Whole-Genome Resequencing Analysis Reveals Insights into Sex Determination and Gene Loci Associated with Sex Differences in Procambarus clarkii
Abstract
1. Introduction
2. Results
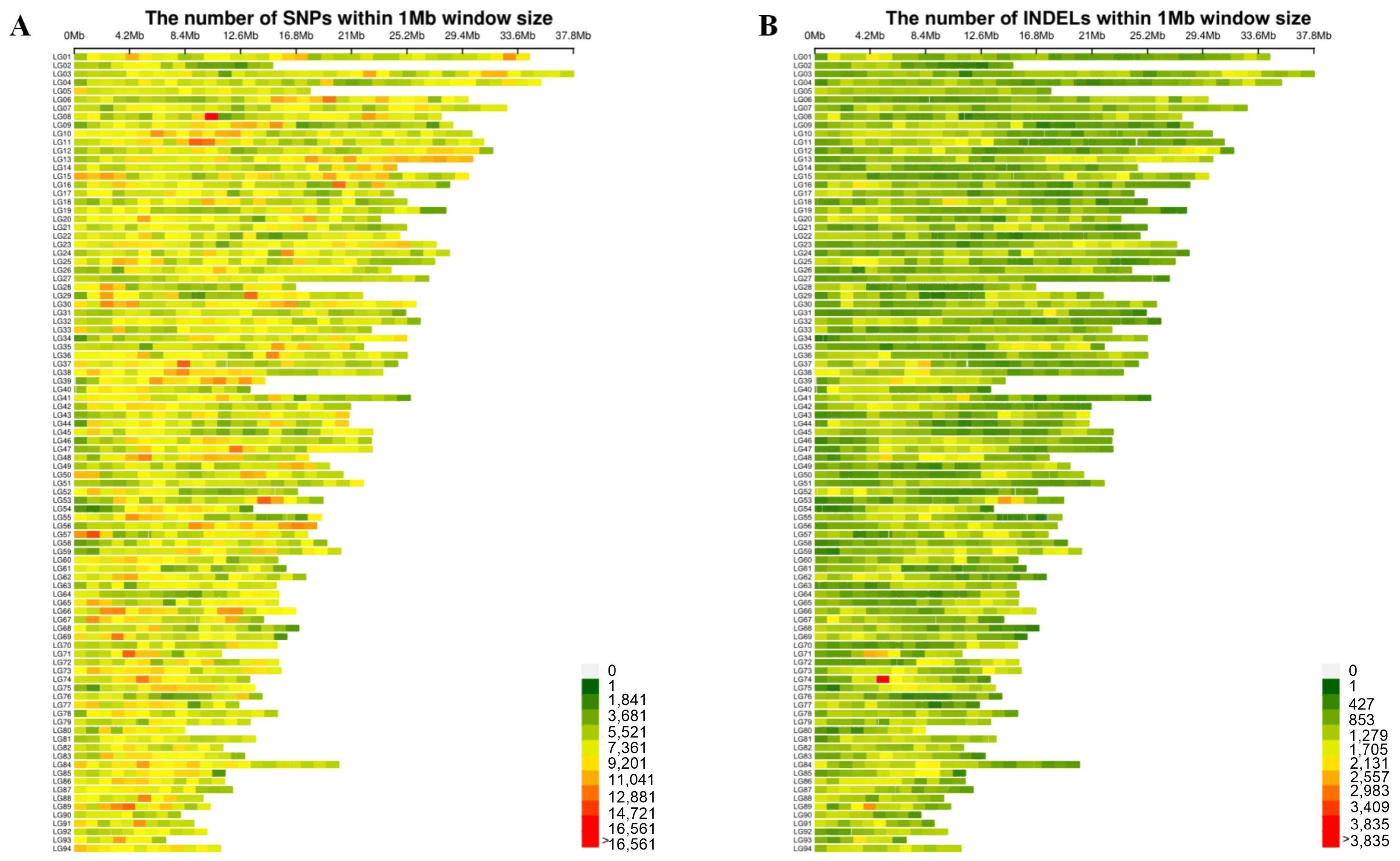
2.1. Resequencing Data and SNP Calling
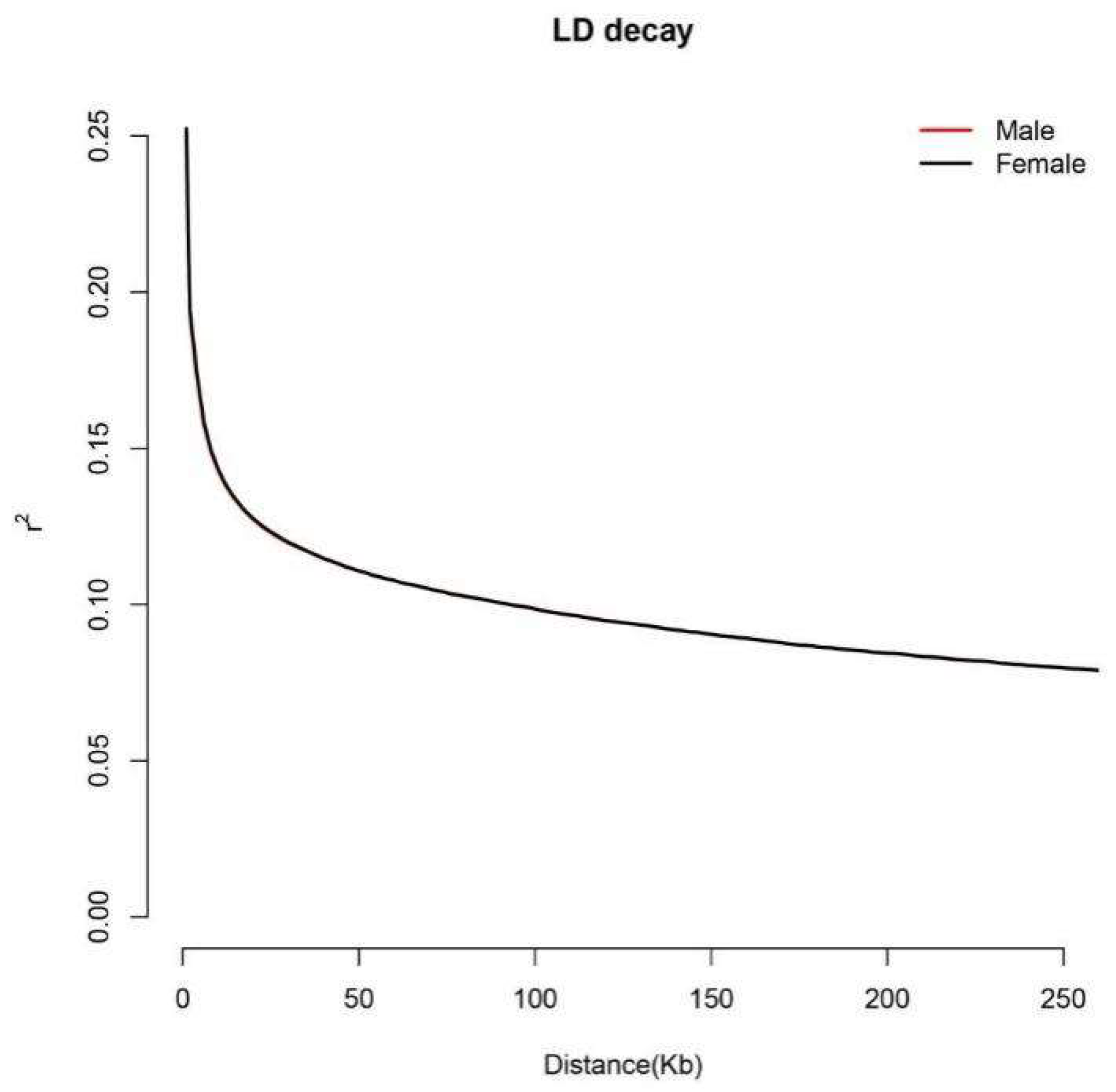
2.2. Comparison of LD
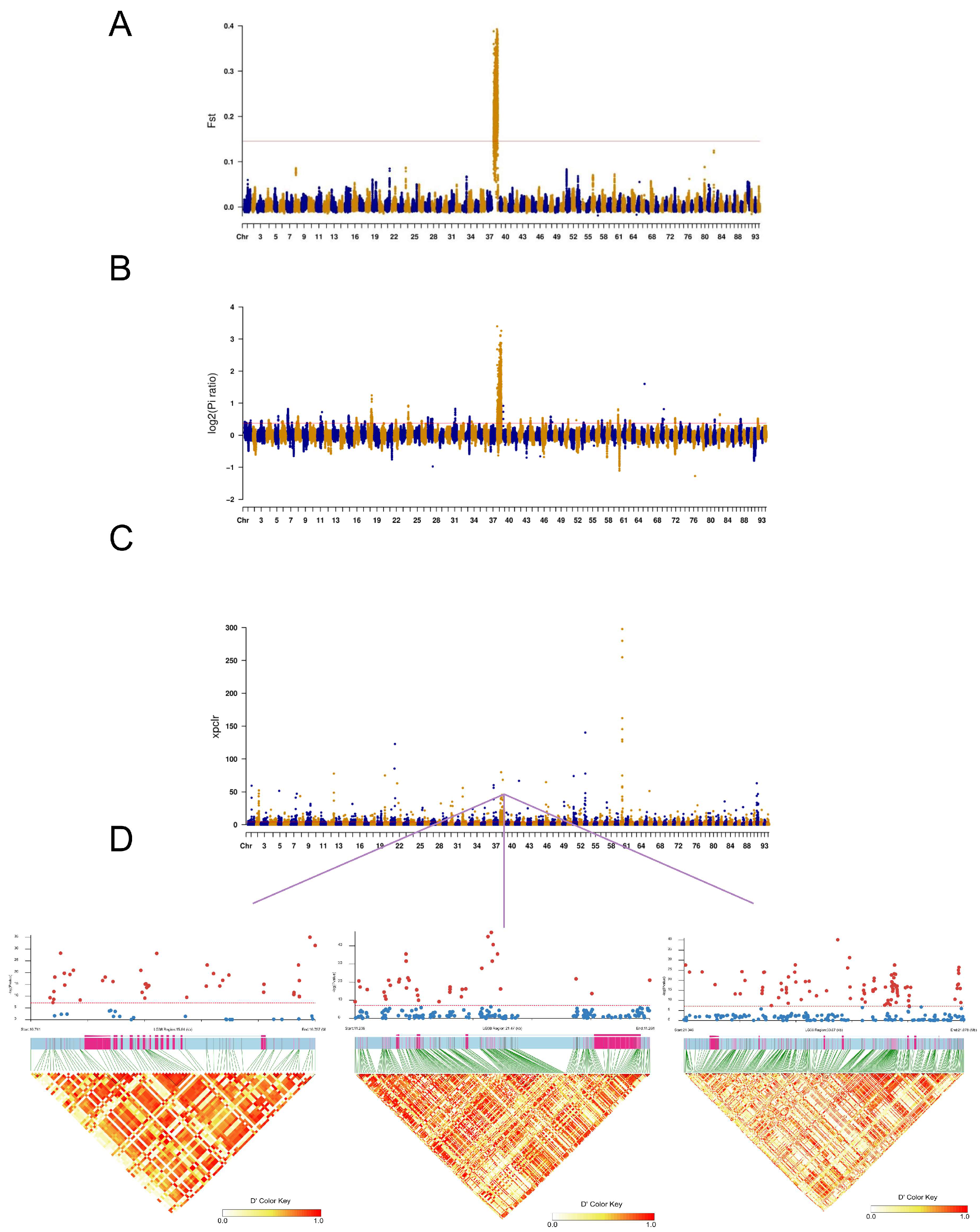
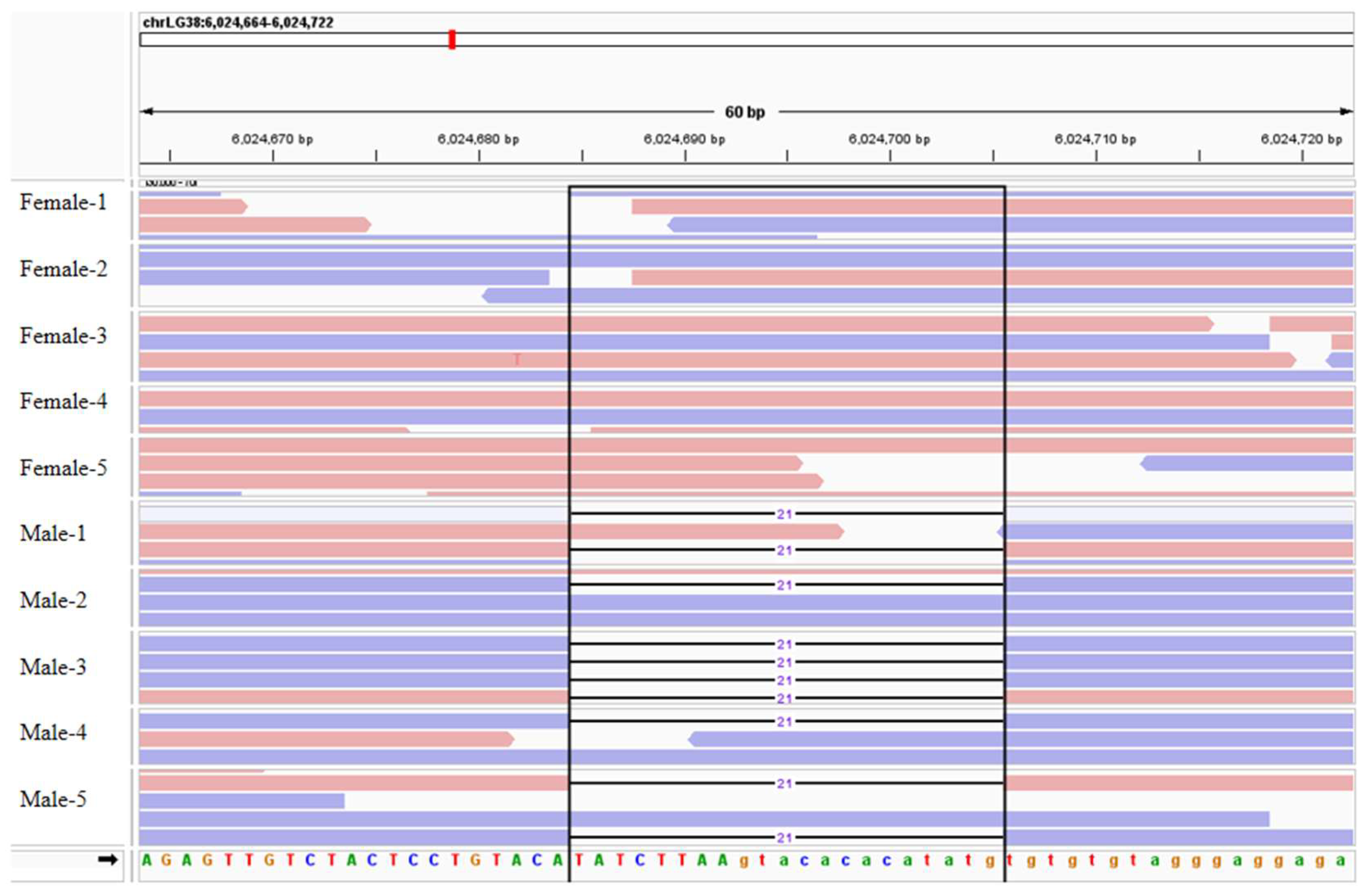
2.3. Identification and Mapping of Sex-Linked Variant
2.4. Selection Signatures Between Male and Female Populations
2.5. Genome-Wide Association Studies
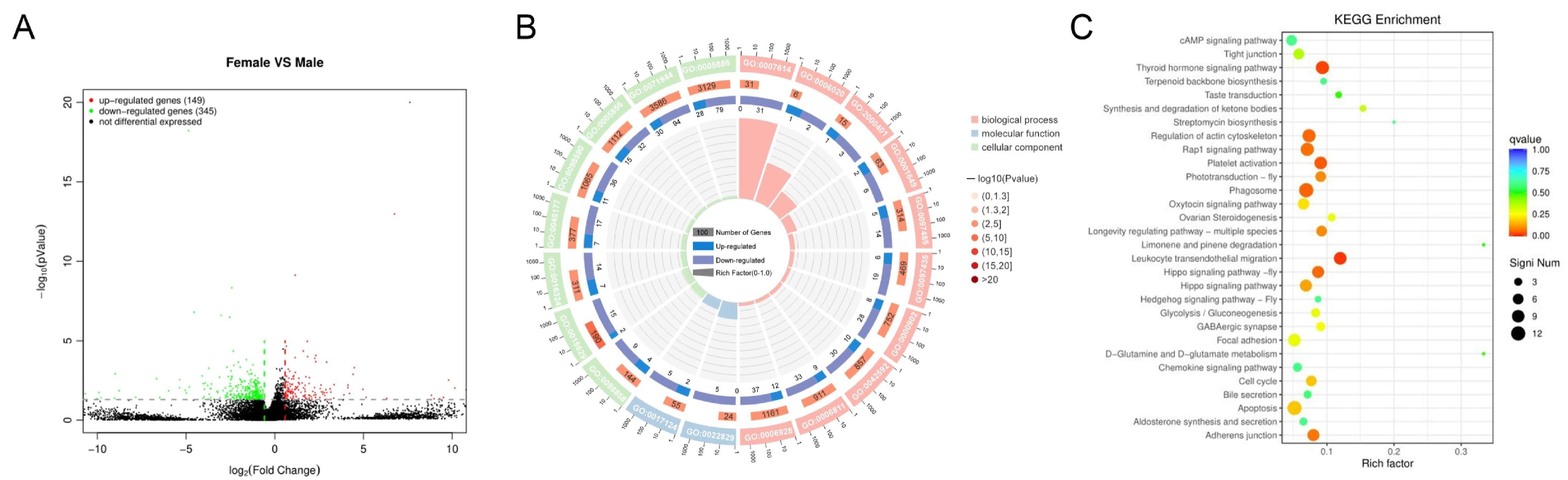
2.6. Functional Annotation of Candidate Genes by GO and KEGG Pathway Analysis
2.7. Major Candidate Genes Associated with Growth and Developmental Traits
2.8. Major Candidate Genes Associated with Immunity
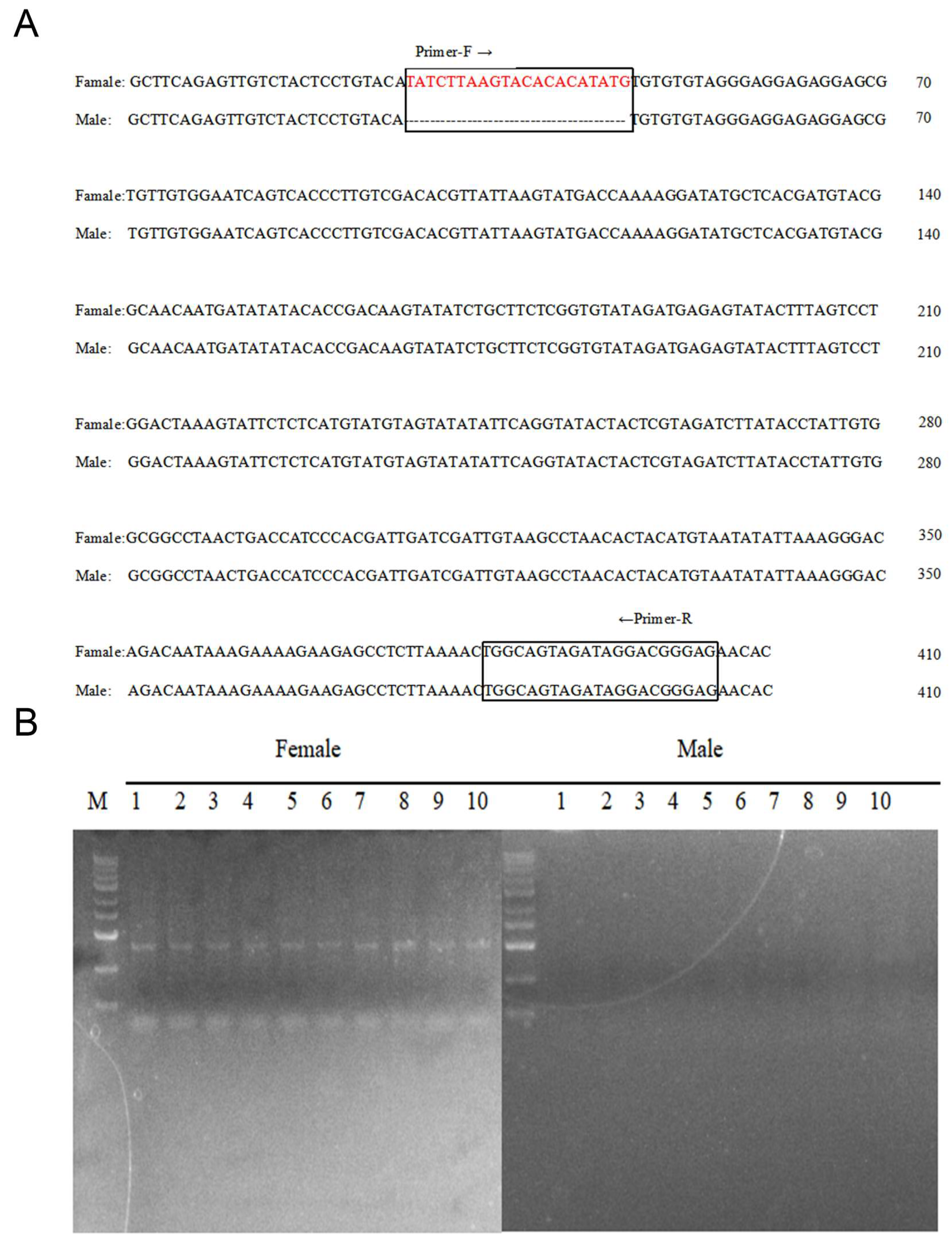
2.9. Development and Verification of Sex-Specific Markers
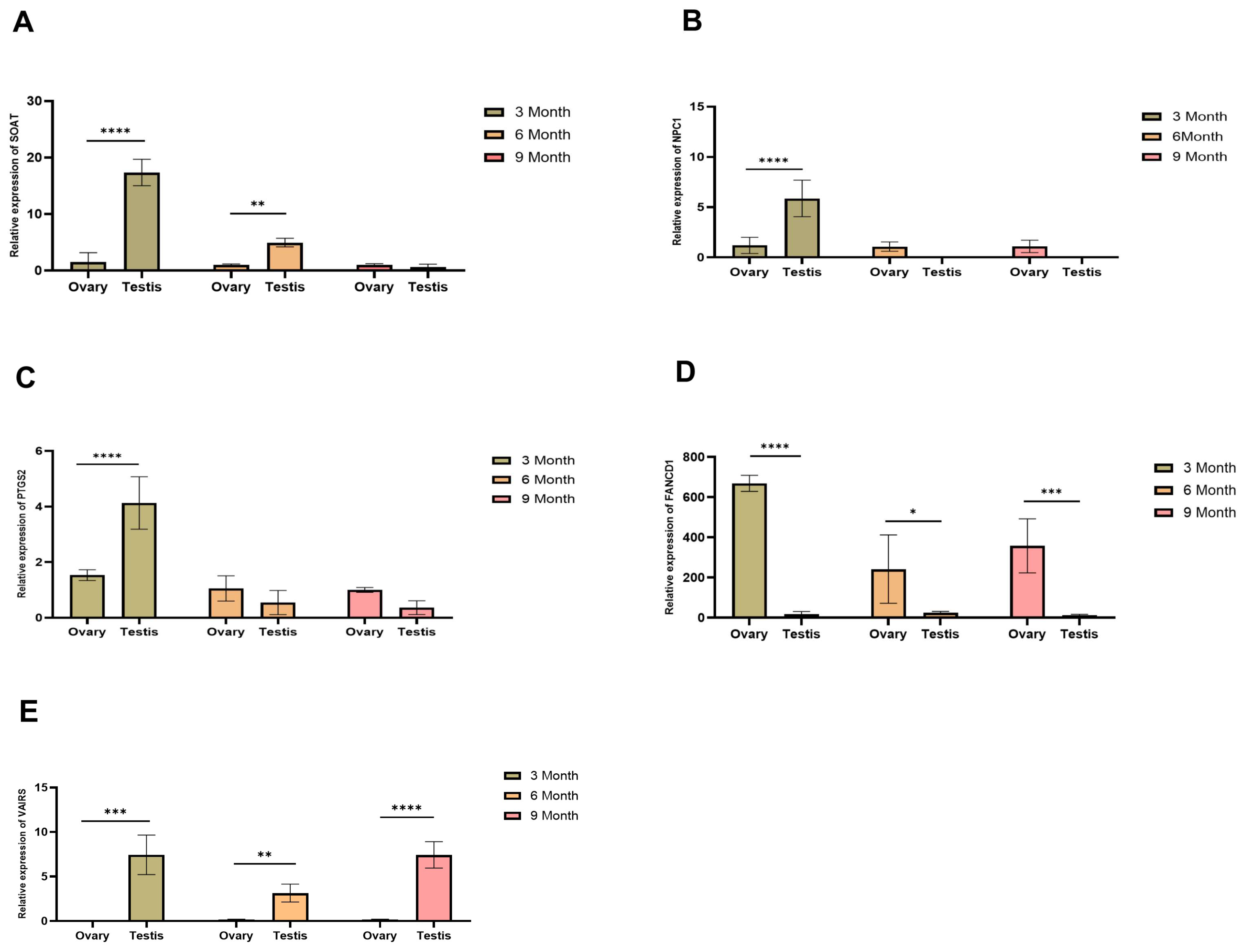
2.10. Expression of Candidate Genes in the Gonads
2.11. Transcriptome Analysis
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Sample Collection and Whole Genome Re-Sequencing
4.3. Linkage Disequilibrium (LD)
4.4. Genome-Wide Selective Sweep Analysis
4.5. Genome-Wide Association Study (GWAS)
4.6. Validation PCR Amplification of Male Chromosome-Specific Fragments
4.7. Functional Annotation of Candidate Genes for Sexdetermination
4.8. RNA-Seq Analysis
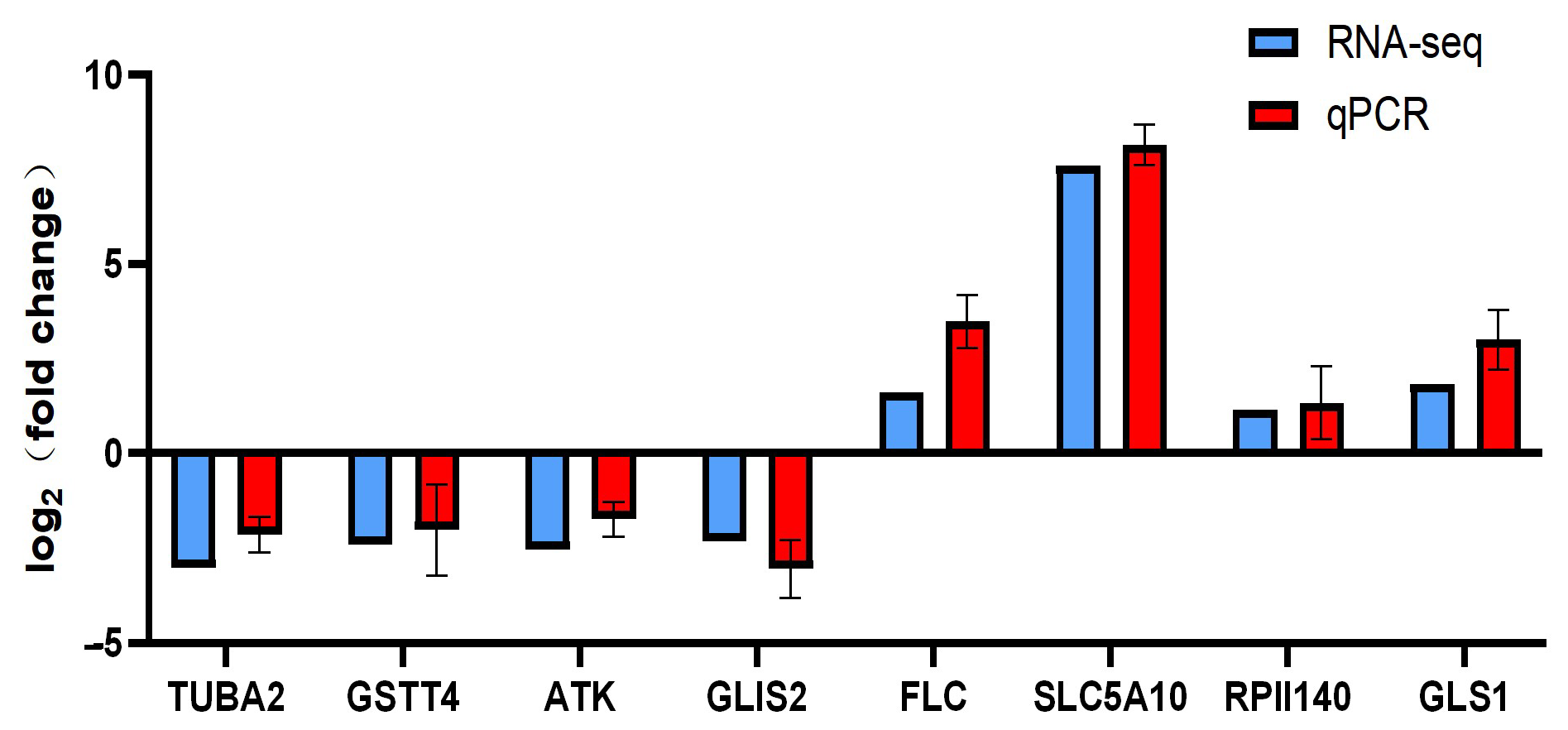
4.9. Validation of Candidate Genes by RT-qPCR
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ye, Z.; Bishop, T.; Wang, Y.; Shahriari, R.; Lynch, M. Evolution of sex determination in crustaceans. Mar. Life Sci. Technol. 2023, 5, 1–11. [Google Scholar] [CrossRef]
- Heath, B.; Laura, R.; Doris, B. Sex Determination, Sex Chromosomes, and Karyotype Evolution in Insects. J. Hered. 2017, 108, 78–93. [Google Scholar]
- Irwin, D.E. Sex chromosomes and speciation in birds and other ZW systems. Mol. Ecol. 2018, 27, 3831–3851. [Google Scholar] [CrossRef]
