Cartilage Intermediate Layer Protein 2 Aggravates Hepatic Lipid Accumulation and Inflammation Through the IRE1α/XBP1 Pathway
Abstract
1. Introduction
2. Result
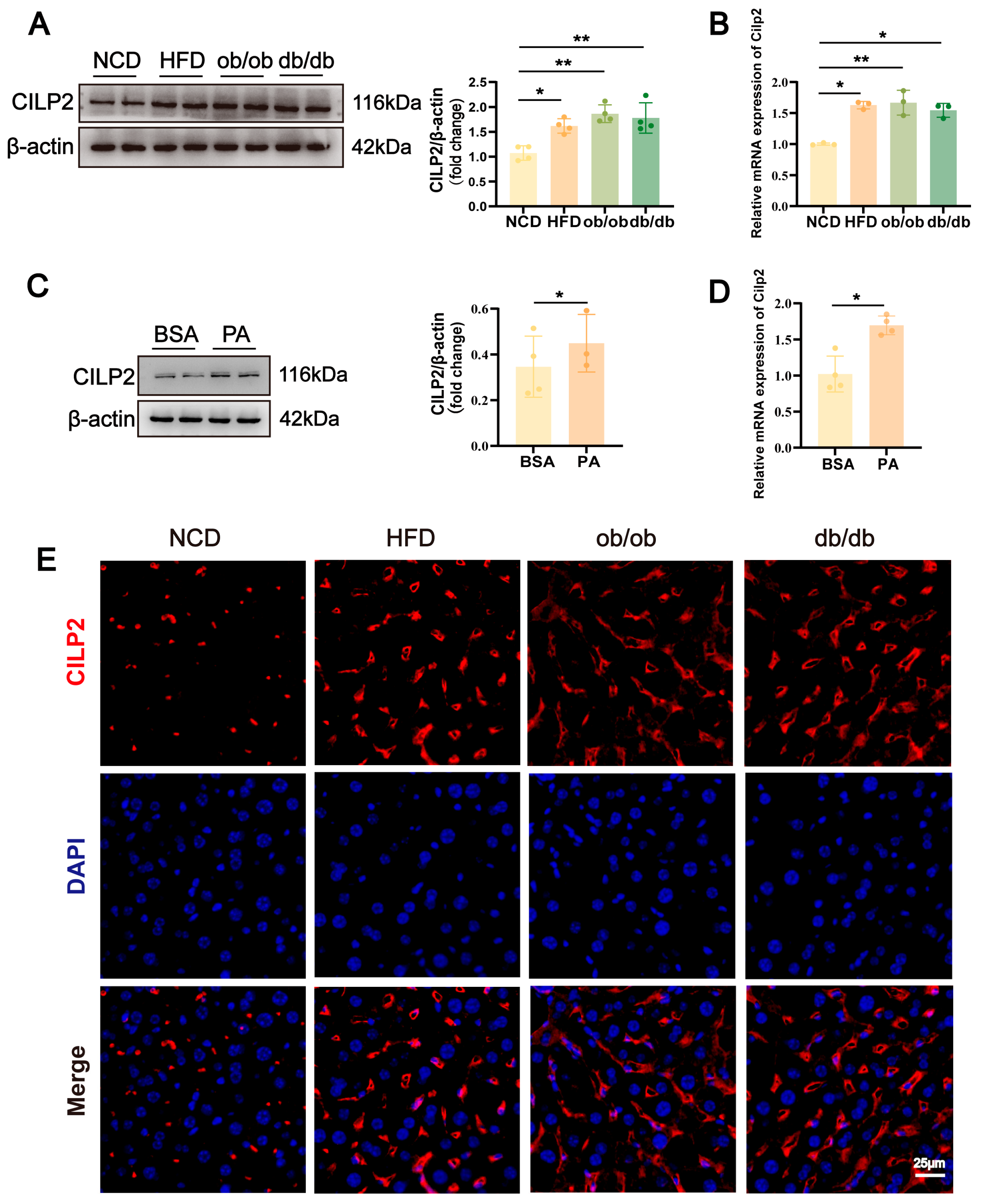
2.1. CILP2 Upregulated in Steatotic Liver and Hepatocytes
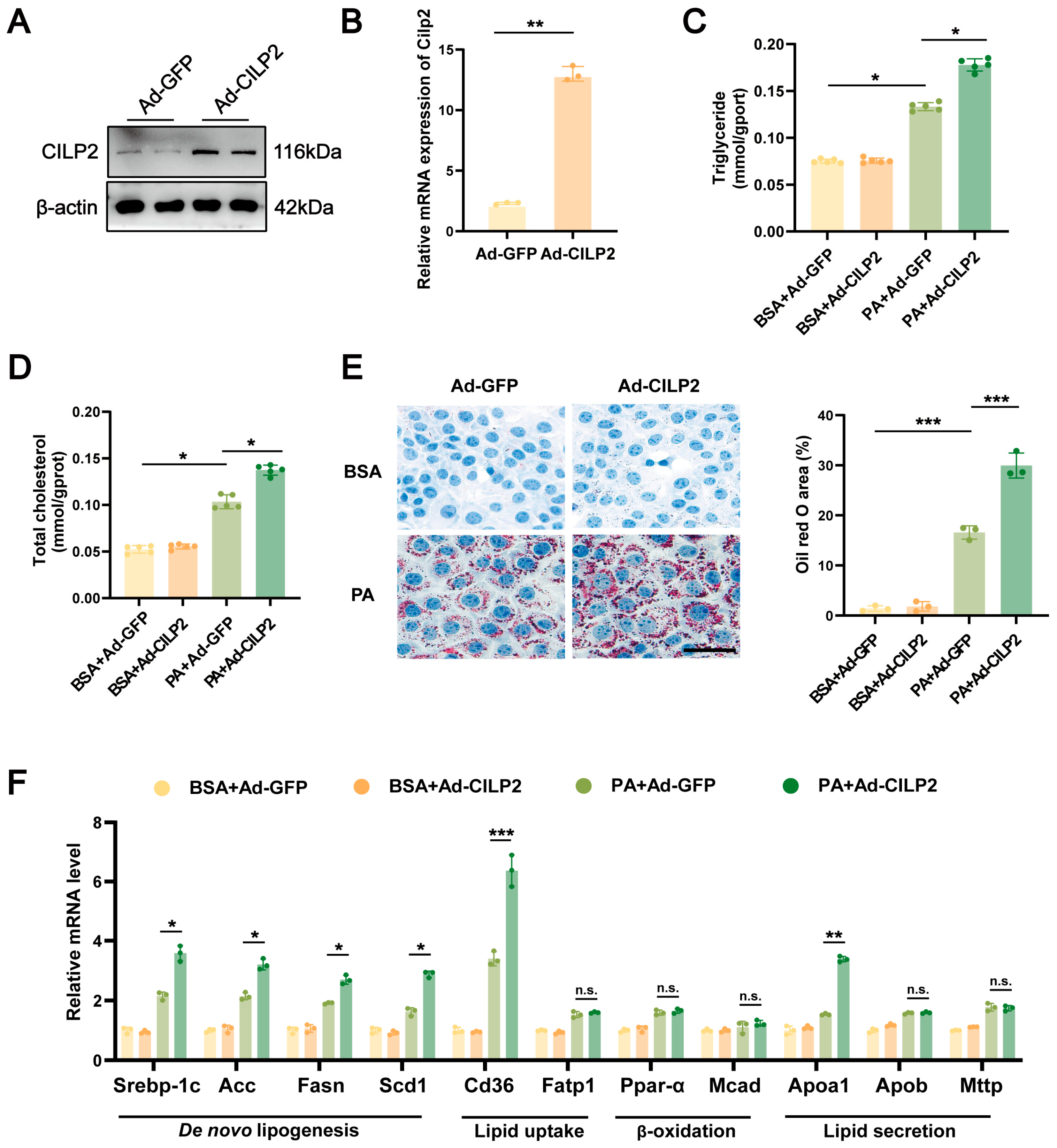
2.2. CILP2 Overexpression Enhanced Lipid Accumulation in Hepatocytes
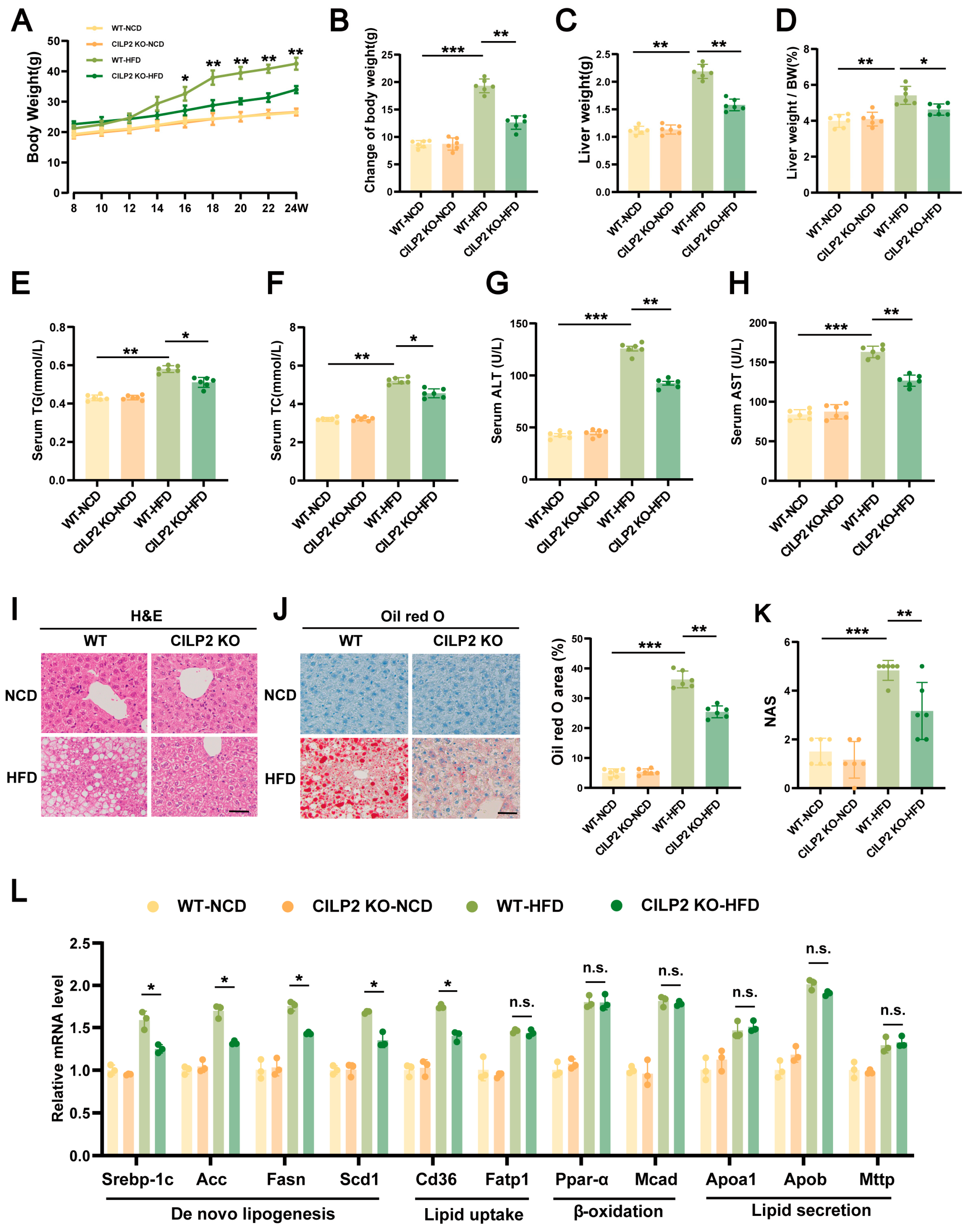
2.3. CILP2 KO Reduced Body Weight and Improved IR and Hepatic Steatosis in HFD-Fed Mice
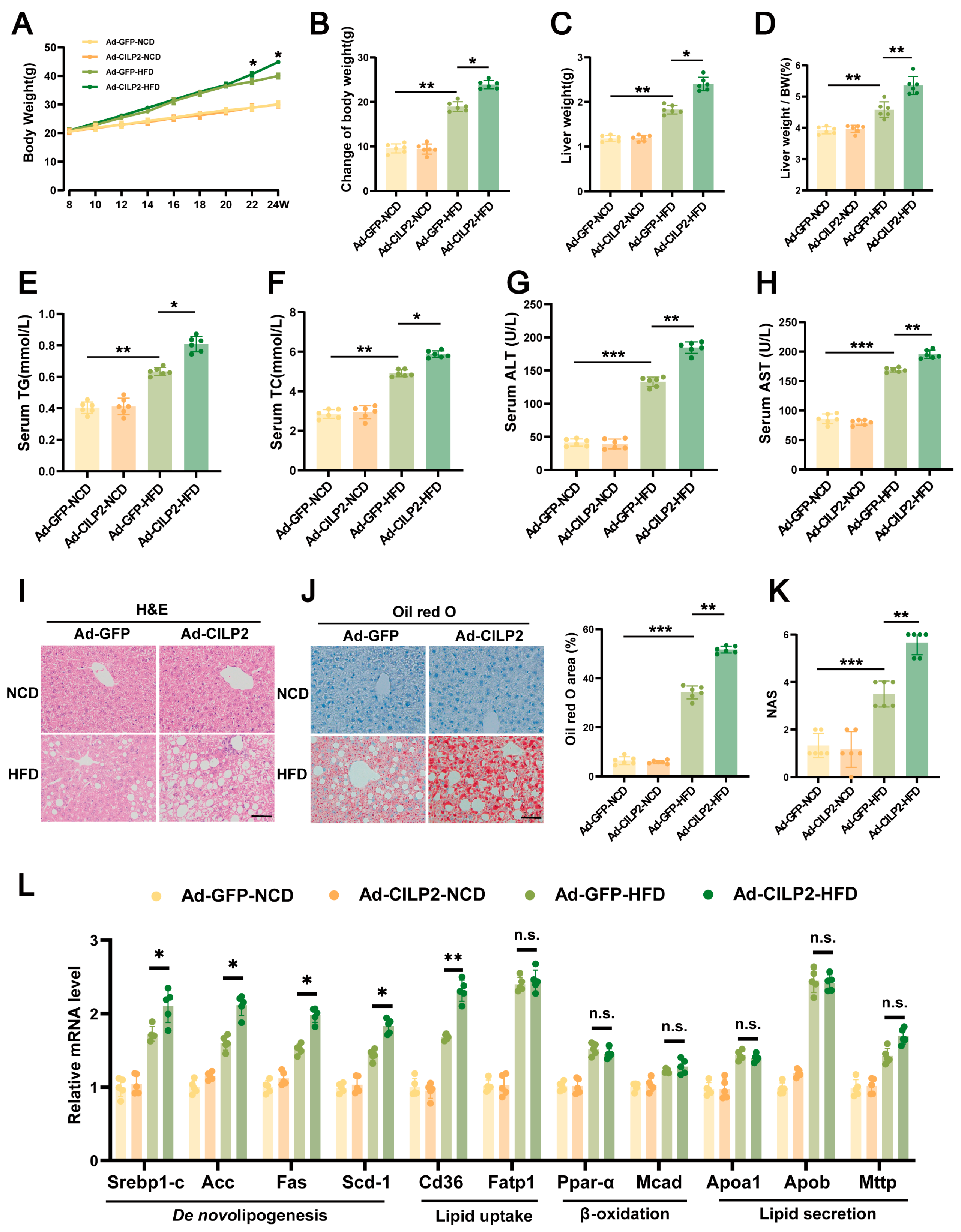
2.4. CILP2 Overexpression Increased Body Weight and Promoted IR and Hepatic Steatosis in HFD-Fed Mice
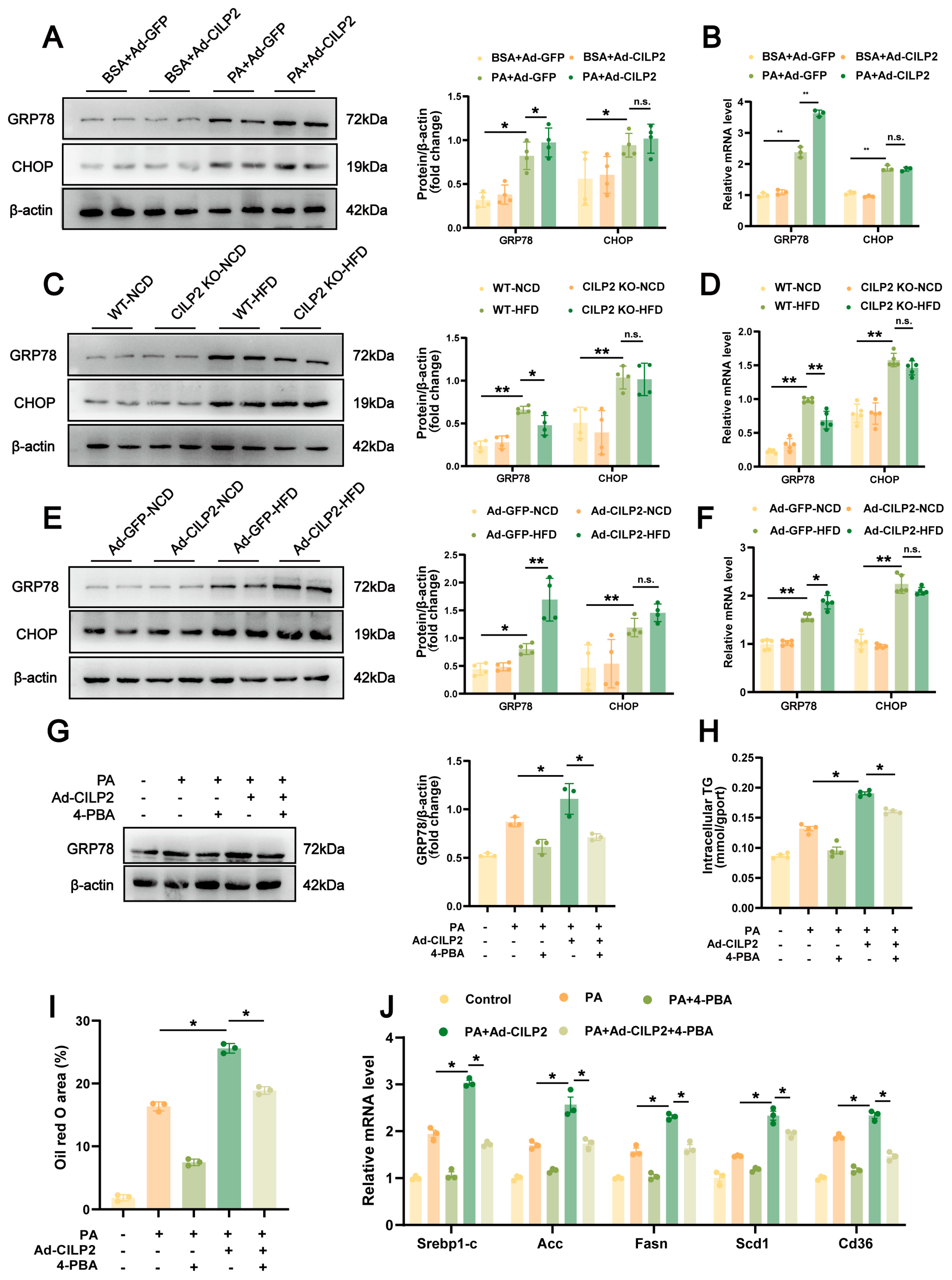
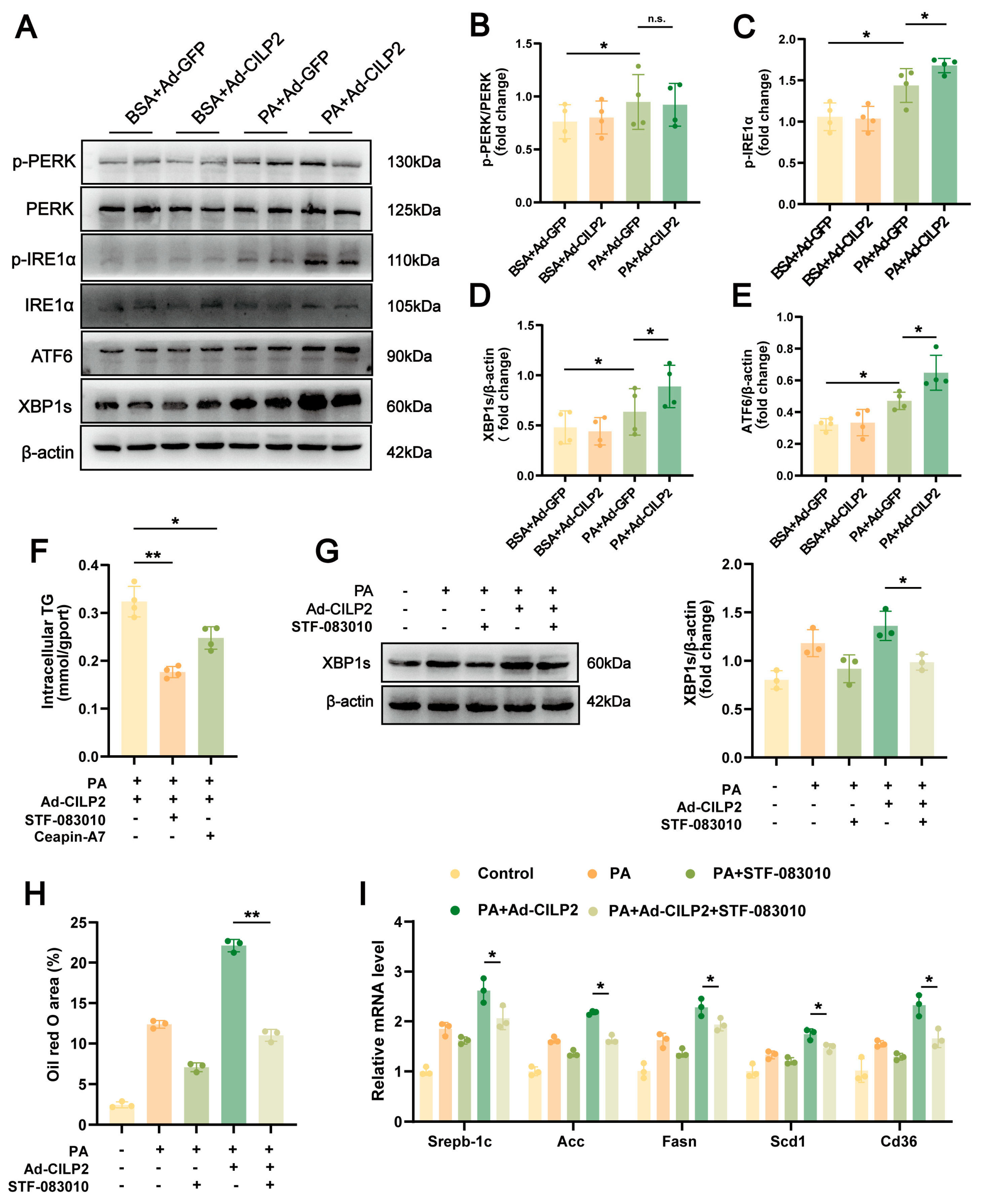
2.5. ER Stress in CILP2-Mediated Hepatic Lipid Accumulation
2.6. CILP2 Enhanced Hepatic Lipid Accumulation by Promoting the IRE1α/XBP1 Pathway
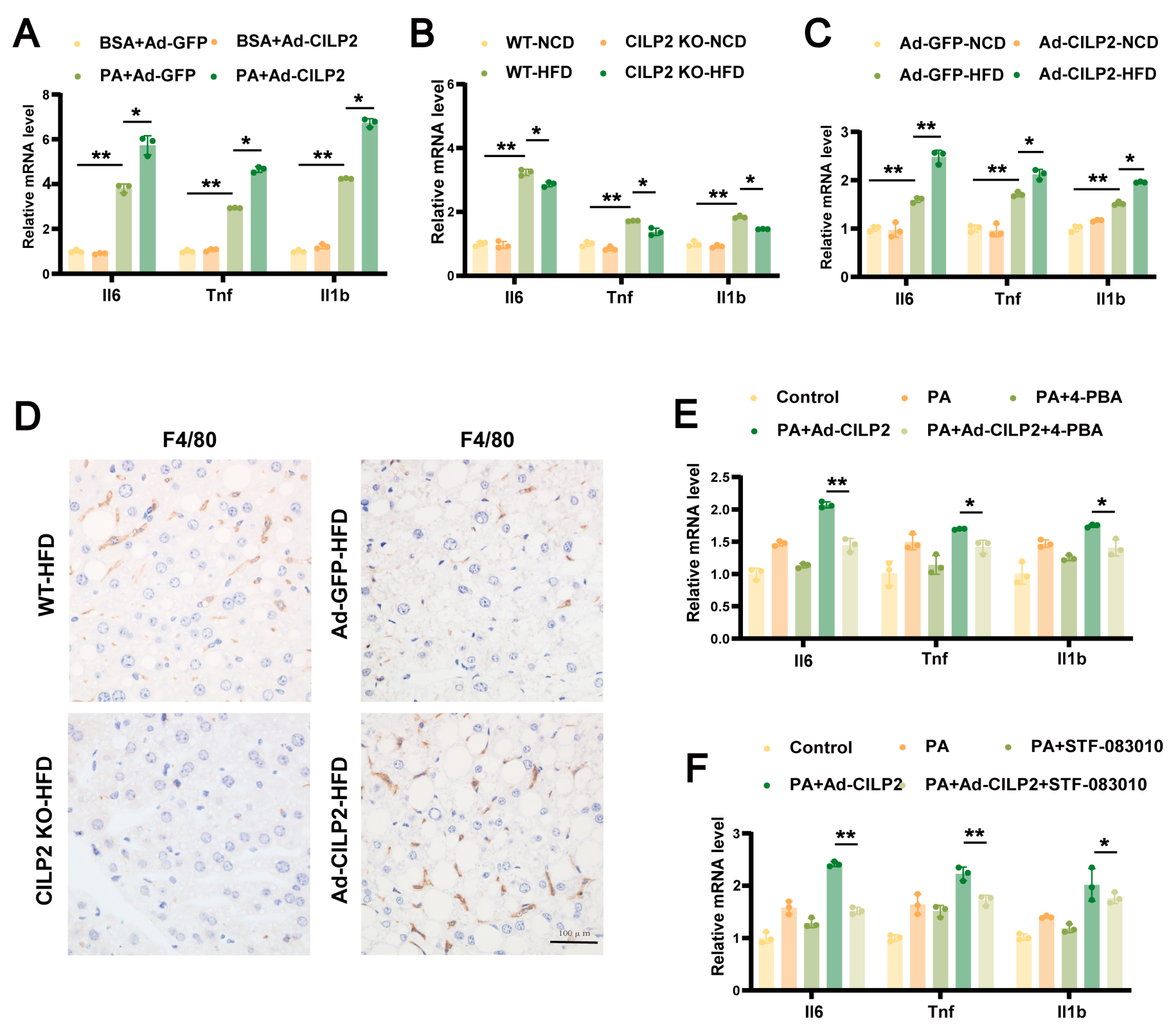
2.7. CILP2 Regulated Hepatic Inflammatory Responses Through ER Stress
3. Discussion
4. Materials and Methods
4.1. Animal Models and Treatment
4.2. Cell Culture and Treatment
4.3. In Vivo Experiment
4.4. Histological Analysis
4.5. Immunohistochemistry and Immunofluorescence
4.6. Western Blot Analysis
4.7. RNA Extraction and Quantitative RT-PCR Analysis (qRT-PCR)
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| MASLD | Metabolic dysfunction-associated fatty liver disease |
| MASH | Metabolic dysfunction-associated steatohepatitis |
| ER | Endoplasmic reticulum |
