 1991, 60, 1617.
- Cherkasov, A.; Jonsson, M. J. Chem. Inf. Comp. Sci. 1998, 38, 1151. [CrossRef]
- Cherkasov, A.; Jonsson, M. J. Chem. Inf. Comp. Sci. 1999, 39, 1057. [CrossRef]
- Cherkasov, A.R.; Jonsson, M.; Galkin, V.I. J. Mol. Graph. Model. 1999, 17, 28. [CrossRef]
- Cherkasov, A.; Jonsson, M. J. Chem. Inf. Comp. Sci. 2000, 40, 1222. [CrossRef]
- Cherkasov, A.; Sprous, D.; Chen, R. J. Phys. Chem. A. 2003, 107, 9695.
- Galkin, V.I.; Cherkasov, A.R.; Cherkasov, R.A. Phosphorus, Silicon, Sulphur 1999, 146, 329.
- Byvalov, E.; Fechner, U.; Sadowski, J.; Schneider, G. J. Chem. Inf. Comp. Sci. 2003, 43, 1882. [CrossRef]
- Zernov, V.; Balakin, K.V.; Ivaschenko, A.A.; Savchuk, N.P.; Pletnev, I.V. J. Chem. Inf. Comp. Sci. 2003, 43, 2048. [CrossRef]
- Anzali, S.; Barenickel, G.; Cezanne, B.; Krug, M.; Filimonov, D.; Poroikob, V. J. Med. Chem. 2001, 44, 2432. [CrossRef]
- Murcia-Soler, M.; Perez-Gimenez, F.; Garcia-March, F.J.; Salabert-Salvador, M.T.; Diaz-Villanueva, W.; Castro-Bleda, M.J. J. Chem. Inf. Comp. Sci. 2003, 43, 1688. [CrossRef]
- Frimurer, T.M.; Bywater, R.; Naerum, L.; Lauritsen, L.N.; Brunak, S. J. Chem. Inf. Comp. Sci. 2000, 40, 1315. [CrossRef]
- Sadowski, J.; Kubinyi, H. J. Med. Chem. 1998, 41, 3325. [CrossRef]
- Galvez, J.; de Julian-Ortiz, J.V.; Garcia-Domenech, R. J. Mol. Graph. Model. 2001, 20, 84. [CrossRef]
- Ajay, A.; Walters, W.P.; Murcko, M.A. J. Med. Chem. 1998, 41, 3314. [CrossRef]
- Jaen-Oltra, J.; Salabert-Salvador, M.T.; Garcia-March, F.J.; Perez-Gimenez, F.; Tomas-Vert, F. J. Med. Chem. 2000, 43, 1143. [CrossRef] [PubMed]
- Garcia-Domenech, R.; de Julian-Ortiz, J.V. J. Chem. Inf. Comp. Sci. 1998, 38, 445. [CrossRef]
- Tomas-Vert, F.; Perez-Gimenez, F.; Salabert-Salvador, M.T.; Garcia-March, F.J.; Jaen-Oltra, J. J. Molec. Struct. (Theochem). 2000, 504, 249. [CrossRef]
- Mishra, R.K.; Garcia-Domenech, R.; Galvez, J. J. Chem. Inf. Comp. Sci. 2001, 41, 387. [CrossRef]
- Cronin, M.T.D.; Aptula, A.O.; Dearden, J.C.; Duffy, J.C.; Netzeva, T.I.; Patel, H.; Rowe, P.H.; Schultz, T.W.; Worth, A.P.; Voutzoulidis, K.; Schuurmann, G. J. Chem. Inf. Comp. Sci. 2002, 42, 869. [CrossRef]
- Molina, E.; Diaz, H.G.; Gonzalez, M.P.; Rodriguez, E.; Uriarte, E. J. Chem. Inf. Comp. Sci. 2004, 44, 515. [CrossRef]
- Gozalbez, R.; Galvez, J.; Moreno, A.; Garcia-Domenech, R. J. Pharm. Pharmacol. 1999, 51, 111. [CrossRef]
- Molecular Operational Environment; Chemical Computation Group Inc.: Montreal, Canada, 2004.
- Zupan, J.; Gasteiger, J. Neural Networks in Chemistry and Drug Design, 2nd Ed. ed; Wiley: New York, 1999. [Google Scholar]
- Dastidar, S.D.; Ganguly, K.; Chaudhuri, K.; Chakrabarty, A.N. Int. J. Antimicrob. Agents 2000, 14, 249.
- Annadurai, S.; Basu, S.; Ray, S.; Dastidar, S.D.; Chakrabarty, A.N. Indian J. Exp. Biol. 1998, 36, 86.
- Dastidar, S.D.; Saha, P.K.; Sanymat, B.; Chakrabarty, A.N. J. Appl. Bact. 1976, 42, 209. [CrossRef]
- Dastidar, S.D.; Jairaj, J.; Mookerjee, M.; Chakrabarty, A.N. Acta Microbiol. Immun. Hung. 1997, 44, 241.
- Molnar, J.; Mandi, Y.; Kiraly, J. Acta Microbiol. Immun. Hung. 1976, 23, 45.
- Kristiansen, J.E. Acta Path. Microbioil. Scand. Sect. B. 1979, 87, 317.
- Kristiansen, J.E.; Mortensen, I. Pramocol. Toxicol. 1987, 60, 100.
- Dastidar, S.D.; Chaudhuri, K.; Annadurai, S.; Ray, S.; Mookerjee, M.; Chakrabarty, A.N. J. Chemother. 1995, 7, 201. [CrossRef]
- Dash, S.K.; Dastidar, S.D.; Chaudhuri, K. Ind. J. Exp. Biol. 1977, 15, 324.
- Dastidar, S.D.; Mondal, U.; Niyogi, S.; Chakrabarty, A.N. Ind. J. Med. Res. 1986, 84, 142.
- Drug Data Report 2000, 22, 530.
- ChemIDPlus database:. http://chem.sis.nlm.nih.gov/chemidplus/.
- Halgren, T.A. J. Comp. Chem. 1996, 17, 490. [CrossRef]
- SNNS: Stuttgart Neural Network Simulator, Version 4.0; University of Stuttgart: Stuttgart, 1995.
© 2005 by MDPI (http://www.mdpi.org). Reproduction is permitted for noncommercial purposes.
Share and Cite
Cherkasov, A. Inductive QSAR Descriptors. Distinguishing Compounds with Antibacterial Activity by Artificial Neural Networks. Int. J. Mol. Sci. 2005, 6, 63-86. https://doi.org/10.3390/i6010063
Cherkasov A. Inductive QSAR Descriptors. Distinguishing Compounds with Antibacterial Activity by Artificial Neural Networks. International Journal of Molecular Sciences. 2005; 6(1):63-86. https://doi.org/10.3390/i6010063
Chicago/Turabian StyleCherkasov, Artem. 2005. "Inductive QSAR Descriptors. Distinguishing Compounds with Antibacterial Activity by Artificial Neural Networks" International Journal of Molecular Sciences 6, no. 1: 63-86. https://doi.org/10.3390/i6010063
APA StyleCherkasov, A. (2005). Inductive QSAR Descriptors. Distinguishing Compounds with Antibacterial Activity by Artificial Neural Networks. International Journal of Molecular Sciences, 6(1), 63-86. https://doi.org/10.3390/i6010063