- Ezaz, T.; Moritz, B.; Waters, P.; Marshall Graves, J.A.; Georges, A.; Sarre, S.D. The ZW sex microchromosomes of an Australian dragon lizard share no homology with those of other reptiles or birds. Chromosome Res. 2009, 17, 965–973. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Hui, M.; Liu, Y.; Song, C.; Li, X.; Li, Y.; Liu, L.; Shi, G.; Wang, S.; Li, F.; et al. High-density linkage mapping aided by transcriptomics documents ZW sex determination system in the Chinese mitten crab Eriocheir sinensis. Heredity 2015, 115, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Devlin, R.H.; Nagahama, Y. Sex determination and sex differentiation in fish: An overview of genetic, physiological, and environmental influences. Aquaculture 2002, 208, 191–364. [Google Scholar] [CrossRef]
- Ventura, T.; Aflalo, E.D.; Weil, S.; Kashkush, K.; Sagi, A. Isolation and characterization of a female-specific DNA marker in the giant freshwater prawn Macrobrachium rosenbergii. Heredity 2011, 107, 456–461. [Google Scholar] [CrossRef]
- Tom, L.; Ohad, R.; Rivka, M.; Shahar, D.; Dudu, A.; Anna, A.; Sklarz, M.Y.; Vered, C.-C.; Kobi, B.; Assaf, S.; et al. Production of WW males lacking the masculine Z chromosome and mining the Macrobrachium rosenbergii genome for sex-chromosomes. Sci. Rep. 2019, 9, 12408. [Google Scholar] [CrossRef]
- Jan, S.; Debbie, R.; Ilse, V.; Brad, A.; John, B.; Marnik, V. High-density linkage maps and sex-linked markers for the black tiger shrimp (Penaeus monodon). Genetics 2008, 179, 917–925. [Google Scholar]
- Pan, N.; Wang, M.; Zhong, L.; Bian, W.; Chen, X.; Zhang, S. Identification of male-specific SNP markers and development of rapid PCR-based genetic sex identification method in channel catfish (Ictalurus punctatus). Aquaculture 2022, 547, 737535. [Google Scholar] [CrossRef]
- Fu, Y.; Luo, L.; Wang, S.; Yu, Y.; Wang, Y.; Gao, Z. Identification of sex-specific markers using genome re-sequencing in the blunt snout bream (Megalobrama amblycephala). BMC Genom. 2024, 25, 963. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Lai, J.; Liang, W.; Ye, S.; Li, J.; Zhou, J.; Zhang, Y.; Peng, S.; Zhan, H.; Zheng, P.; et al. Identification of sex-specific DNA markers in the army fish (Spinibarbus hollandi) by whole genome re-sequencing method. Aquaculture 2024, 583, 740605. [Google Scholar] [CrossRef]
- Ai, C.H.; Zhu, Z.X.; Huang, D.D.; Yang, G.; De, L.T.; Bai, Y.; Liang, X.Y.; Xiong, Y.Y.; Lin, Y.L.; Lin, H.R.; et al. Identification of SNPs and candidate genes associated with early growth in orange-spotted grouper (Epinephelus coioides) by a genome-wide association study. Aquaculture 2023, 565, 739129. [Google Scholar] [CrossRef]