| SREBP-1c | Sterol regulatory element-binding protein-1c |
| FASN | Fatty acid synthase |
| ACC | Acetyl-CoA carboxylase |
| SCD1 | Stearoyl-CoA desaturase 1 |
| CD36 | Cluster of differentiation 36 |
| IL-6 | Interleukin-6 |
| TNFα | Tumor necrosis factorα |
| IL-1β | Interleukin-1β |
| CILP2 | Cartilage intermediate layer protein 2 |
| TG | Triglyceride |
| TC | Total cholesterol |
| AST | Aspartate amino transferase |
| ALT | Alanine amino transferase |
| PERK | Protein kinase RNA-like endoplasmic reticulum kinase |
| eIF2α | Eukaryotic translation initiation factor 2α |
| ATF4 | Activating transcription factor 4 |
| IRE1α | Inositol-requiring enzyme 1α |
| XBP1 | X-box binding protein 1 |
| ATF6 | Activating transcription factor 6 |
| GRP78 | Glucose-regulated protein 78 |
| CHOP | C/EBP homologous protein |
| PA | Palmitic acid |
| BSA | Bovine serum albumin |
| GTT | Glucose tolerance test |
| ITT | Insulin tolerance test |
| NAS | Non-alcoholic fatty liver disease activity score |
| IR | Insulin resistance |
| HFD | High fat diet |
| NCD | Normal chow diet |
| AUC | Area under the curve |
References
- Xu, X.; Poulsen, K.L.; Wu, L.; Liu, S.; Miyata, T.; Song, Q.; Wei, Q.; Zhao, C.; Lin, C.; Yang, J. Targeted therapeutics and novel signaling pathways in non-alcohol-associated fatty liver/steatohepatitis (NAFL/NASH). Signal Transduct. Target. Ther. 2022, 7, 287. [Google Scholar] [CrossRef] [PubMed]