- Chen, J.; Shao, X.; Ye, S.; Xiao, J.; Zou, Y.; Ye, K.; Zhang, L.; Wang, Z.; Xiao, S.; Cai, M. Development of sex-specific markers in Spinyhead croaker, Collichthys lucidus. Aquaculture 2022, 547, 737424. [Google Scholar] [CrossRef]
- Lin, A.; Xiao, S.; Xu, S.; Ye, K.; Lin, X.; Sun, S.; Wang, Z. Identification of a male-specific DNA marker in the large yellow croaker (Larimichthys crocea). Aquaculture 2017, 480, 116–122. [Google Scholar] [CrossRef]
- Fang, S.; Zhang, Y.; Shi, X.; Zheng, H.; Li, S.; Zhang, Y.; Fazhan, H.; Waiho, K.; Tan, H.; Ikhwanuddin, M.; et al. Identification of male-specific SNP markers and development of PCR-based genetic sex identification technique in crucifix crab (Charybdis feriatus ) with implication of an XX/XY sex determination system. Genomics 2020, 112, 404–411. [Google Scholar] [CrossRef]
- Shi, X.; Waiho, K.; Li, X.; Ikhwanuddin, M.; Miao, G.; Lin, F.; Zhang, Y.; Li, S.; Zheng, H.; Liu, W.; et al. Female-specific SNP markers provide insights into a WZ/ZZ sex determination system for mud crabs Scylla paramamosain, S. tranquebarica and S. serrata with a rapid method for genetic sex identification. BMC Genom. 2018, 19, 981. [Google Scholar] [CrossRef]
- Wang, Y.; Hong, D.; Yao, J.; Tan, H.; Wang, S.; Li, J.; Luo, Y.; Wang, D.; Liu, S. Comparative transcriptome preliminary reveals the molecular mechanism of the growth rate of Procambarus clarkii. Reprod. Breed. 2021, 1, 204–209. [Google Scholar] [CrossRef]
- FAO. The State of World Fisheries and Aquaculture; FAO: Rome, Italy, 2022. [Google Scholar]
- Fisheries Department of Ministry of Agriculture. China Fishery Statical Yearbook: 2025; China Agriculture Press: Beijing, China, 2025. [Google Scholar]
- Jin, S.; Jacquin, L.; Xiong, M.; Li, R.; Lek, S.; Li, W.; Zhang, T. Reproductive pattern and population dynamics of commercial red swamp crayfish (Procambarus clarkii) from China: Implications for sustainable aquaculture management. PeerJ 2019, 7, e6214. [Google Scholar] [CrossRef]
- Peng, B.; Tan, Y.-F.; Peng, G.-H.; Xiong, L.-J.; Bai, X.-F. Path Analysis of Effects of Phenotypic Traits Attributes on Abdomen Meat Weight of Red Swamp Crayfish Procambarus Clarkia; CABI: Wallingford, UK, 2021. [Google Scholar]
- Huang, P.; Guo, W.; Wang, Y.; Xiong, Y.; Ge, S.; iGong, G.; Lin, Q.; Xu, Z.; Gui, J.; Mei, J. Genome-wide association study reveals the genetic basis of growth trait in yellow catfish with sexual size dimorphism. Genomics 2022, 114, 110380. [Google Scholar] [CrossRef]
- Perez, R.C.; Ibarra, A. Heritabilities and genetic correlations of size traits at harvest size in sexually dimorphic Pacific white shrimp (Litopenaeus vannamei) grown in two environments. Aquac. Res. 2003, 34, 1079–1085. [Google Scholar] [CrossRef]
- Wu, C.; Xiang, J.J.D.; Biology, R. Genetic determination and exogenous influence in sex differentiation in crustacean. Dev. Reprod. Biol. 2002, 11, 88–94. [Google Scholar]
- Salvadori, S.; Coluccia, E.; Deidda, F.; Cau, A.; Cannas, R.; Lobina, C.; Sabatini, A.; Deiana, A.M. Karyotype, ribosomal genes, and telomeric sequences in the crayfish Procambarus clarkii (Decapoda: Cambaridae). J. Crustac. Biol. 2014, 34, 525–531. [Google Scholar] [CrossRef]
- Liao, M.; Xu, M.; Hu, R.; Xu, Z.; Bonvillain, C.; Li, Y.; Li, X.; Luo, X.; Wang, J.; Wang, J.; et al. The chromosome-level genome assembly of the red swamp crayfish Procambarus clarkii. Sci. Data 2024, 11, 885. [Google Scholar] [CrossRef]
- Ventura, T.; Manor, R.; Aflalo, E.D.; Weil, S.; Rosen, O.; Sagi, A. Timing Sexual Differentiation: Full Functional Sex Reversal Achieved Through Silencing of a Single Insulin-Like Gene in the Prawn, Macrobrachium rosenbergii. Biol. Reprod. 2012, 86, 90. [Google Scholar] [CrossRef]
- Ma, K.; Feng, J.; Lin, J.; Li, J. The complete mitochondrial genome of Macrobrachium nipponense. Gene 2011, 487, 160–165. [Google Scholar] [CrossRef]
- Yu, H.-P.J.O. Five Species of the Genus Macrobrachium (Crustacea, Decapoda, Palaemonidae) from Taiwan; Occasional Papers of the Zoological Laboratory, Faculty of Agriculture, Kyushu University: Fukuoka, Japan, 1972; Volume 3, pp. 45–55. [Google Scholar]
- Sun, R.; Li, Y. A sex-reversing factor: Insulin-like androgenic gland hormone in decapods. Rev. Aquac. 2021, 13, 1352–1366. [Google Scholar] [CrossRef]
- Geyer, J.; Döring, B.; Meerkamp, K.; Ugele, B.; Bakhiya, N.; Fernandes, C.F.; Godoy, J.R.; Glatt, H.; Petzinger, E. Cloning and functional characterization of human sodium-dependent organic anion transporter (SLC10A6). J. Biol. Chem. 2007, 282, 19728–19741. [Google Scholar] [CrossRef] [PubMed]