- Rinella, M.E.; Rinella, M.E.; Lazarus, J.V.; Ratziu, V.; Francque, S.M.; Sanyal, A.J.; Kanwal, F.; Romero, D.; Abdelmalek, M.F.; Anstee, Q.M.; et al. A multisociety Delphi consensus statement on new fatty liver disease nomenclature. J. Hepatol. 2023, 79, 1542–1556. [Google Scholar] [CrossRef]
- Jung, T.W.; Choi, K.M. Pharmacological Modulators of Endoplasmic Reticulum Stress in Metabolic Diseases. Int. J. Mol. Sci. 2016, 17, 192. [Google Scholar] [CrossRef]
- Lebeaupin, C.; Vallée, D.; Hazari, Y.; Hetz, C.; Chevet, E.; Bailly-Maitre, B. Endoplasmic reticulum stress signalling and the pathogenesis of non-alcoholic fatty liver disease. J. Hepatol. 2018, 69, 927–947. [Google Scholar] [CrossRef]
- Jiang, D.; Niwa, M.; Koong, A.C. Targeting the IRE1α-XBP1 branch of the unfolded protein response in human diseases. Semin. Cancer Biol. 2015, 33, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Shan, B.; Dai, J.; Xia, Z.; Cai, J.; Chen, T.; Lv, S.; Feng, Y.; Zheng, L.; Wang, Y.; et al. Dual role for inositol-requiring enzyme 1α in promoting the development of hepatocellular carcinoma during diet-induced obesity in mice. Hepatology 2018, 68, 533–546. [Google Scholar] [CrossRef]
- Lee, A.H.; Scapa, E.F.; Cohen, D.E.; Glimcher, L.H. Regulation of hepatic lipogenesis by the transcription factor. XBPScience 2008, 320, 1492–1496. [Google Scholar] [CrossRef]
- Hotamisligil, G.S. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell 2010, 140, 900–917. [Google Scholar] [CrossRef]
- Wu, T.; Zhang, Q.; Wu, S.; Hu, W.; Zhou, T.; Li, K.; Liu, D.; Gu, H.F.; Zheng, H.; Zhu, Z.; et al. CILP-2 is a novel secreted protein and associated with insulin resistance. J. Mol. Cell Biol. 2019, 11, 1083–1094. [Google Scholar] [CrossRef]
- Willer, C.J.; Sanna, S.; Jackson, A.U.; Scuteri, A.; Bonnycastle, L.L.; Clarke, R.; Heath, S.C.; Timpson, N.J.; Najjar, S.S.; Stringham, H.M.; et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 2008, 40, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Dreja, T.; Jovanovic, Z.; Rasche, A.; Kluge, R.; Herwig, R.; Tung, Y.C.; Joost, H.G.; Yeo, G.S.; Al-Hasani, H. Diet-induced gene expression of isolated pancreatic islets from a polygenic mouse model of the metabolic syndrome. Diabetologia 2010, 53, 309. [Google Scholar] [CrossRef]
- Luptáková, L.; Benčová, D.; Siváková, D.; Cvíčelová, M. Association of CILP2 and ACE Gene Polymorphisms with Cardiovascular Risk Factors in Slovak Midlife Women. Biomed. Res. Int. 2013, 2013, 634207. [Google Scholar] [CrossRef] [PubMed]
- Tai, E.S.; Sim, X.L.; Ong, T.H.; Wong, T.Y.; Saw, S.M.; Aung, T.; Kathiresan, S.; Orho-Melander, M.; Ordovas, J.M.; Tan, J.T.; et al. Polymorphisms at newly identified lipid-associated loci are associated with blood lipids and cardiovascular disease in an Asian Malay population. J. Lipid Res. 2009, 50, 514–520. [Google Scholar] [CrossRef]
- Boonvisut, S.; Nakayama, K.; Makishima, S.; Watanabe, K.; Miyashita, H.; Lkhagvasuren, M.; Kagawa, Y.; Iwamoto, S. Replication analysis of genetic association of the NCAN-CILP2 region with plasma lipid levels and non-alcoholic fatty liver disease in Asian and Pacific ethnic groups. Lipids Health Dis. 2016, 15, 8. [Google Scholar] [CrossRef]