- Grosser, G.; Fietz, D.; Günther, S.; Bakhaus, K.; Schweigmann, H.; Ugele, B.; Brehm, R.; Petzinger, E.; Bergmann, M.; Geyer, J. Cloning and functional characterization of the mouse sodium-dependent organic anion transporter Soat (Slc10a6). J. Steroid Biochem. Mol. Biol. 2013, 138, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Fietz, D.; Bakhaus, K.; Wapelhorst, B.; Grosser, G.; Günther, S.; Alber, J.; Döring, B.; Kliesch, S.; Weidner, W.; Galuska, C.E. Membrane transporters for sulfated steroids in the human testis-cellular localization, expression pattern and functional analysis. PLoS ONE 2013, 8, e62638. [Google Scholar] [CrossRef]
- Winkler, M.B.L.; Nel, L.; Frain, K.M.; Dedic, E.; Olesen, E.; Pedersen, B.P. Sterol uptake by the NPC system in eukaryotes: A Saccharomyces cerevisiae perspective. FEBS Lett. 2021, 596, 160–179. [Google Scholar] [CrossRef]
- Wang, C.; Ma, Z.; Scott, M.P.; Huang, X. The cholesterol trafficking protein NPC1 is required for Drosophila spermatogenesis. Dev. Biol. 2011, 351, 146–155. [Google Scholar] [CrossRef]
- Ke, X.-X.; Chao, H.; Abbas, M.N.; Kausar, S.; Gul, I.; Ji, H.; Yang, L.; Cui, H. Niemann-Pick type C1 regulates cholesterol transport and metamorphosis in silkworm, Bombyx mori (Dazao). Int. J. Biol. Macromol. 2020, 152, 525–534. [Google Scholar] [CrossRef]
- Jiang, S.; Zhang, W.; Xiong, Y.; Zhang, M.; Yuan, H.; Niu, Y.; Qiao, H.; Fu, H. NPC Intracellular Cholesterol Transporter 1 Regulates Ovarian Maturation and Molting in Female Macrobrachium nipponense. Int. J. Mol. Sci. 2024, 25, 6049. [Google Scholar] [CrossRef]
- lAhmad, M.F.; Marjanu, H.E.; Norazilah, M.J.; Muhammad, A.A.; Saiful, E.S.; Ani, A.Z.; Nao, S.; Abdul, K.A.K. Unravelling the role of HAS2, GREM1, and PTGS2 gene expression in cumulus cells: Implications for human oocyte development competency—A systematic review and integrated bioinformatic analysis. Front. Endocrinol. 2024, 15, 1274376. [Google Scholar]
- Frungieri, M.B.; Gonzalez-Calvar, S.I.; Parborell, F.; Albrech, T.M.; Mayerhofer, A.; Calandra, R.S. Cyclooxygenase-2 and prostaglandin F2 alpha in Syrian hamster Leydig cells: Inhibitory role on luteinizing hormone/human chorionic gonadotropin-stimulated testosterone production. Endocrinology 2006, 147, 4476–4485, Correction in Endocrinology 2024, 165, bqae005. [Google Scholar] [CrossRef]