- Li, Q.; Pu, D.; Xia, X.; Liu, H.; Li, L. Serum Concentrations of Cartilage Intermediate Layer Protein 2 Were Higher in Overweight and Obese Subjects. Biomed. Res. Int. 2022, 2022, 6290064. [Google Scholar] [CrossRef]
- Hu, W.; Li, K.; Han, H.; Geng, S.; Zhou, B.; Fan, X.; Xu, S.; Yang, M.; Liu, H.; Yang, G.; et al. Circulating Levels of CILP2 Are Elevated in Coronary Heart Disease and Associated with Atherosclerosis. Oxid. Med. Cell Longev. 2020, 2020, 1871984. [Google Scholar] [CrossRef]
- Tubbs, E.; Theurey, P.; Vial, G.; Bendridi, N.; Bravard, A.; Chauvin, M.A.; Ji-Cao, J.; Zoulim, F.; Bartosch, B.; Ovize, M.; et al. Mitochondria-associated endoplasmic reticulum membrane (MAM) integrity is required for insulin signaling and is implicated in hepatic insulin resistance. Diabetes 2014, 63, 3279–3294. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Garcia-Carbonell, R.; Yamachika, S.; Zhao, P.; Dhar, D.; Loomba, R.; Kaufman, R.J.; Saltiel, A.R.; Karin, M. ER Stress Drives Lipogenesis and Steatohepatitis via Caspase-2 Activation of S1P. Cell 2018, 175, 133–145.e15. [Google Scholar] [CrossRef]
- Liu, C.; Zhou, B.; Meng, M.; Zhao, W.; Wang, D.; Yuan, Y.; Zheng, Y.; Qiu, J.; Li, Y.; Li, G.; et al. FOXA3 induction under endoplasmic reticulum stress contributes to non-alcoholic fatty liver disease. J. Hepatol. 2021, 75, 150–162. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Kaufman, R.J. The role of ER stress in lipid metabolism and lipotoxicity. J. Lipid Res. 2016, 57, 1329–1338. [Google Scholar] [CrossRef]
- Shreya, S.; Grosset, C.F.; Jain, B.P. Unfolded Protein Response Signaling in Liver Disorders: A 2023 Updated Review. Int. J. Mol. Sci. 2023, 24, 14066. [Google Scholar] [CrossRef]
- Wang, S.; Chen, Z.; Lam, V.; Han, J.; Hassler, J.; Finck, B.N.; Davidson, N.O.; Kaufman, R.J. IRE1α-XBP1s induces PDI expression to increase MTP activity for hepatic VLDL assembly and lipid homeostasis. Cell Metab. 2012, 16, 473–486. [Google Scholar] [CrossRef]
- Mao, T.; Shao, M.; Qiu, Y.; Huang, J.; Zhang, Y.; Song, B.; Wang, Q.; Jiang, L.; Liu, Y.; Han, J.D.; et al. PKA phosphorylation couples hepatic inositol-requiring enzyme 1α to glucagon signaling in glucose metabolism. Proc. Natl. Acad. Sci. USA 2011, 108, 15852–15857. [Google Scholar] [CrossRef]
- Yu, X.; Ren, L.P.; Wang, C.; Zhu, Y.J.; Xing, H.Y.; Zhao, J.; Song, G.Y. Role of X-Box Binding Protein-1 in Fructose-Induced De Novo Lipogenesis in HepG2 Cells. Chin. Med. J. 2018, 131, 2310–2319. [Google Scholar] [CrossRef]
- Guo, B.; Li, Z. Endoplasmic reticulum stress in hepatic steatosis and inflammatory bowel diseases. Front. Genet. 2014, 5, 242. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Shen, H.; Li, X.; Wang, H. Endoplasmic reticulum stress in innate immune cells—A significant contribution to non-alcoholic fatty liver disease. Front. Immunol. 2022, 13, 951406. [Google Scholar] [CrossRef]