- Sirianni, R.; Chimento, A.; De, L.A.; Zolea, F.; Carpino, A.; Rago, V.; Maggiolini, M.; Andò, S.; Pezzi, V. Inhibition of cyclooxygenase-2 down-regulates aromatase activity and decreases proliferation of Leydig tumor cells. J. Biol. Chem. 2009, 284, 28905–28916. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Marí, A.; Postlethwait, J.H. The Role of Fanconi Anemia/BRCA Genes in Zebrafish Sex Determination. Methods Cell Biol. 2011, 105, 461–490. [Google Scholar]
- Papadaki, M.; Le, N.S.; Mylonas, C.C.; Sarropoulou, E. Exploring the Fanconi Anemia Gene Expression and Regulation by MicroRNAs in Gilthead Seabream (Sparus aurata) at Different Gonadal Development Stages. Mar. Biotechnol. 2025, 27, 74. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.; Wang, H.; Cao, Z.; Su, N.; Wang, Y.; Zheng, Y. Deficiency of ValRS-m Causes Male Infertility in Drosophila melanogaster. Int. J. Mol. Sci. 2024, 25, 7489. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del, A.G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.11–11.10.33. [Google Scholar] [CrossRef]
- Yin, L.; Zhang, H.; Tang, Z.; Xu, J.; Yin, D.; Zhang, Z.; Yuan, X.; Zhu, M.; Zhao, S.; Li, X.; et al. rMVP: A Memory-efficient, Visualization-enhanced, and Parallel-accelerated tool for Genome-Wide Association Study. Genom. Proteom. Bioinform. 2021, 19, 619–628. [Google Scholar] [CrossRef]
- Zhang, C.; Dong, S.-S.; Xu, J.-Y.; He, W.-M.; Yang, T.-L. PopLDdecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 2019, 35, 1786–1788. [Google Scholar] [CrossRef]
- Lu, K.; Wei, L.; Li, X.; Wang, Y.; Wu, J.; Liu, M.; Zhang, C.; Chen, Z.; Xiao, Z.; Jian, H.; et al. Whole-genome resequencing reveals Brassica napus origin and genetic loci involved in its improvement. Nat. Commun. 2019, 10, 1154. [Google Scholar] [CrossRef]
- Zhou, X.; Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012, 44, 821–824. [Google Scholar] [CrossRef]
- Thorvaldsdóttir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.K.; Mangalam, A.; Dwivedi, S.; Naik, S.J.B. Primer premier: Program for design of degenerate primers from a protein sequence. BioTechniques 1998, 24, 318–319. [Google Scholar] [CrossRef]
- Dong, S.-S.; He, W.-M.; Ji, J.-J.; Zhang, C.; Guo, Y.; Yang, T.-L. LDBlockShow: A fast and convenient tool for visualizing linkage disequilibrium and haplotype blocks based on variant call format files. Brief. Bioinform. 2020, 22, bbaa227. [Google Scholar] [CrossRef]