- Horn, P.; Tacke, F. Unboxing the cell-type specific contribution of endoplasmic reticulum stress to NASH pathophysiology-myeloid X-box-binding protein 1 as a driver of steatohepatitis and fibrosis. Transl. Gastroenterol. Hepatol. 2022, 8, 14. [Google Scholar] [CrossRef]
- Wei, D.; Tian, X.; Zhu, L.; Wang, H.; Sun, C. USP14 governs CYP2E1 to promote nonalcoholic fatty liver disease through deubiquitination and stabilization of HSP90AA1. Cell Death Dis. 2023, 14, 566. [Google Scholar] [CrossRef] [PubMed]
- Nissar, A.U.; Sharma, L.; Mudasir, M.A.; Nazir, L.A.; Umar, S.A.; Sharma, P.R.; Vishwakarma, R.A.; Tasduq, S.A. Chemical chaperone 4-phenyl butyric acid (4-PBA) reduces hepatocellular lipid accumulation and lipotoxicity through induction of autophagy. J. Lipid Res. 2017, 58, 1855–1868. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Huo, Z.; Luan, H.; Huang, Y.; Shen, Y.; Sheng, L.; Liang, J.; Wu, F. Scutellarin ameliorates hepatic lipid accumulation by enhancing autophagy and suppressing IRE1α/XBP1 pathway. Phytother. Res. 2022, 36, 433–447. [Google Scholar] [CrossRef]
- Xue, F.; Lu, J.; Buchl, S.C.; Sun, L.; Shah, V.H.; Malhi, H.; Maiers, J.L. Coordinated signaling of activating transcription factor 6α and inositol-requiring enzyme 1α regulates hepatic stellate cell-mediated fibrogenesis in mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2021, 320, G864–G879. [Google Scholar] [CrossRef] [PubMed]