- Buchfink, B.; Reuter, K.; Drost, H.G. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods 2021, 18, 366–368. [Google Scholar] [CrossRef] [PubMed]
- Kolberg, L.; Raudvere, U.; Kuzmin, I.; Adler, P.; Vilo, J.; Peterson, H. g: Profiler—Interoperable web service for functional enrichment analysis and gene identifier mapping (2023 update). Nucleic Acids Res. 2023, 51, W207–W212. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]

| Samples | Rawreads | Rawbases | Cleanreads | Cleanbases | Cleanrate | CleanQ20 | CleanQ30 | Depth | GC |
|---|---|---|---|---|---|---|---|---|---|
| GTC-1 | 193,152,830 | 28,972,924,500 | 186,218,432 | 27,535,480,258 | 95.04% | 98.85% | 96.41% | 10.07 | 43.85% |
| GTC-2 | 179,197,382 | 26,879,607,300 | 170,202,402 | 25,055,113,518 | 93.21% | 98.94% | 96.69% | 9.16 | 43.75% |
| GTC-3 | 199,069,488 | 29,860,423,200 | 191,042,044 | 28,206,309,962 | 94.46% | 98.88% | 96.53% | 10.31 | 43.78% |
| GTD-1 | 204,217,290 | 30,632,593,500 | 197,851,494 | 29,284,902,859 | 95.60% | 98.03% | 95.31% | 10.71 | 43.25% |
| GTD-2 | 207,191,432 | 31,078,714,800 | 199,160,526 | 29,396,689,564 | 94.59% | 98.86% | 96.43% | 10.75 | 43.80% |
| GTD-3 | 207,876,526 | 31,181,478,900 | 199,619,274 | 29,481,122,292 | 94.55% | 97.94% | 95.07% | 10.78 | 43.36% |
| MTC-1 | 204,264,918 | 30,639,737,700 | 196,684,036 | 29,067,170,758 | 94.87% | 97.97% | 95.09% | 10.63 | 43.17% |
| MTC-2 | 209,304,784 | 31,395,717,600 | 197,546,716 | 29,030,992,365 | 92.47% | 98.05% | 95.34% | 10.61 | 43.34% |
| MTC-3 | 199,314,512 | 29,897,176,800 | 193,014,748 | 28,582,595,654 | 95.60% | 97.98% | 95.17% | 10.45 | 43.00% |
| MTD-1 | 199,842,842 | 29,976,426,300 | 193,234,426 | 28,637,856,155 | 95.53% | 97.94% | 95.02% | 10.47 | 43.32% |
| MTD-2 | 190,860,038 | 28,629,005,700 | 181,756,682 | 26,747,994,213 | 93.43% | 98.91% | 96.61% | 9.78 | 43.89% |
| MTD-3 | 176,952,476 | 26,542,871,400 | 167,846,118 | 24,758,608,426 | 93.28% | 98.84% | 96.42% | 9.05 | 43.91% |
| Samples | TotalReads | TotalBases | MappedReads | MappedBases | MappedReadsRate | MapDepth | Cov1X | Cov3X |
|---|---|---|---|---|---|---|---|---|
| GTC-1 | 186,218,432 | 27,535,480,258 | 184,092,905 | 27,218,647,300 | 0.9886 | 9.83 | 0.8709 | 0.7751 |
| GTC-2 | 170,202,402 | 25,055,113,518 | 168,392,658 | 24,785,923,669 | 0.9894 | 8.95 | 0.8665 | 0.7631 |
| GTC-3 | 191,042,044 | 28,206,309,962 | 188,921,743 | 27,890,640,114 | 0.9889 | 10.07 | 0.8747 | 0.7851 |
| GTD-1 | 197,851,494 | 29,284,902,859 | 195,729,280 | 28,968,754,192 | 0.9893 | 10.46 | 0.8745 | 0.7873 |
| GTD-2 | 199,160,526 | 29,396,689,564 | 197,025,587 | 29,078,708,101 | 0.9893 | 10.5 | 0.8773 | 0.7911 |
| GTD-3 | 199,619,274 | 29,481,122,292 | 197,353,506 | 29,143,704,531 | 0.9886 | 10.52 | 0.8744 | 0.7887 |
| MTC-1 | 196,684,036 | 29,067,170,758 | 194,515,299 | 28,744,047,991 | 0.989 | 10.38 | 0.8734 | 0.7868 |
| MTC-2 | 197,546,716 | 29,030,992,365 | 195,583,429 | 28,739,457,302 | 0.9901 | 10.38 | 0.8721 | 0.7855 |
| MTC-3 | 193,014,748 | 28,582,595,654 | 191,074,431 | 28,293,389,832 | 0.9899 | 10.22 | 0.869 | 0.7794 |
| MTD-1 | 193,234,426 | 28,637,856,155 | 191,005,460 | 28,305,426,423 | 0.9885 | 10.22 | 0.8742 | 0.7849 |
| MTD-2 | 181,756,682 | 26,747,994,213 | 179,857,259 | 26,465,480,807 | 0.9895 | 9.56 | 0.8732 | 0.7773 |
| MTD-3 | 167,846,118 | 24,758,608,426 | 166,078,347 | 24,495,385,469 | 0.9895 | 8.84 | 0.8631 | 0.7563 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Li, J.; Chen, Y.; Wang, Y.; Liu, S. Whole-Genome Resequencing Analysis Reveals Insights into Sex Determination and Gene Loci Associated with Sex Differences in Procambarus clarkii. Int. J. Mol. Sci. 2026, 27, 938. https://doi.org/10.3390/ijms27020938
Li J, Chen Y, Wang Y, Liu S. Whole-Genome Resequencing Analysis Reveals Insights into Sex Determination and Gene Loci Associated with Sex Differences in Procambarus clarkii. International Journal of Molecular Sciences. 2026; 27(2):938. https://doi.org/10.3390/ijms27020938
Chicago/Turabian StyleLi, Jian, Yitian Chen, Yude Wang, and Shaojun Liu. 2026. "Whole-Genome Resequencing Analysis Reveals Insights into Sex Determination and Gene Loci Associated with Sex Differences in Procambarus clarkii" International Journal of Molecular Sciences 27, no. 2: 938. https://doi.org/10.3390/ijms27020938
APA StyleLi, J., Chen, Y., Wang, Y., & Liu, S. (2026). Whole-Genome Resequencing Analysis Reveals Insights into Sex Determination and Gene Loci Associated with Sex Differences in Procambarus clarkii. International Journal of Molecular Sciences, 27(2), 938. https://doi.org/10.3390/ijms27020938