- Kleiner, D.E.; Brunt, E.M.; Van Natta, M.; Behling, C.; Contos, M.J.; Cummings, O.W.; Ferrell, L.D.; Liu, Y.C.; Torbenson, M.S.; Unalp-Arida, A.; et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 2005, 41, 1313–1321. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Chen, S.; Dong, L.; Shan, Y.; Chen, Z.; Xia, Y.; Liu, J.; Liu, D.; Yang, G.; Yang, M.; Li, K. Cartilage Intermediate Layer Protein 2 Aggravates Hepatic Lipid Accumulation and Inflammation Through the IRE1α/XBP1 Pathway. Int. J. Mol. Sci. 2026, 27, 1213. https://doi.org/10.3390/ijms27031213
Chen S, Dong L, Shan Y, Chen Z, Xia Y, Liu J, Liu D, Yang G, Yang M, Li K. Cartilage Intermediate Layer Protein 2 Aggravates Hepatic Lipid Accumulation and Inflammation Through the IRE1α/XBP1 Pathway. International Journal of Molecular Sciences. 2026; 27(3):1213. https://doi.org/10.3390/ijms27031213
Chicago/Turabian StyleChen, Siqi, Lun Dong, Yingying Shan, Zhili Chen, Yitao Xia, Jiaxin Liu, Dongfang Liu, Gangyi Yang, Mengliu Yang, and Ke Li. 2026. "Cartilage Intermediate Layer Protein 2 Aggravates Hepatic Lipid Accumulation and Inflammation Through the IRE1α/XBP1 Pathway" International Journal of Molecular Sciences 27, no. 3: 1213. https://doi.org/10.3390/ijms27031213
APA StyleChen, S., Dong, L., Shan, Y., Chen, Z., Xia, Y., Liu, J., Liu, D., Yang, G., Yang, M., & Li, K. (2026). Cartilage Intermediate Layer Protein 2 Aggravates Hepatic Lipid Accumulation and Inflammation Through the IRE1α/XBP1 Pathway. International Journal of Molecular Sciences, 27(3), 1213. https://doi.org/10.3390/ijms27031213




